Genome-Wide Identification, Molecular Characterization, and Expression Analysis of the HSP70 and HSP90 Gene Families in Thamnaconus septentrionalis
Abstract
1. Introduction
2. Results
2.1. Identification and Characteristics of HSP70 and HSP90 Genes in T. septentrionalis
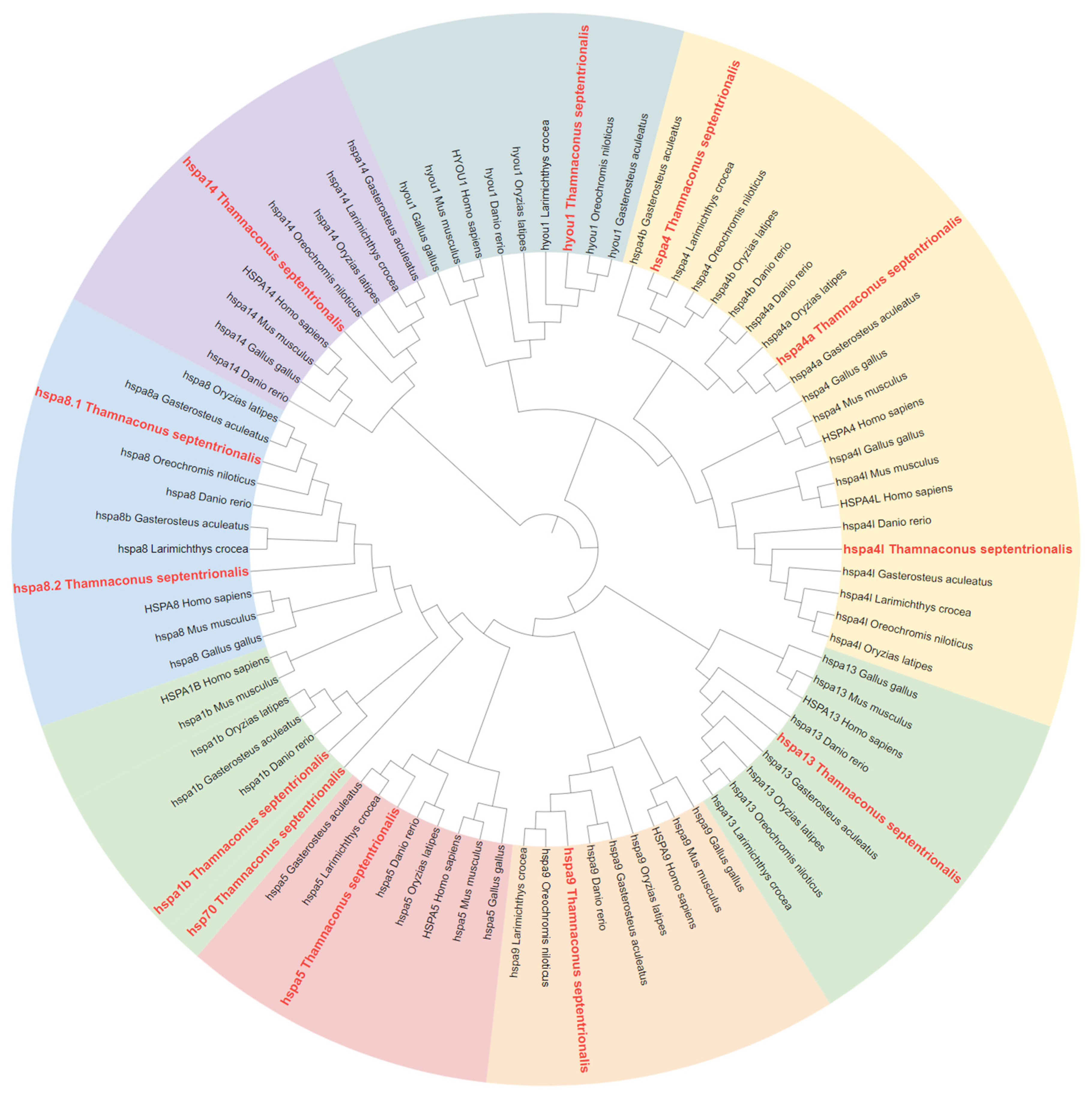
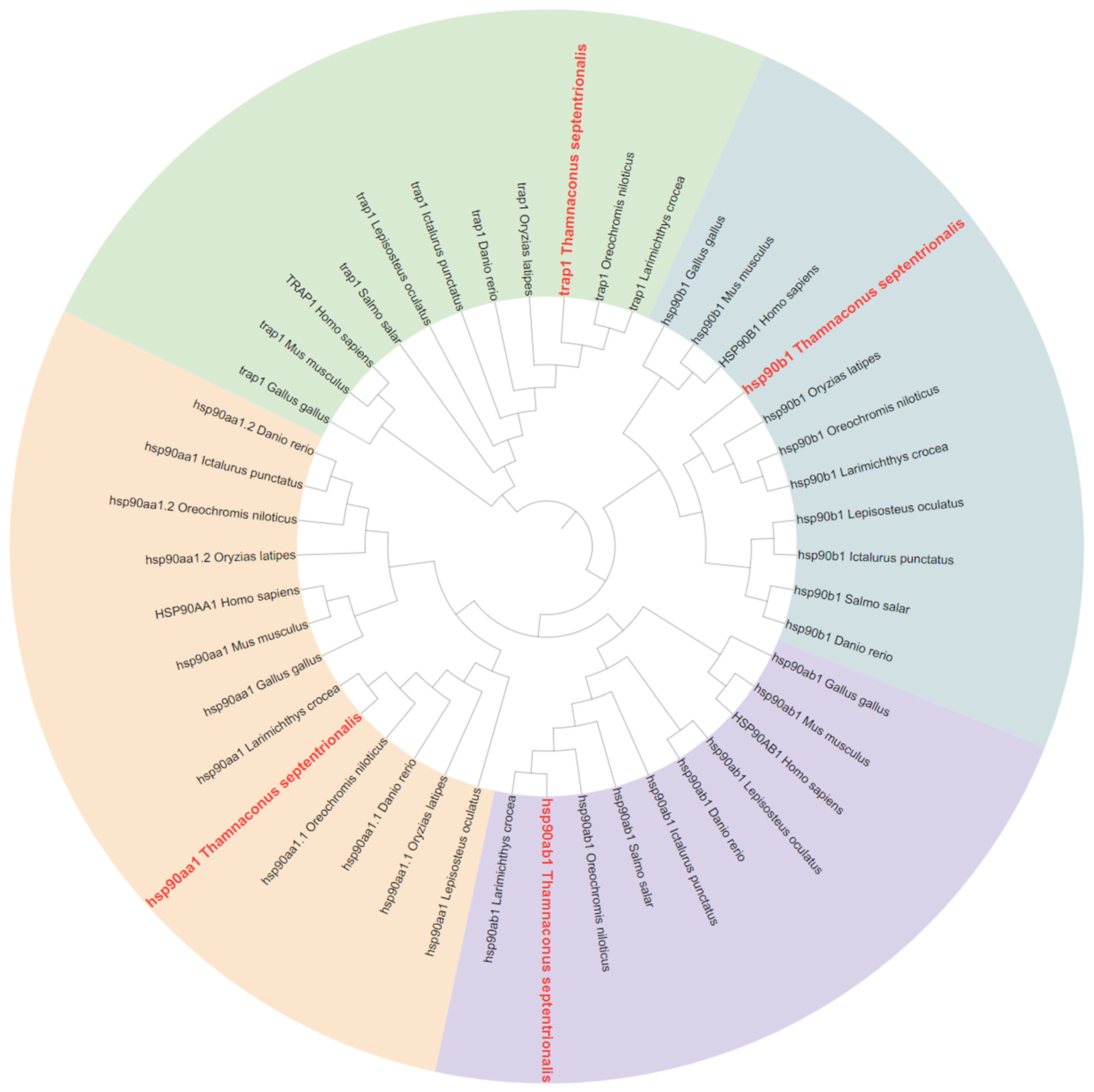
2.2. Phylogenetic Analysis of HSP70 and HSP90 Genes
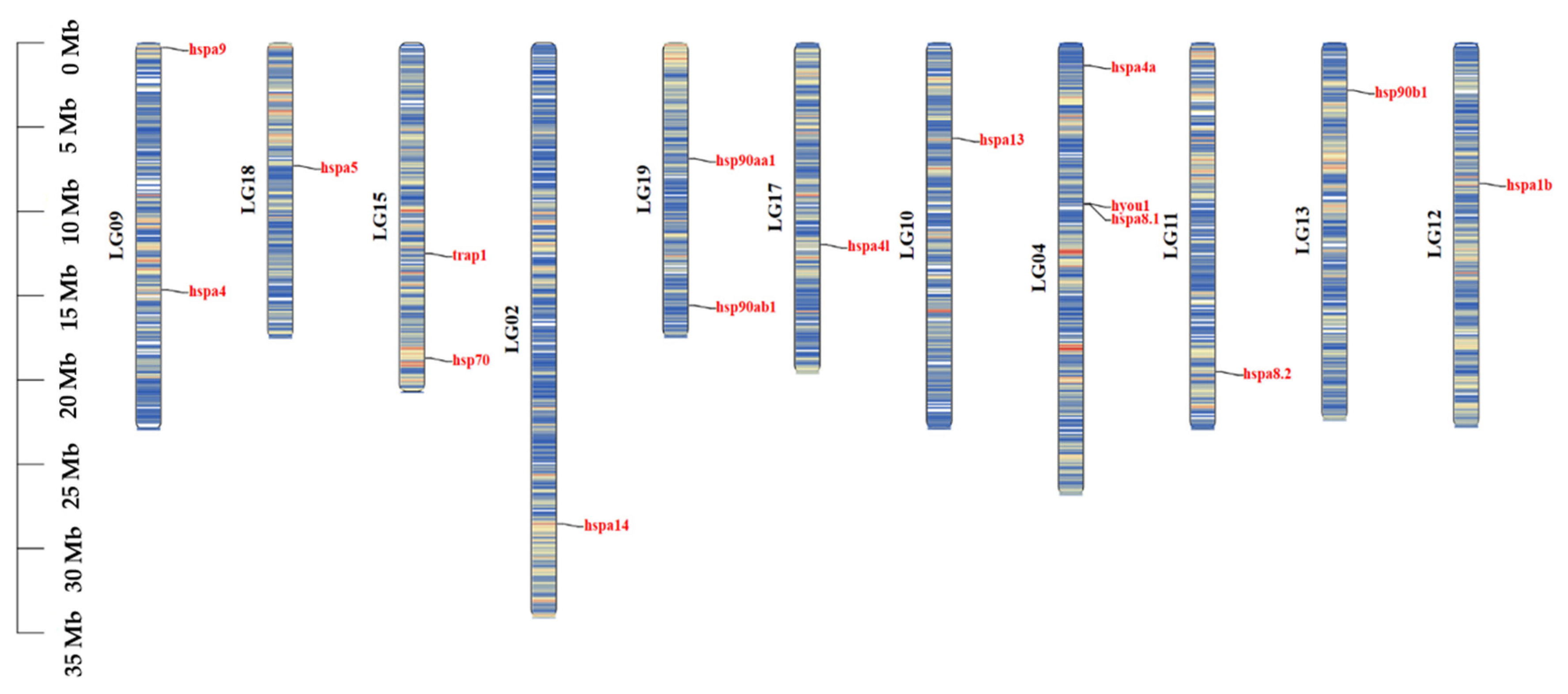
2.3. Gene Structure and Chromosome Distribution Analysis
2.4. Motif and Conserved Domain Analysis
2.5. Protein Structure Prediction and Subcellular Localization
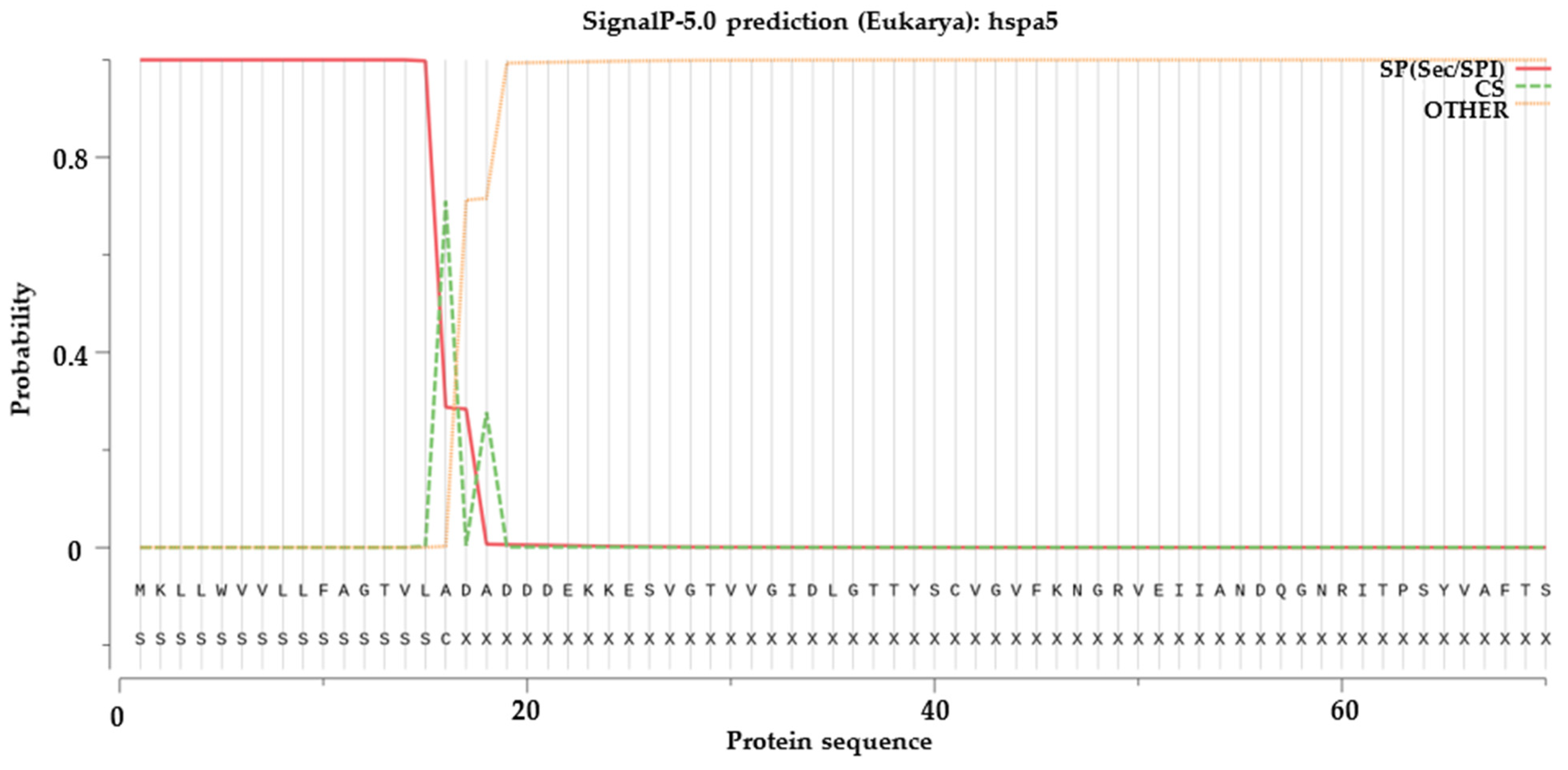
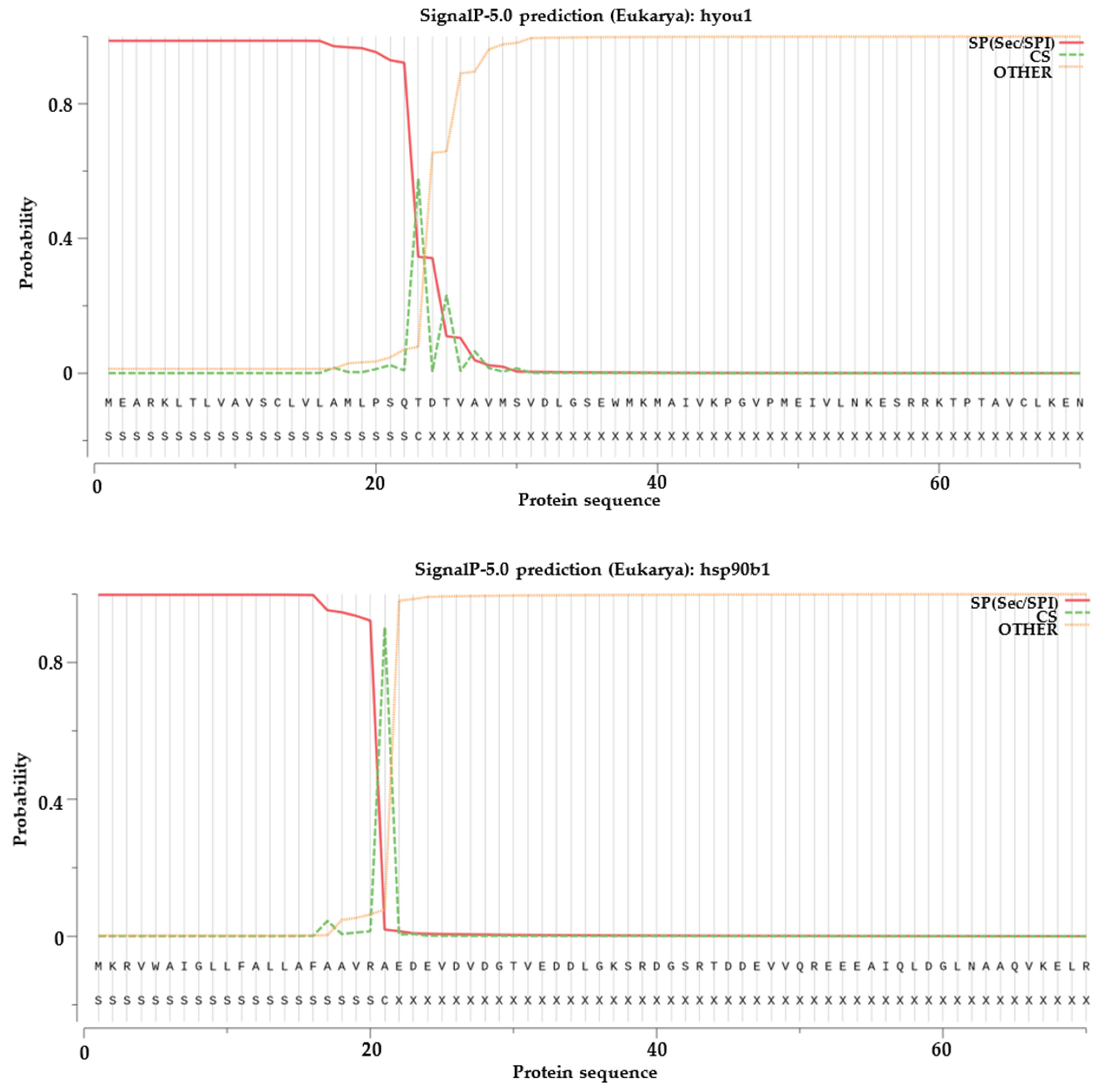
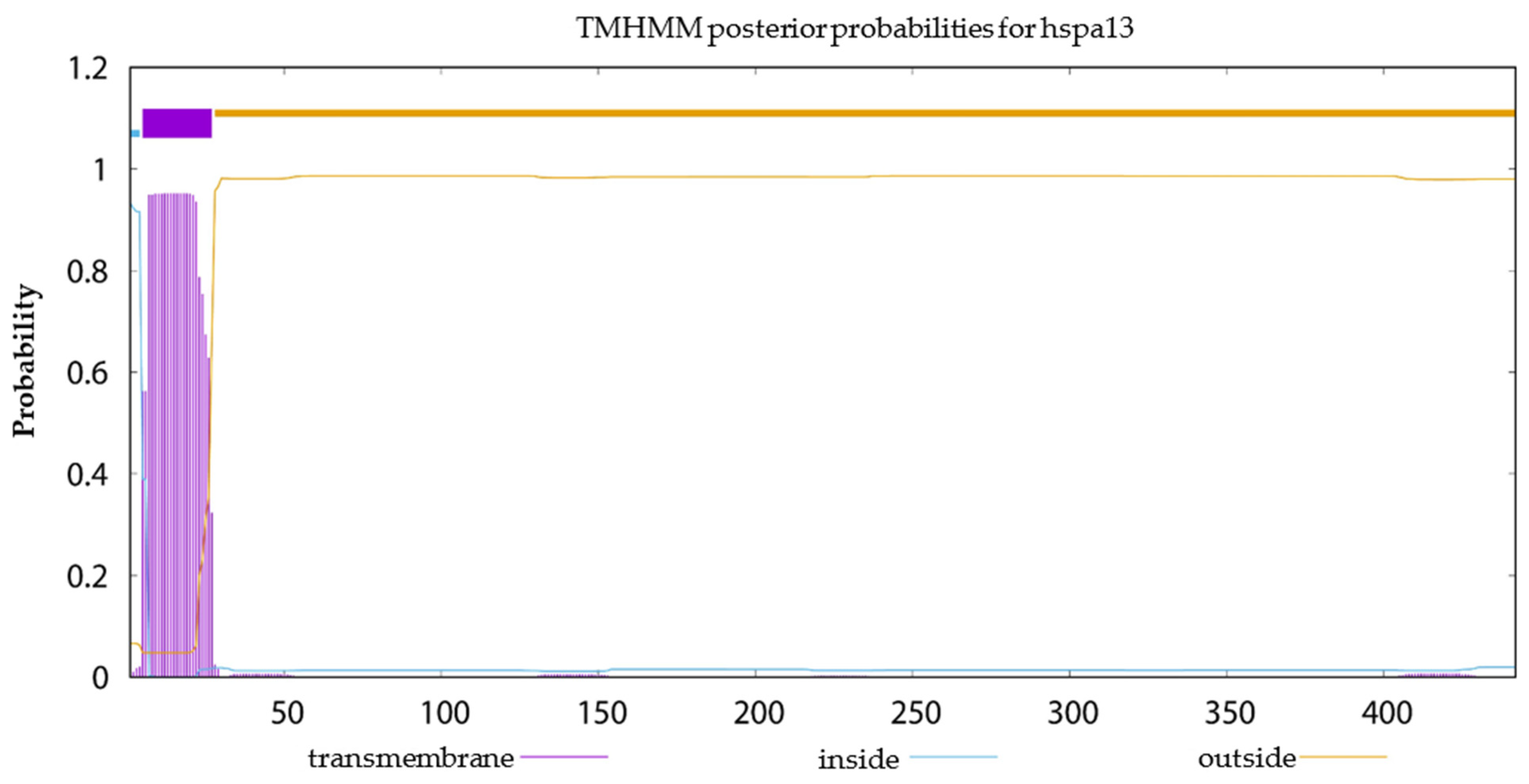
2.6. Signal Peptide Prediction, Transmembrane Structural Domain Prediction and Selection Test on Duplicated HSP70 and HSP90 Gene Pairs
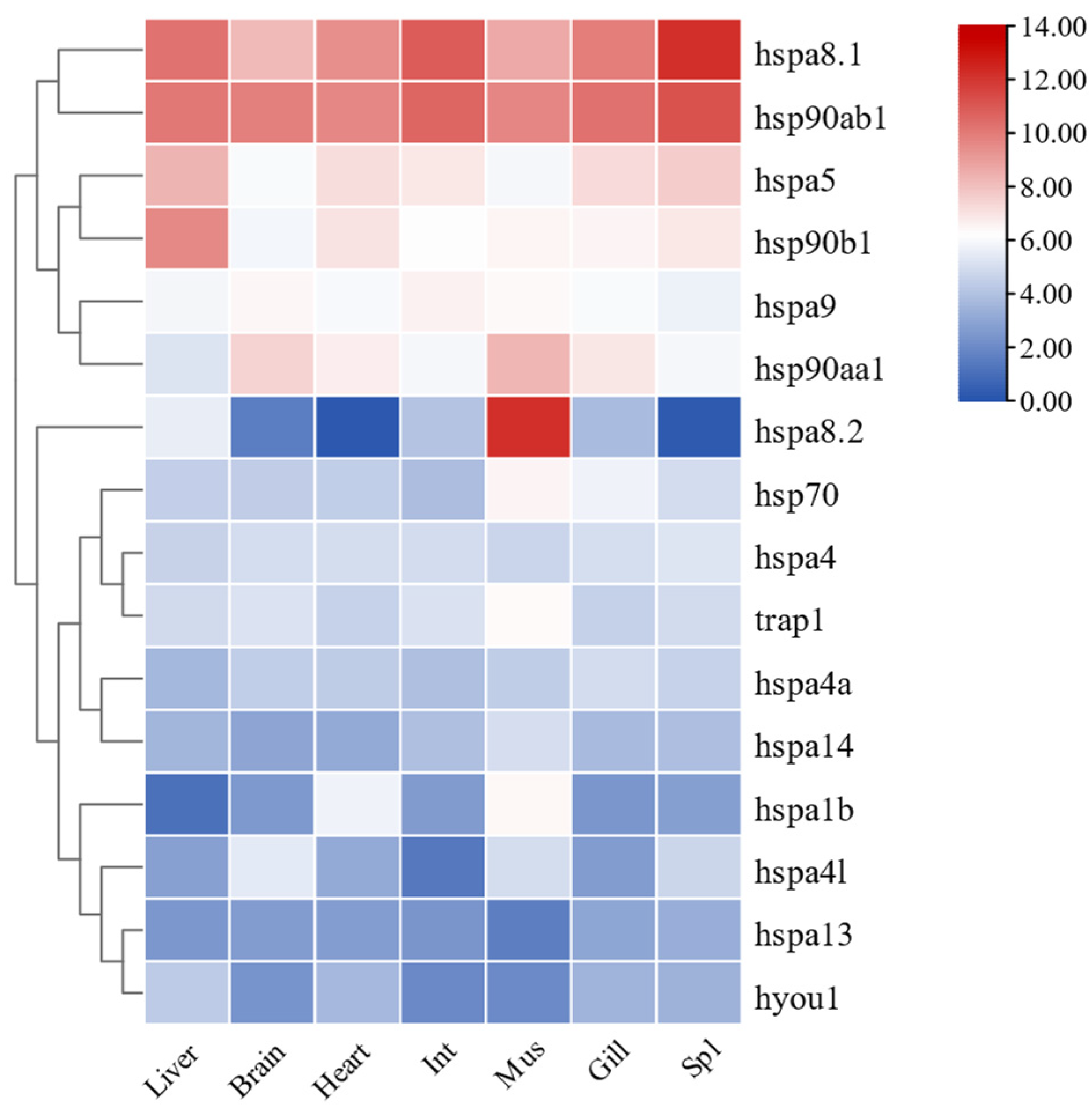
2.7. Expression Patterns of HSP70 and HSP90 Genes in Tissues of T. septentrionalis
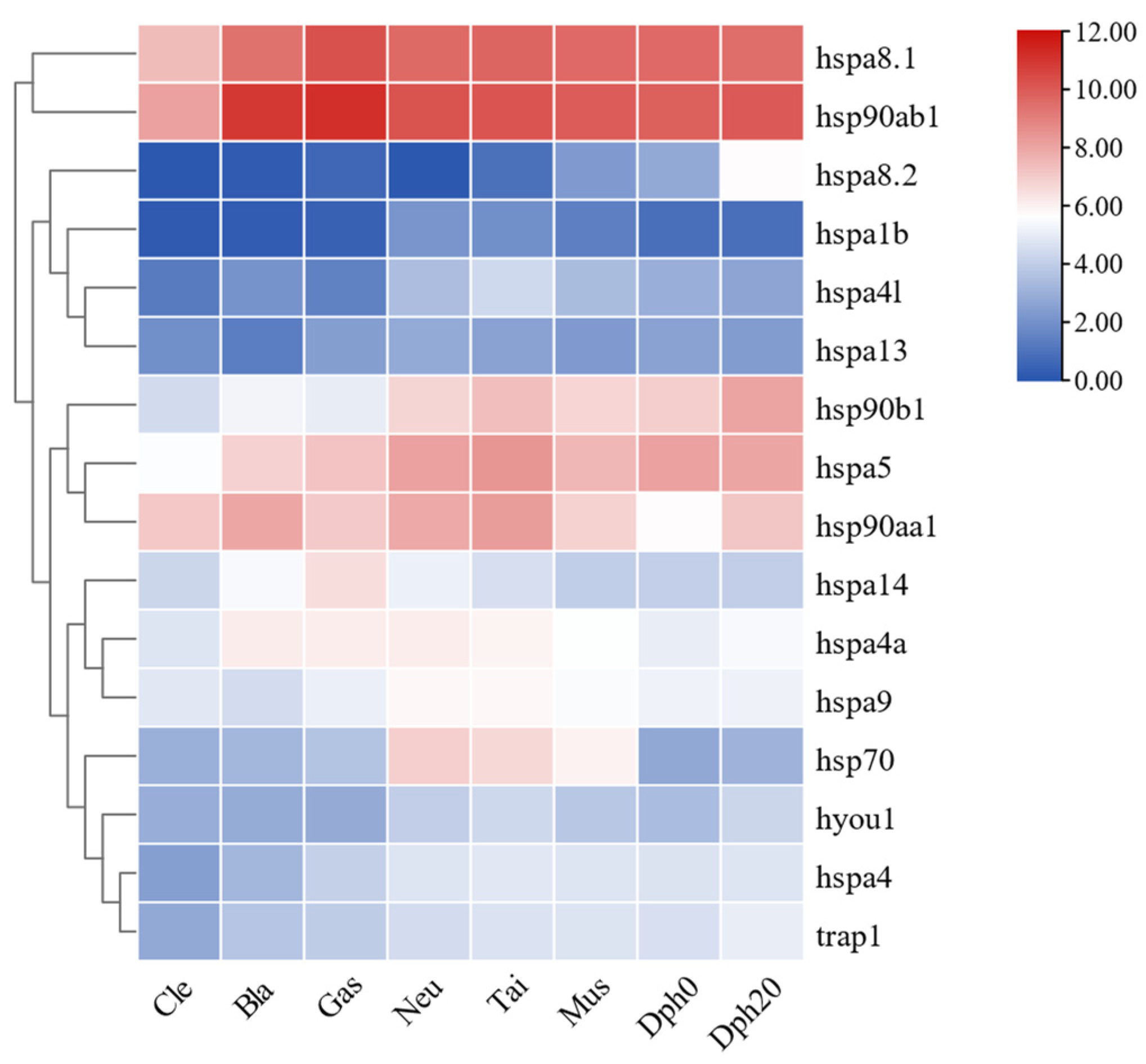
2.8. Expression Patterns of HSP70 and HSP90 Genes during Different Developmental Stages of T. septentrionalis
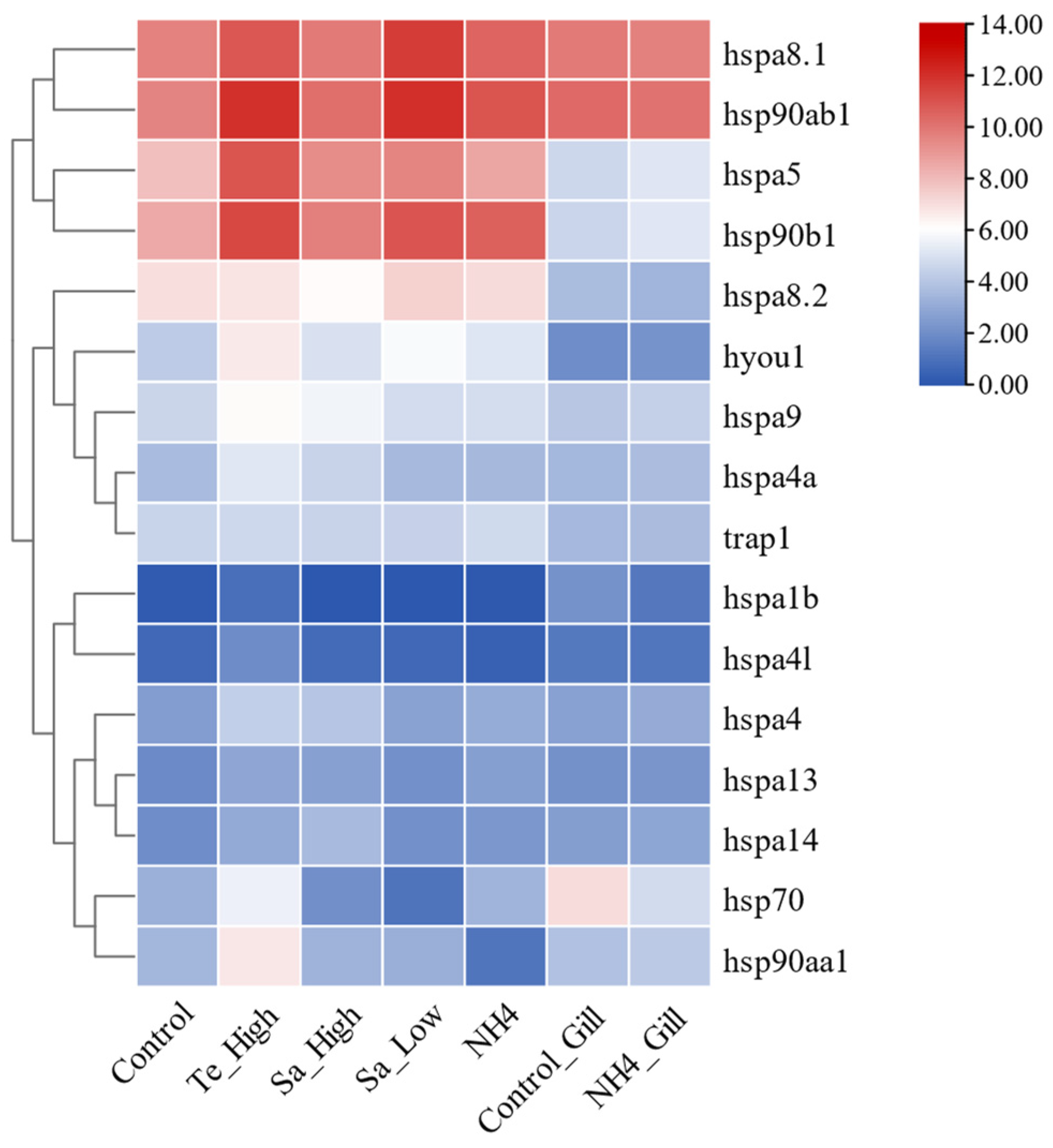
2.9. Expression Patterns of HSP70 and HSP90 Genes in Abiotic Stresses of T. septentrionalis
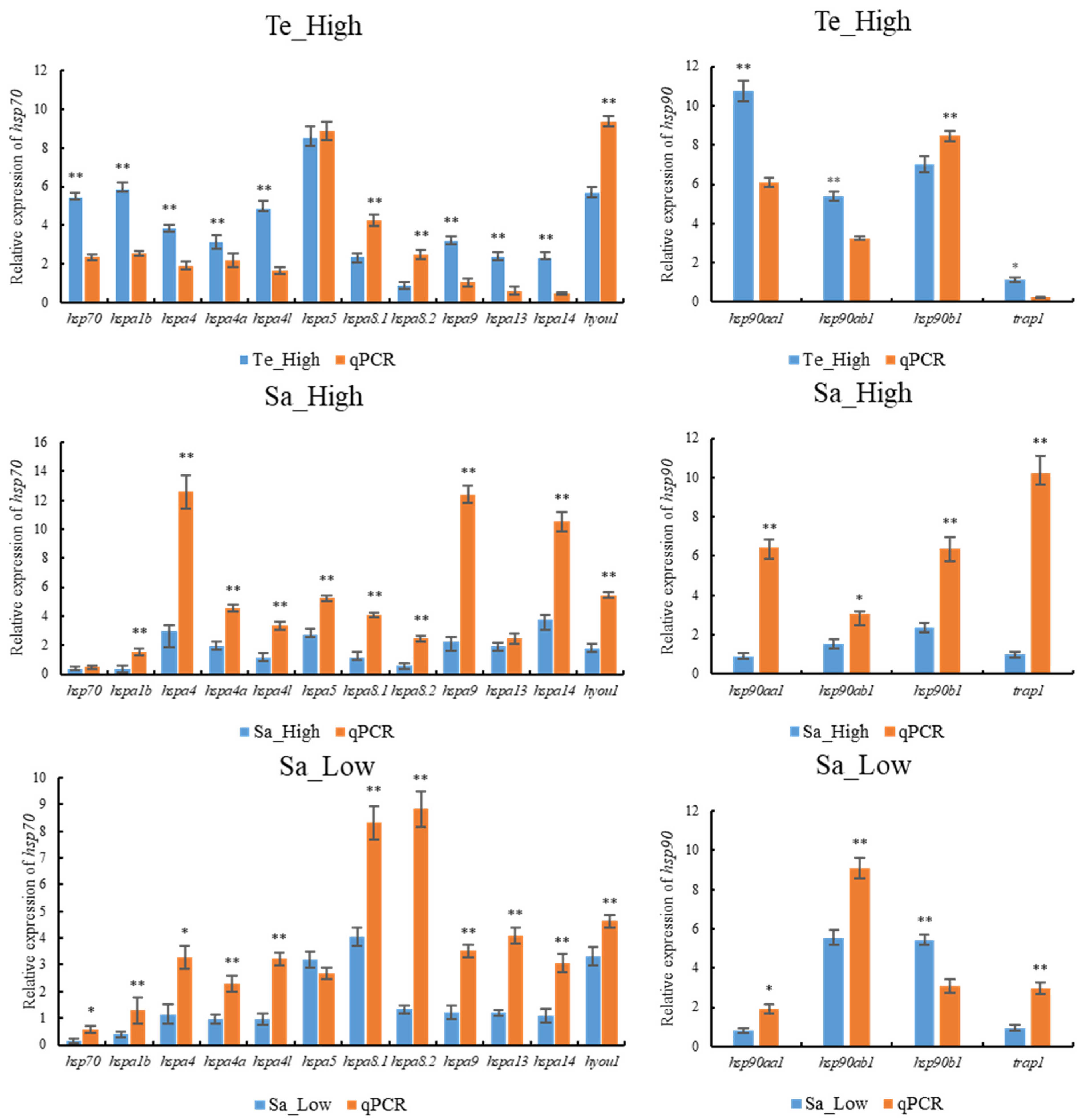
2.10. qPCR Validation of HSP70 and HSP90 Genes Expression Patterns under Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Identification and Nomenclature of HSP70 and HSP90 Gene Family Members in T. septentrionalis
4.2. Physicochemical Property Analysis and Phylogenetic Analysis
4.3. Gene Structure, Chromosome Distribution Analysis, Motif, and Conserved Domain Analysis
4.4. Protein Structure and Subcellular Localization
4.5. Signal Peptides Prediction, Transmembrane Structural Domain Prediction and Selection Test of Duplicated HSP70 and HSP90 Genes
4.6. Expression Profiles of HSP70 and HSP90 Genes in Tissues of T. septentrionalis
4.7. Expression Profiles of HSP70 and HSP90 Genes during Different Developmental Stages of T. septentrionalis
4.8. Expression Profiles of HSP70 and HSP90 Genes under Abiotic Stresses
4.9. Quantitative Real-Time PCR (qPCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eid, I.I.; Bhassu, S.; Goh, Z.H.; Khoo, L.T.; Tan, G.Y.A. Molecular characterization and gene evolution of the heat shock protein 70 gene in snakehead fish with different tolerances to temperature. Biochem. Syst. Ecol. 2016, 66, 137–144. [Google Scholar] [CrossRef]
- Zheng, W.; Xu, X.; Chen, Y.; Wang, J.; Zhang, T.; Zechen, E.; Chen, S.; Liu, Y. Genome-Wide Identification, Molecular Characterization, and Involvement in Response to Abiotic and Biotic Stresses of the HSP70 Gene Family in Turbot (Scophthalmus maximus). Int. J. Mol. Sci. 2023, 24, 6025. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.; Biemar, F.; Müller, F.; Iyengar, A.; Prunet, P.; Maclean, N.; Martial, J.A.; Muller, M. Cloning and expression analysis of an inducible HSP70 gene from tilapia fish. FEBS Lett. 2000, 474, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.; Hageman, J.; Vos, M.; Kubota, H.; Tanguay, R.; Bruford, E.; Cheetham, M.; Chen, B.; Hightower, L. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 2009, 14, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Genest, O.; Wickner, S.; Doyle, S. Hsp90 and Hsp70 chaperones: Collaborators in protein remodeling. J. Biol. Chem. 2019, 294, 2109–2120. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Bansal, A.; Hashimoto-Torii, K. HSP70 and HSP90 in neurodegenerative diseases. Neurosci. Lett. 2020, 716, 134678. [Google Scholar] [CrossRef] [PubMed]
- Hangzo, H.; Banerjee, B.; Saha, S.; Saha, N. Ammonia stress under high environmental ammonia induces Hsp70 and Hsp90 in the mud eel, Monopterus Cuchia. Fish Physiol. Biochem. 2017, 43, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Padmini, E. Physiological adaptations of stressed fish to polluted environments: Role of heat shock proteins. Rev. Environ. Contam. Toxicol. 2010, 206, 1–27. [Google Scholar] [PubMed]
- Ma, F.; Luo, L.; Wang, Q. Hsp60/10 and sHsp families of heat shock protein genes in rainbow trout (Oncorhynchus mykiss) and their expression under heat stress. Aquac. Int. 2022, 30, 1–18. [Google Scholar] [CrossRef]
- Sun, Y.; Wen, H.; Tian, Y.; Mao, X.; Li, X.; Li, J.; Hu, Y.; Liu, Y.; Li, J.; Li, Y. HSP90 and HSP70 Families in Lateolabrax maculatus: Genome-Wide Identification, Molecular Characterization, and Expression Profiles in Response to Various Environmental Stressors. Front. Physiol. 2021, 12, 784803. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Jin, S.; Shou, C.; Han, Z. Hsp70 Gene Family in Sebastiscus marmoratus: The Genome-Wide Identification and Transcriptome Analysis under Thermal Stress. Genes 2023, 14, 1779. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Sun, S.; Gao, T.; Han, Z. The Hsp70 Gene Family in Boleophthalmus pectinirostris: Genome-Wide Identification and Expression Analysis under High Ammonia Stress. Animals 2019, 9, 36. [Google Scholar] [CrossRef]
- Umam, K.; Tang, C.-H.; Lee, T.-H. The roles of heat shock protein 70 (HSP70) and heat shock protein 90 (HSP90) in osmoregulatory and metabolic tissues of milkfish Chanos chanos under osmotic stress. Sci. Bus. Exch. 2013, 6, 1058. [Google Scholar]
- Shi, Q.; Yu, C.; Zhu, D.; Li, S.; Wen, X. Effects of dietary Sargassum horneri on resisting hypoxia stress, which changes blood biochemistry, antioxidant status, and hepatic HSP mRNA expressions of juvenile black sea bream Acanthopagrus schlegelii. J. Appl. Phycol. 2020, 32, 3457–3466. [Google Scholar] [CrossRef]
- Bian, L.; Li, F.; Ge, J.; Wang, P.; Chang, Q.; Zhang, S.; Li, J.; Liu, C.; Liu, K.; Liu, X.; et al. Chromosome-level genome assembly of the greenfin horse-faced filefish (Thamnaconus septentrionalis) using Oxford Nanopore PromethION sequencing and Hi-C technology. Mol. Ecol. Resour. 2020, 20, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Kho, K.; Chen, Z.; Kim, J.; Kim, Y.; Zhang, Z. Cryopreservation of filefish (Thamnaconus septentrionalis Gunther, 1877) sperm. Aquac. Res. 2004, 35, 1429–1433. [Google Scholar] [CrossRef]
- Zhan, B.; Lou, D.; Zhong, J. Evaluation of Thamnaconus septentrionalis Resources and their Rational Utilization. Aquac. Fish. 1986, 4, 409–418. (In Chinese) [Google Scholar]
- Li, G.; Wang, Q.; Zhang, K. Artificial breeding technology of Thamnaconus septentrionalis. China Fish. 2021, 552, 82–84. (In Chinese) [Google Scholar]
- Xue, M. Early Development and Growth of Thamnaconus septentrionali; Shanghai Ocean University: Shanghai, China. (In Chinese)
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.; Frank, M.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Szalay, M.; Kovács, I.; Korcsmáros, T.; Böde, C.; Csermely, P. Stress-induced rearrangements of cellular networks: Consequences for protection and drug design. FEBS Lett. 2007, 581, 3675–3680. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Yang, J.; QI, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm 2022, 3, e161. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Nillegoda, N.; Mayer, M.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Hoter, A.; El-Sabban, M.; Naim, H. The HSP90 Family: Structure, Regulation, Function, and Implications in Health and Disease. Int. J. Mol. Sci. 2018, 19, 2560. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.; Sullivan, W.; Toft, D. The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperone complex. J. Biol. Chem. 2002, 277, 38294–38304. [Google Scholar] [CrossRef] [PubMed]
- Brocchieri, L.; De Macario, E.; Macario, A. hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. Bmc Evol. Biol. 2008, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Li, C.; Xie, Y.; Liu, S.; Zhang, J.; Yao, J.; Jiang, C.; Li, Y.; Liu, Z. Genome-wide identification of Hsp70 genes in channel catfish and their regulated expression after bacterial infection. Fish Shellfish Immunol. 2016, 49, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Xu, H.; Han, Z. Genome-Wide Identification of Hsp70 Genes in the Large Yellow Croaker (Larimichthys crocea) and Their Regulated Expression Under Cold and Heat Stress. Genes 2018, 9, 590. [Google Scholar] [CrossRef]
- Elicker, K.; Hutson, L. Genome-wide analysis and expression profiling of the small heat shock proteins in zebrafish. Gene 2007, 403, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Franck, E.; Madsen, O.; Van Rheede, T.; Ricard, G.; Huynen, M.; de Jong, W. Evolutionary diversity of vertebrate small heat shock proteins. J. Mol. Evol. 2004, 59, 792–805. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Gho, H.; Nguyen, M.; Kim, S.; An, G. Genome-wide expression analysis of HSP70 family genes in rice and identification of a cytosolic HSP70 gene highly induced under heat stress. Funct. Integr. Genom. 2013, 13, 391–402. [Google Scholar] [CrossRef]
- Wang, X.; Komatsu, S. Plant subcellular proteomics: Application for exploring optimal cell function in soybean. J. Proteom. 2016, 143, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.; Canniff, N.; Guay, K.; Hebert, D. The Role of Endoplasmic Reticulum Chaperones in Protein Folding and Quality Control. Prog. Mol. Subcell. Biol. 2021, 59, 27–50. [Google Scholar] [PubMed]
- Moore, R.; Purugganan, M. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [PubMed]
- Kulmuni, J.; Wurm, Y.; Pamilo, P. Comparative genomics of chemosensory protein genes reveals rapid evolution and positive selection in ant-specific duplicates. Heredity 2013, 110, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Conant, G.; Wolfe, K. Turning a hobby into a job: How duplicated genes find new functions. Nat. Rev. Genet. 2008, 9, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Voldoire, E.; Brunet, F.; Naville, M.; Volff, J.; Galiana, D. Expansion by whole genome duplication and evolution of the Sox gene family in teleost fish. PLoS ONE 2017, 12, e0180936. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ding, N.; Lu, T.; Zhao, J.; Wang, Z.; Jiang, P.; Liu, S.; Wang, X.; Liu, B.; Li, L. Evolutionary Contribution of Duplicated Genes to Genome Evolution in the Ginseng Species Complex. Genome Biol. Evol. 2021, 13, evab051. [Google Scholar] [CrossRef] [PubMed]
- Holub, E. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pan, C.; Wang, Y.; Ye, L.; Wu, J.; Chen, L.; Zou, T.; Lu, G. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. Bmc Genom. 2015, 16, 386. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Li, S.; Gao, Y.; Zhan, A. Genome-Wide Identification, Characterization and Expression Analyses of Heat Shock Protein-Related Genes in a Highly Invasive Ascidian Ciona savignyi. Front. Physiol. 2018, 9, 1043. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, J.; Liu, B.; Zhang, L.; Chen, J.; Lu, M. Genome-wide analysis of the Populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. Bmc Genom. 2013, 14, 532. [Google Scholar] [CrossRef]
- Stricher, F.; Macri, C.; Ruff, M.; Muller, S. HSPA8/HSC70 chaperone protein Structure, function, and chemical targeting. Autophagy 2013, 9, 1937–1954. [Google Scholar] [CrossRef]
- Yang, Y.; Han, T.; Xiao, J.; Li, X.; Wang, J. Transcriptome analysis reveals carbohydrate-mediated liver immune responses in Epinephelus akaara. Sci. Rep. 2018, 8, 639. [Google Scholar] [CrossRef]
- Krone, P.; Evans, T.; Lechinger, S. Heat shock gene expression and function during zebrafish embryogenesis. Semin. Cell Dev. Biol. 2003, 14, 267–274. [Google Scholar] [CrossRef]
- Liu, K.; Hao, X.; Wang, Q.; Hou, J.; Lai, X.; Dong, Z.; Shao, C. Genome-wide identification and characterization of heat shock protein family 70 provides insight into its divergent functions on immune response and development of Paralichthys olivaceus. Peerj 2019, 7, e7781. [Google Scholar] [CrossRef]
- Xiao, H.; Wang, H.; He, Q.; Zhou, J.; Du, S. Gene expression and functional analysis of Aha1a and Aha1b in stress response in zebrafish. Comp. Biochem. Physiol. B-BioChemistry Mol. Biol. 2022, 262, 110777. [Google Scholar] [CrossRef]
- Marchin, M.; Kelly, P.; Fang, J. Tracker: Continuous HMMER and BLAST searching. Bioinformatics 2005, 21, 388–389. [Google Scholar] [CrossRef]
- Yu, D.; Lee, D.; Kim, S.; Lee, C.; Song, J.; Kong, E.; Kim, J. Algorithm for Predicting Functionally Equivalent Proteins from BLAST and HMMER Searches. J. Microbiol. Biotechnol. 2012, 22, 1054–1058. [Google Scholar] [CrossRef]
- Bateman, A.; Birney, E.; Durbin, R.; Finn, R.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.; et al. The Pfam protein families database. Nucleic Acids Res. 2000, 28, 263–266. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Bailey, T.; Johnson, J.; Grant, C.; Noble, W. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Guan, J.; Chen, Z.; Zhang, J.; Guan, S.; Zheng, Y.; Wen, H.; Liu, H. Embryonic development of Thamnaconus septentrionali under artificial cultivation conditions. Adv. Mar. Sci. 2011, 29, 498–505. (In Chinese) [Google Scholar]
- Jiang, L.; Zhang, Z.; Wang, Z.; Du, R.; Luan, S.; Wu, Y.; Liu, L. Key technology for early factory breeding of Thamnaconus septentrionali. Fish. Sci. 2021, 40, 801–809. (In Chinese) [Google Scholar]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Gene Name | NCBI Accession Number | Number of Amino Acid | Molecular Weight (Da) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|---|
| hsp70 | PP349927 | 639 | 70,266.65 | 5.44 | 36.44 | 84.98 | −0.423 |
| hspa1b | PP349926 | 639 | 70,106.24 | 5.53 | 37.52 | 83.93 | −0.412 |
| hspa4 | PP349928 | 846 | 95,013.44 | 5.04 | 43.03 | 72.97 | −0.628 |
| hspa4a | PP349923 | 842 | 94,144.84 | 5.17 | 43.54 | 76.28 | −0.534 |
| hspa4l | PP349922 | 832 | 92,954.52 | 5.46 | 41.20 | 79.68 | −0.487 |
| hspa5 | PP349929 | 656 | 72,413.78 | 4.97 | 30.55 | 84.39 | −0.482 |
| hspa8.1 | PP357443 | 515 | 56,702.91 | 5.90 | 36.45 | 81.98 | −0.405 |
| hspa8.2 | PP349925 | 647 | 70,877.18 | 5.27 | 39.45 | 81.82 | −0.422 |
| hspa9 | PP349930 | 688 | 74,170.99 | 6.24 | 45.59 | 81.79 | −0.374 |
| hspa13 | PP339453 | 442 | 47,831.97 | 5.52 | 40.48 | 102.15 | 0.082 |
| hspa14 | PP349924 | 506 | 54,814.52 | 5.66 | 38.49 | 94.43 | −0.052 |
| hyou1 | PP349931 | 945 | 106,412.40 | 6.03 | 45.14 | 79.47 | −0.527 |
| hsp90aa1 | PP349934 | 725 | 83,238.60 | 5.06 | 35.70 | 82.81 | −0.625 |
| hsp90ab1 | PP349932 | 724 | 83,206.20 | 4.87 | 39.91 | 82.56 | −0.640 |
| hsp90b1 | PP349933 | 801 | 91,991.32 | 4.74 | 38.96 | 79.38 | −0.693 |
| trap1 | PP349935 | 719 | 81,840.08 | 5.99 | 45.54 | 83.99 | −0.440 |
| Gene Name | α Helix | Extended Strand | β Turn | Random Coil | Subcellular Location | Three-Dimensional Structural Quality Parameters |
|---|---|---|---|---|---|---|
| hsp70 | 42.10% | 18.47% | 5.95% | 33.49% | Nucleus | 94.0% |
| hspa1b | 42.57% | 18.94% | 7.20% | 31.30% | Cytosol | 94.5% |
| hspa4 | 42.55% | 14.18% | 3.07% | 40.19% | Cytosol | 90.6% |
| hspa4a | 43.11% | 14.01% | 3.09% | 39.79% | Cytosol | 92.2% |
| hspa4l | 43.63% | 14.06% | 3.25% | 39.06% | Cytosol | 91.9% |
| hspa5 | 43.75% | 18.60% | 6.71% | 30.95% | Endoplasmic reticulum | 93.6% |
| hspa8.1 | 33.79% | 23.30% | 8.54% | 34.37% | Cytosol | 93.2% |
| hspa8.2 | 41.58% | 17.77% | 6.96% | 33.69% | Cytosol | 93.8% |
| hspa9 | 43.75% | 19.77% | 8.14% | 28.34% | mitochondrion | 92.9% |
| hspa13 | 40.27% | 21.27% | 5.66% | 32.81% | Endoplasmic reticulum | 93.7% |
| hspa14 | 34.78% | 24.90% | 5.14% | 35.18% | Cytosol | 93.6% |
| hyou1 | 48.99% | 13.23% | 3.28% | 34.50% | Endoplasmic reticulum | 89.9% |
| hsp90aa1 | 52.55% | 13.66% | 5.38% | 28.41% | Cytosol | 89.5% |
| hsp90ab1 | 50.14% | 13.54% | 4.56% | 31.77% | Cytosol | 90.4% |
| hsp90b1 | 54.68% | 12.36% | 3.75% | 29.21% | Endoplasmic reticulum | 88.2% |
| trap1 | 45.20% | 13.77% | 5.70% | 35.33% | mitochondrion | 88.8% |
| Gene Pair | Ka | Ks | Ka/Ks |
|---|---|---|---|
| hsp70-hspa1b | 0.104125523 | 0.918427 | 0.113374 |
| hspa4-hspa4a | 0.175276849 | 1.508566 | 0.116188 |
| hspa4-hspa4l | 0.307067194 | 1.683617 | 0.182385 |
| hspa4a-hspa4l | 0.285118286 | 1.702043 | 0.167515 |
| hspa8.1-hspa8.2 | 0.06700074 | 1.461276 | 0.045851 |
| hsp90aa1-hsp90ab1 | 0.116635393 | 1.012976 | 0.115141 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Chang, Q.; Fang, Q.; Zhang, Z.; Wu, D.; Bian, L.; Chen, S. Genome-Wide Identification, Molecular Characterization, and Expression Analysis of the HSP70 and HSP90 Gene Families in Thamnaconus septentrionalis. Int. J. Mol. Sci. 2024, 25, 5706. https://doi.org/10.3390/ijms25115706
Chen Y, Chang Q, Fang Q, Zhang Z, Wu D, Bian L, Chen S. Genome-Wide Identification, Molecular Characterization, and Expression Analysis of the HSP70 and HSP90 Gene Families in Thamnaconus septentrionalis. International Journal of Molecular Sciences. 2024; 25(11):5706. https://doi.org/10.3390/ijms25115706
Chicago/Turabian StyleChen, Ying, Qing Chang, Qinmei Fang, Ziyang Zhang, Dan Wu, Li Bian, and Siqing Chen. 2024. "Genome-Wide Identification, Molecular Characterization, and Expression Analysis of the HSP70 and HSP90 Gene Families in Thamnaconus septentrionalis" International Journal of Molecular Sciences 25, no. 11: 5706. https://doi.org/10.3390/ijms25115706
APA StyleChen, Y., Chang, Q., Fang, Q., Zhang, Z., Wu, D., Bian, L., & Chen, S. (2024). Genome-Wide Identification, Molecular Characterization, and Expression Analysis of the HSP70 and HSP90 Gene Families in Thamnaconus septentrionalis. International Journal of Molecular Sciences, 25(11), 5706. https://doi.org/10.3390/ijms25115706







