A “Pro-Asp-Thr” Amino Acid Repeat from Vibrio sp. QY108 Alginate Lyase Exhibits Alginate-Binding Capacity and Enhanced Soluble Expression and Thermostability
Abstract
1. Introduction
2. Results and Discussion
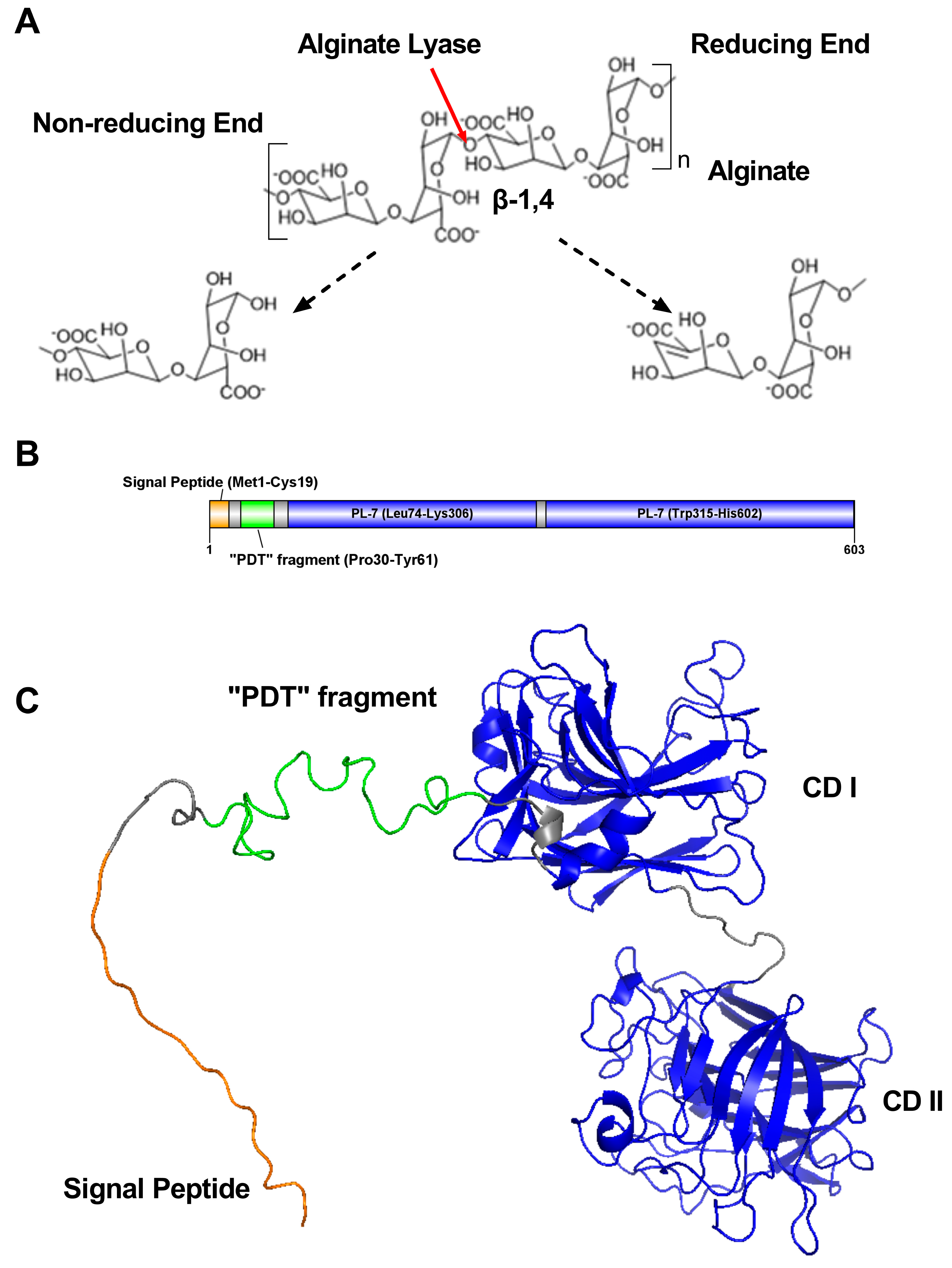
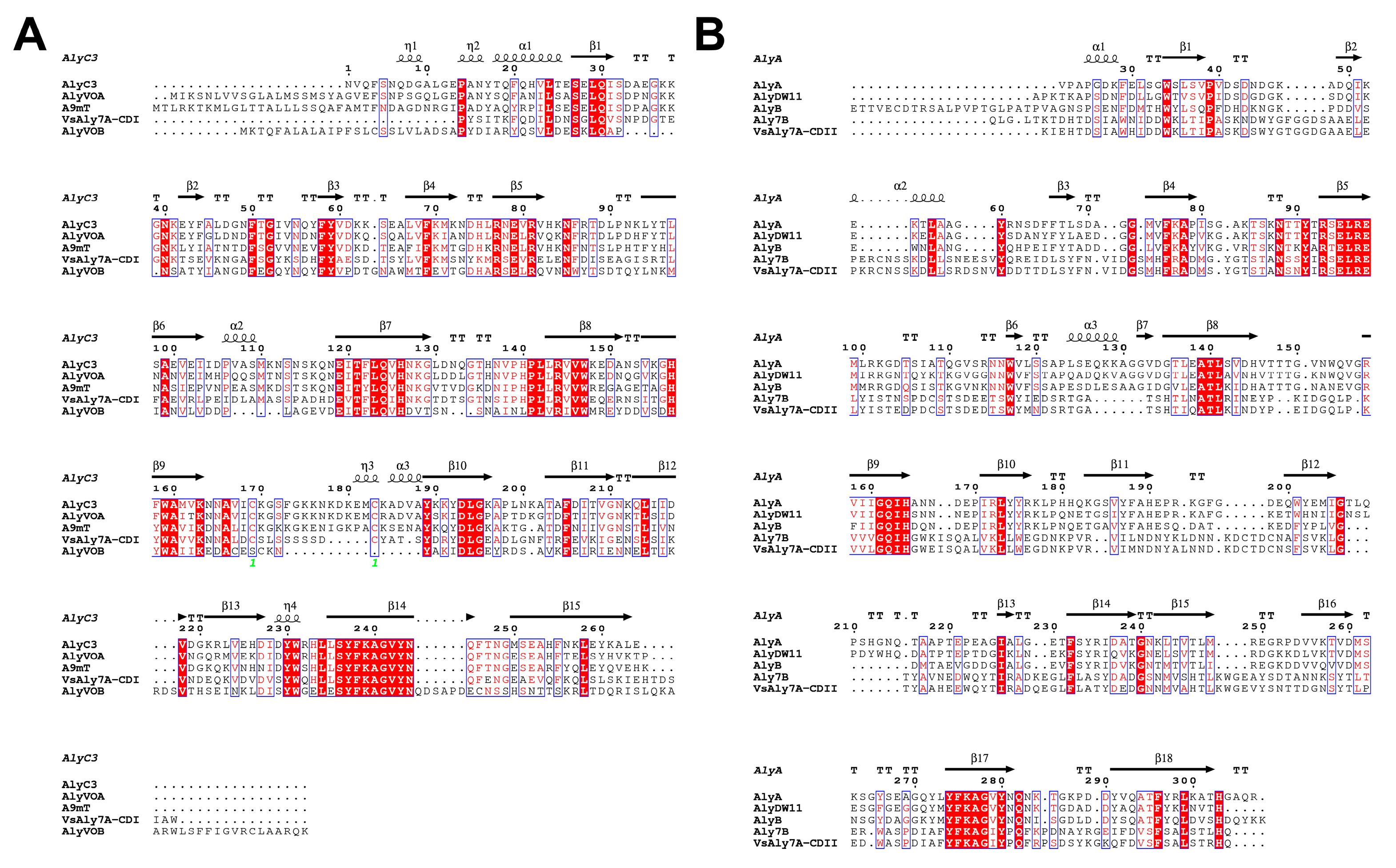
2.1. Alginate-Lyase-Encoding Gene Sequence Analysis
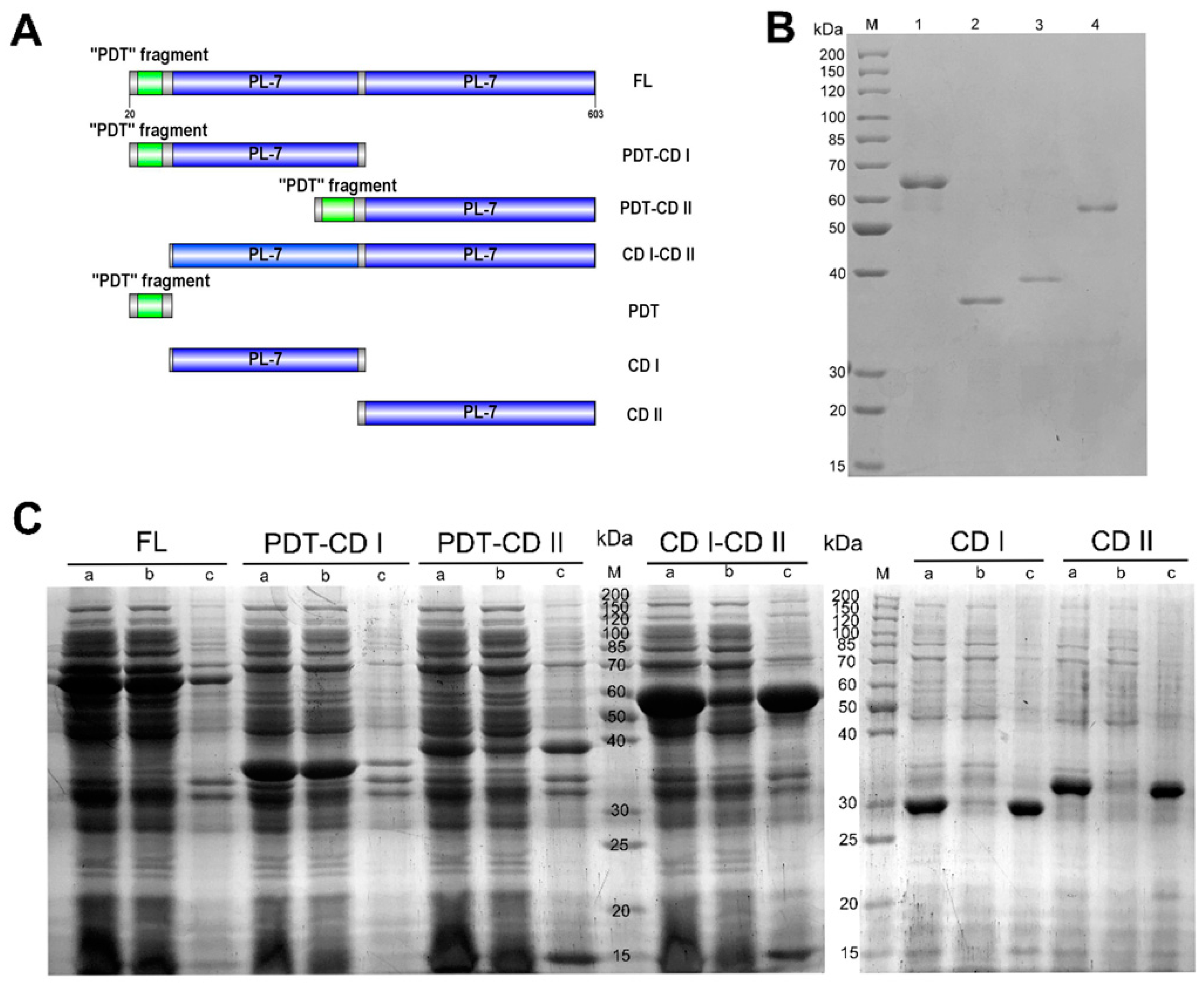
2.2. The Function of Domains in VsAly7A
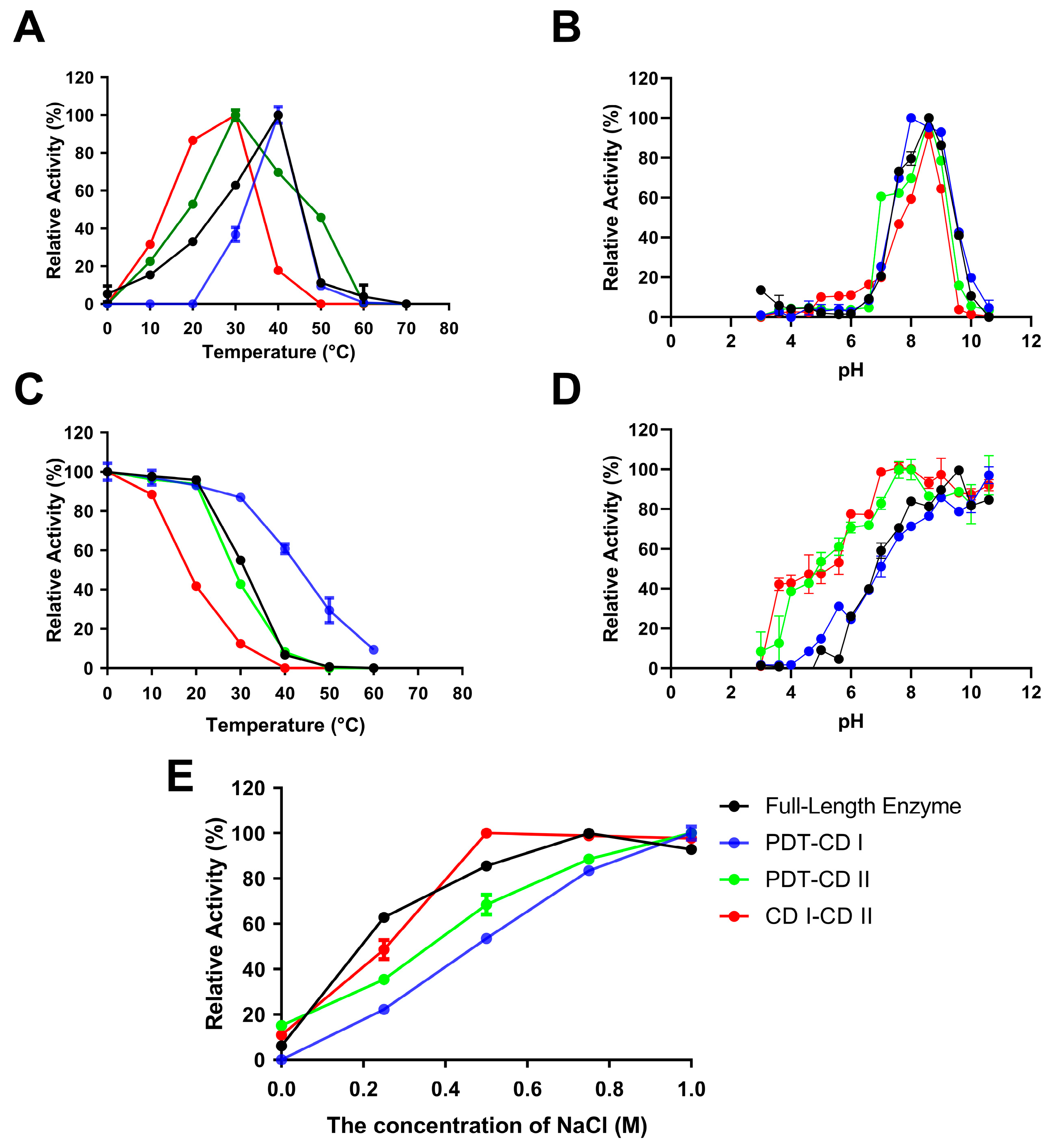
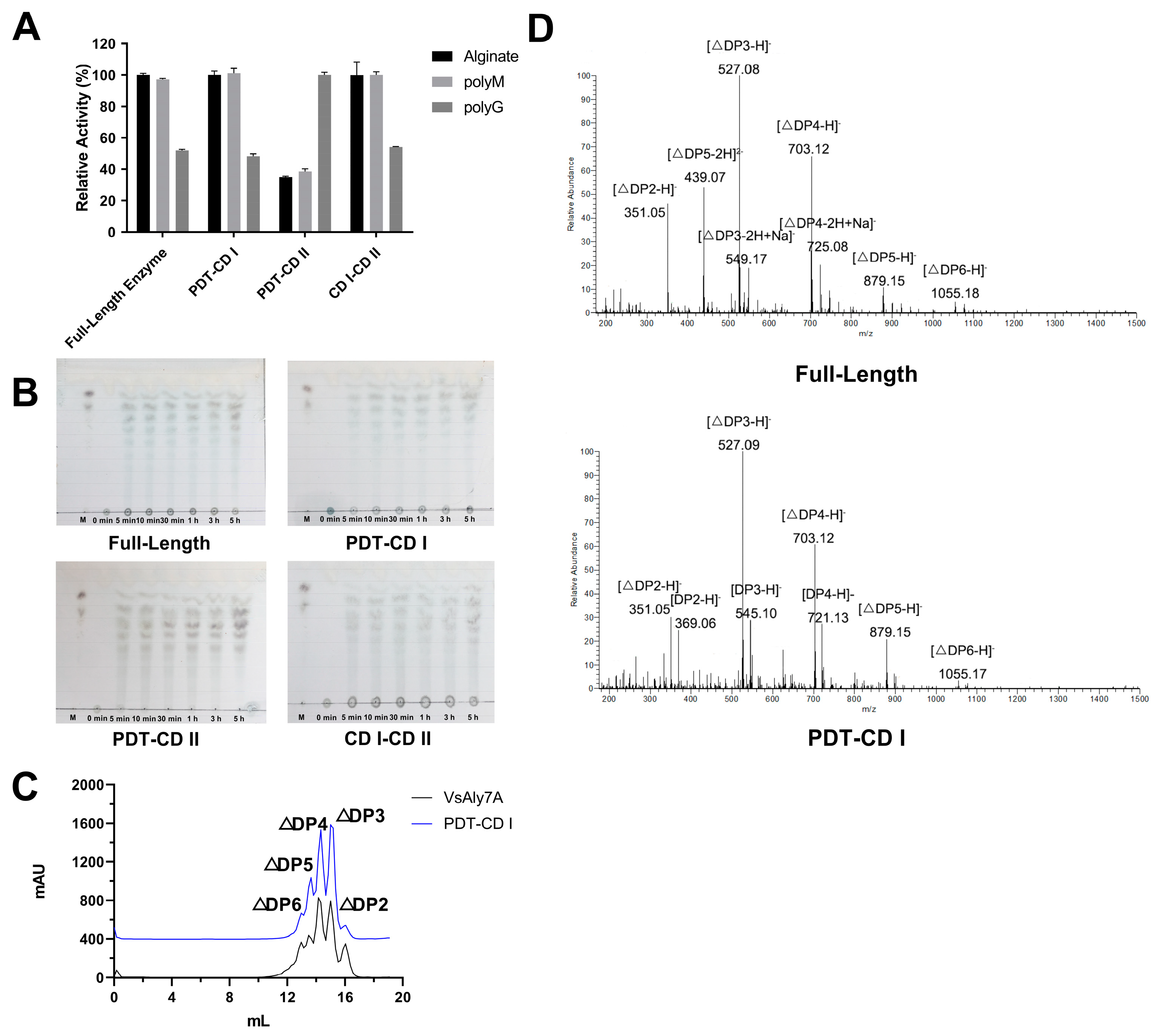
2.3. Biochemical Characterization of VsAly7A and Its Truncated Mutants
2.4. Substrate Specificity and Reaction Products of VsAly7A and Its Truncated Mutants
2.5. “PDT” Fragment: A Novel Alginate-Binding Module
3. Materials and Methods
3.1. Materials and Strains
3.2. Sequence Analysis of Alginate-Lyase-Encoding Gene
3.3. Cloning, Expression, and Purification of Recombinant VsAly7A and the Truncated Mutants
3.4. Enzymatic Activity Assay of Recombinant VsAly7A and Truncated Mutants
3.5. Biochemical Characterization of Recombinant VsAly7A and Truncated Mutants
3.6. Analysis of Reaction Pattern of Recombinant VsAly7A and its Truncated Mutants
3.7. Analysis of End Products of Recombinant VsAly7A and PDT-CD I
3.8. Alginate Gel Bead Preparation and Analysis of Binding Capabilities of Recombinant GST and GST-PDT
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, F.; Wang, P.; Zhang, Y.Z.; Chen, X.L. Diversity of Three-Dimensional Structures and Catalytic Mechanisms of Alginate Lyases. Appl. Environ. Microbiol. 2018, 84, e02040-17. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties and biomedical applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.Y.; Preston, L.A.; Schiller, N.L. ALGINATE LYASE: Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000, 54, 289–340. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Helbert, W.; Poulet, L.; Drouillard, S.; Mathieu, S.; Loiodice, M.; Couturier, M.; Lombard, V.; Terrapon, N.; Turchetto, J.; Vincentelli, R.; et al. Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space. Proc. Natl. Acad. Sci. USA 2019, 116, 6063–6068. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382 Pt 3, 769–781. [Google Scholar] [CrossRef]
- Gilbert, H.J.; Knox, J.P.; Boraston, A.B. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struct. Biol. 2013, 23, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.J.; Williams, S.J. Carbohydrate-active enzymes: Sequences, shapes, contortions and cells. Biochem. Soc. Trans. 2016, 44, 79–87. [Google Scholar] [CrossRef]
- Abbott, D.W.; van Bueren, A.L. Using structure to inform carbohydrate binding module function. Curr. Opin. Struct. Biol. 2014, 28, 32–40. [Google Scholar] [CrossRef]
- Shoseyov, O.; Shani, Z.; Levy, I. Carbohydrate binding modules: Biochemical properties and novel applications. Microbiol. Mol. Biol. Rev. MMBR 2006, 70, 283–295. [Google Scholar] [CrossRef]
- Hashimoto, H. Recent structural studies of carbohydrate-binding modules. Cell. Mol. Life Sci. CMLS 2006, 63, 2954–2967. [Google Scholar] [CrossRef] [PubMed]
- Guillén, D.; Sánchez, S.; Rodríguez-Sanoja, R. Carbohydrate-binding domains: Multiplicity of biological roles. Appl. Microbiol. Biotechnol. 2010, 85, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Ficko-Blean, E.; Boraston, A.B. Insights into the recognition of the human glycome by microbial carbohydrate-binding modules. Curr. Opin. Struct. Biol. 2012, 22, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Armenta, S.; Moreno-Mendieta, S.; Sánchez-Cuapio, Z.; Sánchez, S.; Rodríguez-Sanoja, R. Advances in molecular engineering of carbohydrate-binding modules. Proteins 2017, 85, 1602–1617. [Google Scholar] [CrossRef] [PubMed]
- Tomme, P.; Driver, D.P.; Amandoron, E.A.; Miller, R.C., Jr.; Antony, R.; Warren, J.; Kilburn, D.G. Comparison of a fungal (family I) and bacterial (family II) cellulose-binding domain. J. Bacteriol. 1995, 177, 4356–4363. [Google Scholar] [CrossRef]
- Hervé, C.; Rogowski, A.; Blake, A.W.; Marcus, S.E.; Gilbert, H.J.; Knox, J.P. Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc. Natl. Acad. Sci. USA 2010, 107, 15293–15298. [Google Scholar] [CrossRef] [PubMed]
- Bolam, D.N.; Ciruela, A.; McQueen-Mason, S.; Simpson, P.; Williamson, M.P.; Rixon, J.E.; Boraston, A.; Hazlewood, G.P.; Gilbert, H.J. Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem. J. 1998, 331 Pt 3, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.O.; Choi, E.S.; Lee, H.W.; Hwang, S.H.; Kim, C.S.; Jang, H.W.; Haam, S.J.; Jung, J.K. Enhanced secretion of Bacillus stearothermophilus L1 lipase in Saccharomyces cerevisiae by translational fusion to cellulose-binding domain. Appl. Microbiol. Biotechnol. 2004, 64, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Li, Q.; Zhu, B.; Ni, F.; Sun, Y.; Yao, Z. Effects of module truncation on biochemical characteristics and products distribution of a new alginate lyase with two catalytic modules. Glycobiology 2019, 29, 876–884. [Google Scholar] [CrossRef]
- Zhuang, J.; Zhang, K.; Liu, X.; Liu, W.; Lyu, Q.; Ji, A. Characterization of a Novel PolyM-Preferred Alginate Lyase from Marine Vibrio splendidus OU02. Mar. Drugs 2018, 16, 295. [Google Scholar] [CrossRef]
- Sun, X.M.; Xue, Z.; Sun, M.L.; Zhang, Y.; Zhang, Y.Z.; Fu, H.H.; Zhang, Y.Q.; Wang, P. Characterization of a Novel Alginate Lyase with Two Alginate Lyase Domains from the Marine Bacterium Vibrio sp. C42. Mar. Drugs 2022, 20, 746. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Sun, X.H.; Chen, X.L.; Li, P.Y.; Qin, Q.L.; Zhang, Y.Q.; Xu, F. Synergy of the Two Alginate Lyase Domains of a Novel Alginate Lyase from Vibrio sp. NC2 in Alginate Degradation. Appl. Environ. Microbiol. 2022, 88, e0155922. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Yin, H. Alginate lyase: Review of major sources and classification, properties, structure-function analysis and applications. Bioengineered 2015, 6, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Chen, X.L.; Sun, X.H.; Dong, F.; Li, C.Y.; Li, P.Y.; Ding, H.; Chen, Y.; Zhang, Y.Z.; Wang, P. Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic. J. Biol. Chem. 2020, 295, 16380–16392. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Ning, L.; Jiang, Y.; Ge, L. Biochemical Characterization and Degradation Pattern of a Novel Endo-Type Bifunctional Alginate Lyase AlyA from Marine Bacterium Isoptericola halotolerans. Mar. Drugs 2018, 16, 258. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]


| Protein | Molecular Weight (kDa) | Specific Activity (U/mg) | Specific Activity (U/μmoL) | Vmax (nmol·s−1) | Km (mM) | kcat (s−1) | kcat/Km (mM·s−1) |
|---|---|---|---|---|---|---|---|
| Full-Length | 65.27 | 302.65 | 19,753.38 | 0.61 | 0.31 | 4.35 | 7.02 |
| PDT-CD I | 32.73 | 1955.00 | 63,992.15 | 0.49 | 0.29 | 13.30 | 23.33 |
| PDT-CD II | 39.11 | 70.13 | 2742.81 | 0.87 | 0.42 | 0.46 | 2.88 |
| CD I-CD II | 60.27 | 31.42 | 1893.62 | 0.76 | 4.05 | 0.03 | 0.006 |
| Primers | Sequence (5′ to 3′) | Usage |
|---|---|---|
| Expression of VsAly7A and truncated proteins | ||
| VsAly7A-FL-F | GGAGATATACATATGGGCGGCAGCAGCTCT | Expression of VsAly7A-FL |
| VsAly7A-FL-R | GTGGTGGTGCTCGAGTTGGTGACGGGTGCT | |
| PDT-CD I-F | GGAGATATACATATGGGCGGCAGCAGCTCT | Expression of PDT-CD I |
| PDT-CD I-R | GTGGTGGTGCTCGAGCCACGCGATTGAATC | |
| PDT-CD II-F | GGAGATATACATATGGGCGGCAGCAGCTCT | Expression of PDT-CD II |
| PDT-CD II-O-R | ATCTGTGTGTTCGATAATGTCTTGAAATTTCGTTATGGAGTACG | |
| PDT-CD II-O-F | TAACGAAATTTCAAGACATTATCGAACACACAGATTCAATC | |
| PDT-CD II-R | GTGGTGGTGCTCGAGTTGGTGACGGGTGCT | |
| CD I-CD II-F | GGAGATATACATATGCCGTACTCCATAACG | Expression of CD I-CD II |
| CD I-CD II-R | GTGGTGGTGCTCGAGTTGGTGACGGGTGCT | |
| PDT-F | GGAGATATACATATGGGCGGCAGCAGCTCT | Expression of PDT |
| PDT-R | GTGGTGGTGCTCGAGGCCTGAATTGTCTAA | |
| CD I-F | GGAGATATACATATGCCGTACTCCATAACG | Expression of CD I |
| CD I-R | GTGGTGGTGCTCGAGCCACGCGATTGAATC | |
| CD II-F | GGAGATATACATATGAAAATCGAACACACA | Expression of CD II |
| CD II-R | GTGGTGGTGCTCGAGTTGGTGACGGGTGCT | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Z.; Zhang, F.; Wang, H.; Tang, L.; Yu, W.; Han, F. A “Pro-Asp-Thr” Amino Acid Repeat from Vibrio sp. QY108 Alginate Lyase Exhibits Alginate-Binding Capacity and Enhanced Soluble Expression and Thermostability. Int. J. Mol. Sci. 2024, 25, 5801. https://doi.org/10.3390/ijms25115801
Fu Z, Zhang F, Wang H, Tang L, Yu W, Han F. A “Pro-Asp-Thr” Amino Acid Repeat from Vibrio sp. QY108 Alginate Lyase Exhibits Alginate-Binding Capacity and Enhanced Soluble Expression and Thermostability. International Journal of Molecular Sciences. 2024; 25(11):5801. https://doi.org/10.3390/ijms25115801
Chicago/Turabian StyleFu, Zheng, Fengchao Zhang, Hainan Wang, Luyao Tang, Wengong Yu, and Feng Han. 2024. "A “Pro-Asp-Thr” Amino Acid Repeat from Vibrio sp. QY108 Alginate Lyase Exhibits Alginate-Binding Capacity and Enhanced Soluble Expression and Thermostability" International Journal of Molecular Sciences 25, no. 11: 5801. https://doi.org/10.3390/ijms25115801
APA StyleFu, Z., Zhang, F., Wang, H., Tang, L., Yu, W., & Han, F. (2024). A “Pro-Asp-Thr” Amino Acid Repeat from Vibrio sp. QY108 Alginate Lyase Exhibits Alginate-Binding Capacity and Enhanced Soluble Expression and Thermostability. International Journal of Molecular Sciences, 25(11), 5801. https://doi.org/10.3390/ijms25115801





