Integrating Multiple Database Resources to Elucidate the Gene Flow in Southeast Asian Pig Populations
Abstract
1. Introduction
2. Results
2.1. Genetic Relationships and Population Structure of Yunnan Indigenous Pigs
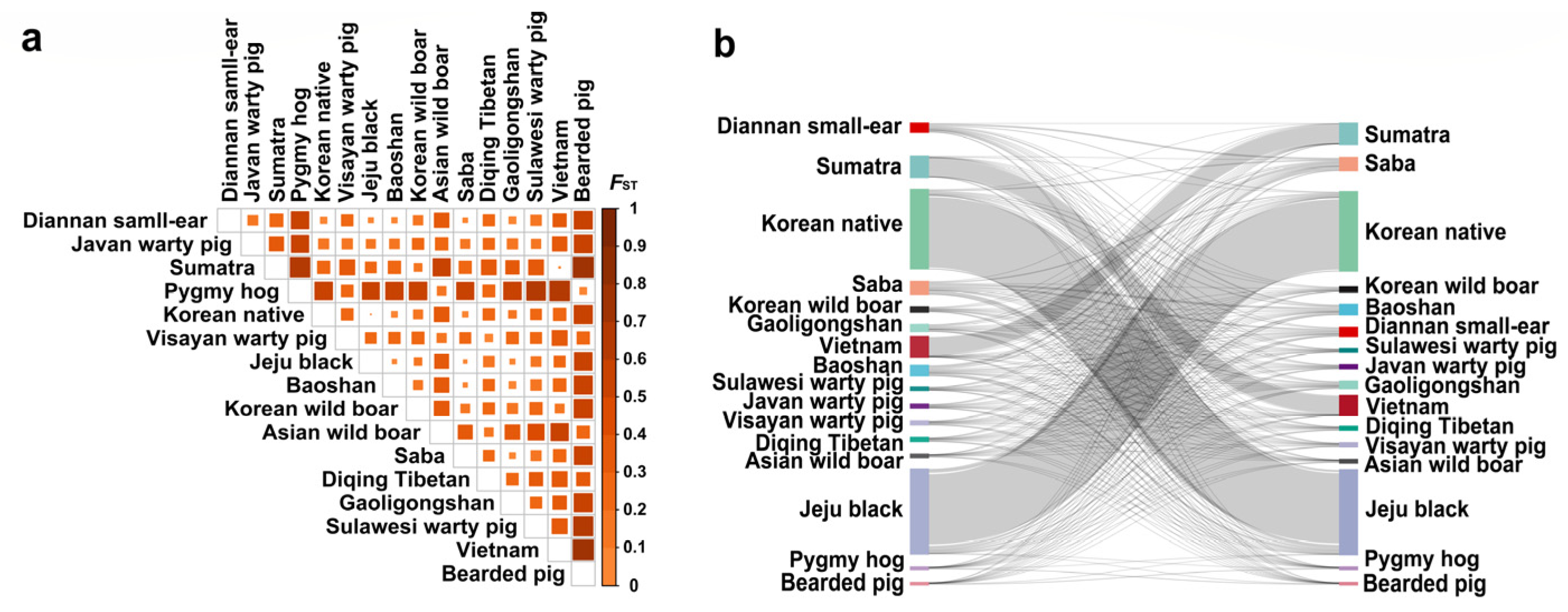
2.2. Genetic Differentiation and Gene Flow Analyses
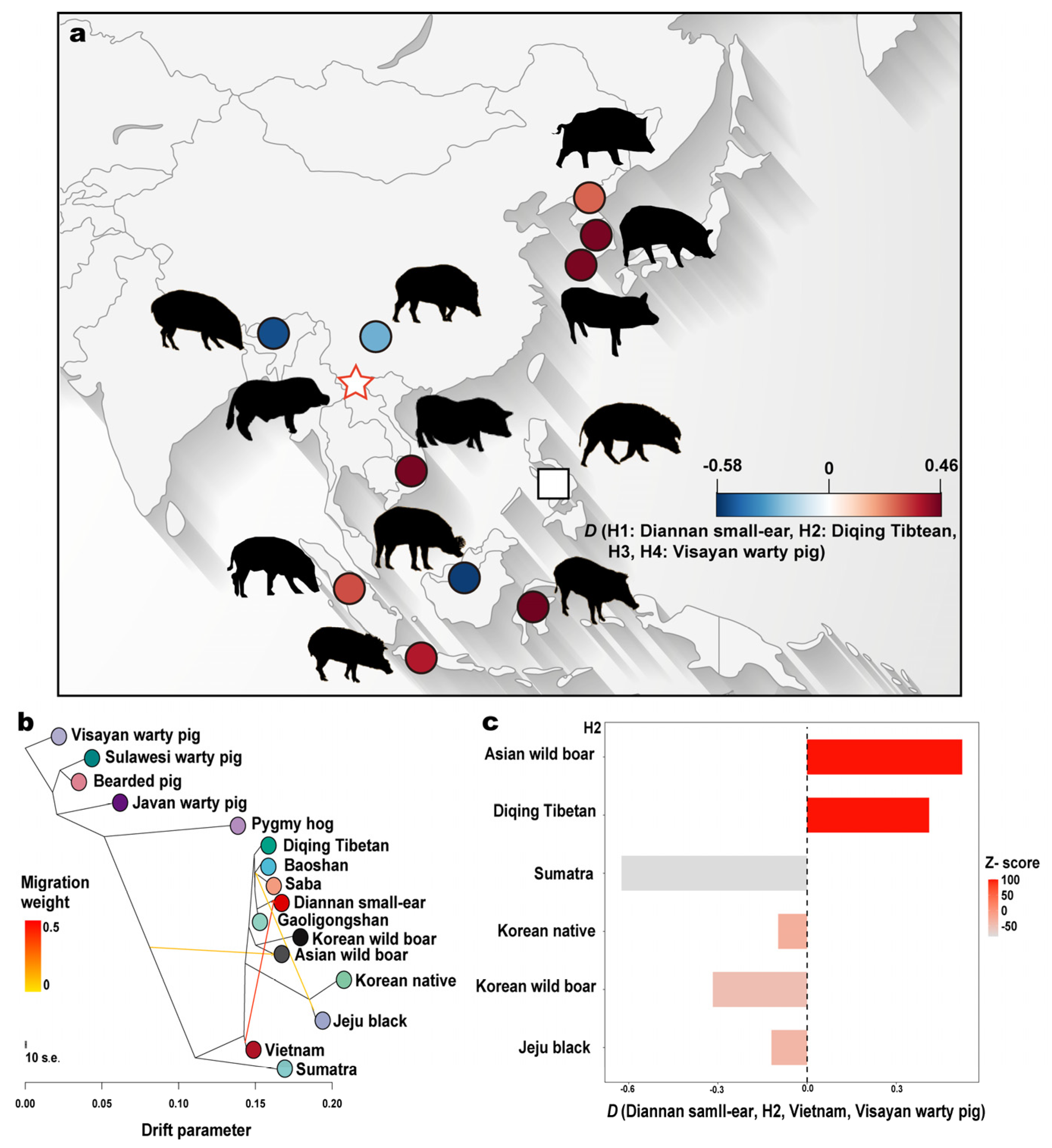
2.3. Migration and Introgression Events Analysis
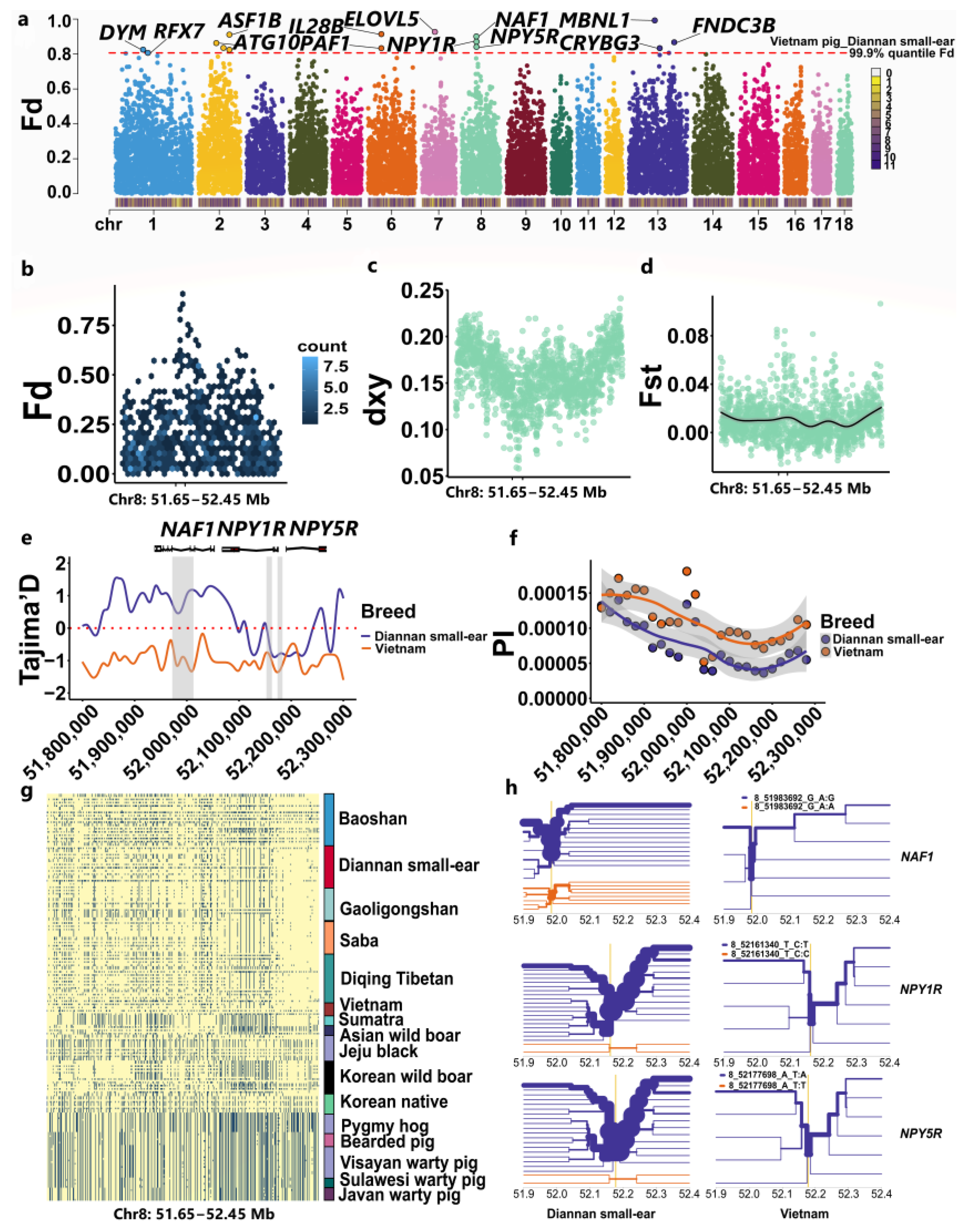
2.4. Multiple-Gene Introgression of Vietnam to Diannan Small-Ear
2.5. Enrichment Analysis of Candidate Genes in Introgression Genes
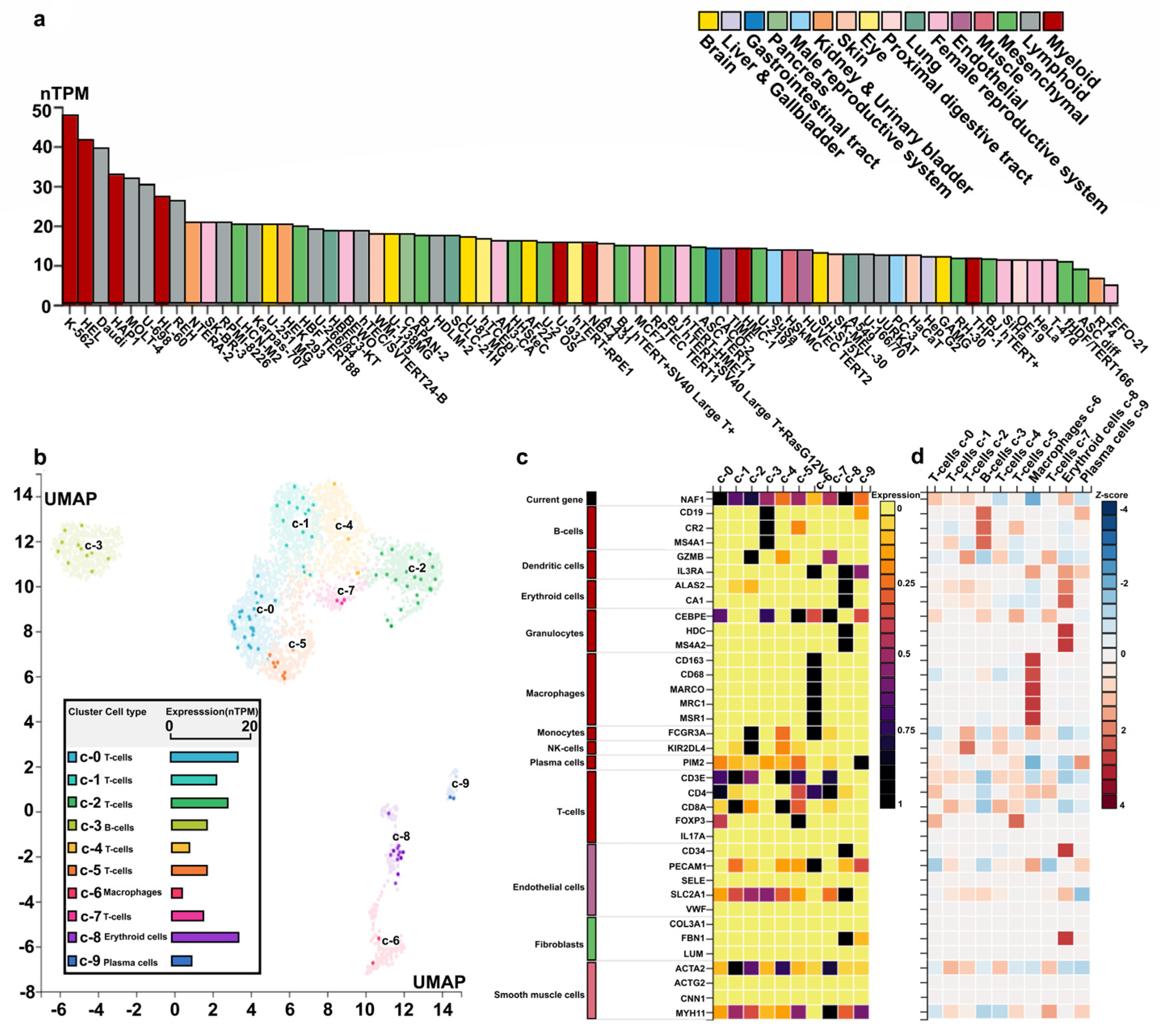
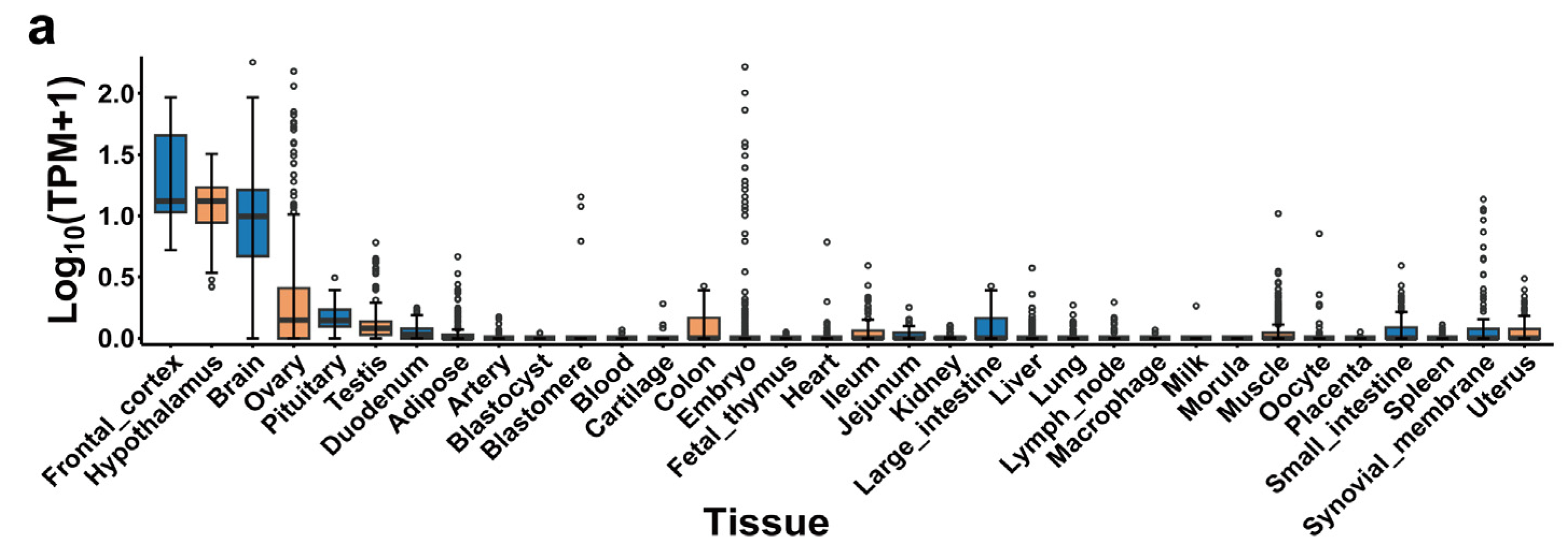
2.6. Cell Expression of the NAF1 Gene in Humans
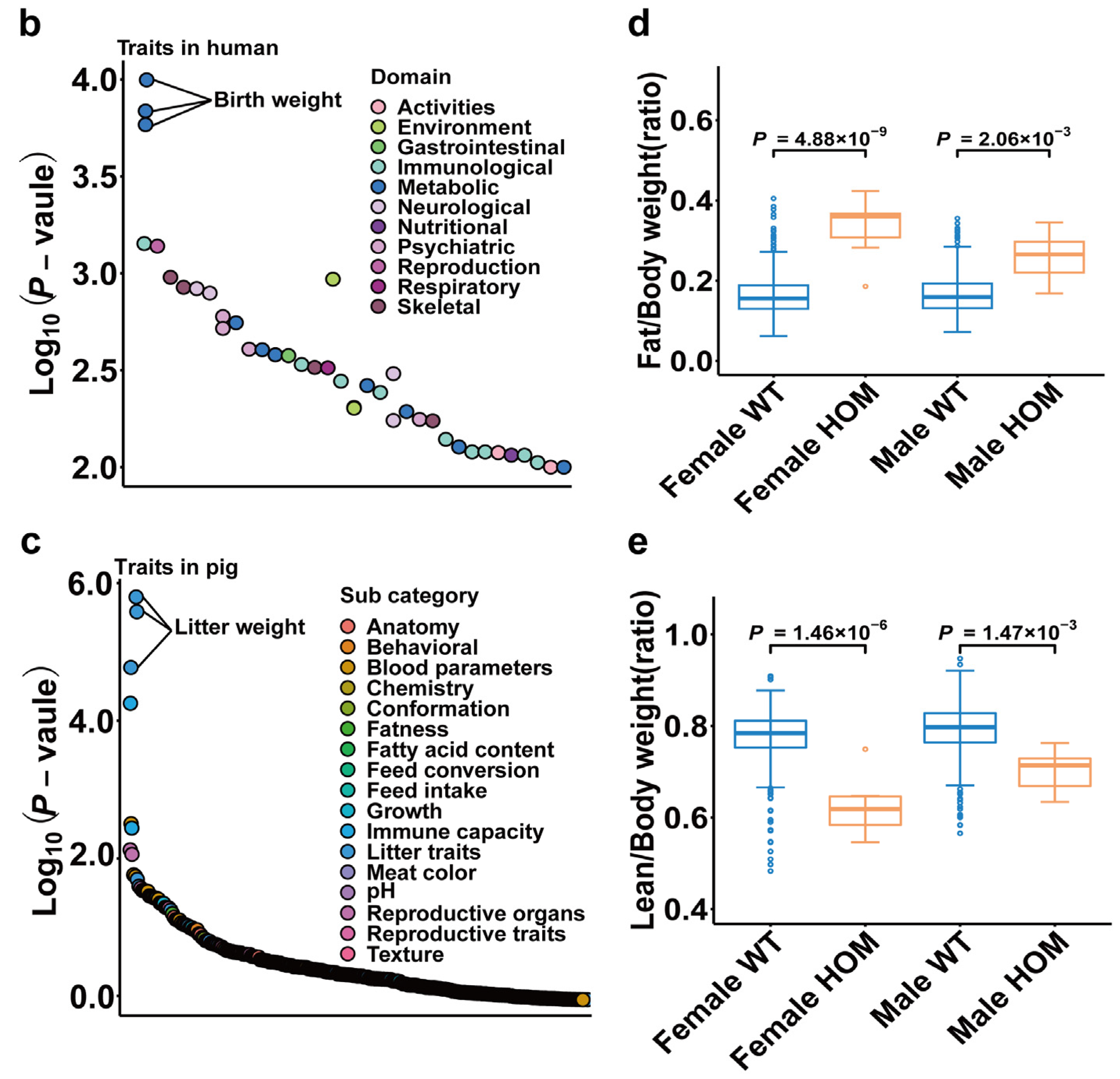
2.7. PigGTEx, Human PheWAS and Mouse Knockouts of NPY5R Gene
3. Discussion
4. Materials and Methods
4.1. Newly Added Samples and Genotypes
4.2. Population Genetic Structure of Yunnan Indigenous Pigs
4.3. Differentiation and Gene Flow Analyses
4.4. Migration Events and Genetic Introgression Analyses
4.5. Genome-Wide Scan for Gene Introgression through fd Statistic
4.6. Gene Annotation and Enrichment Analysis
4.7. RNA Expression in Human Cell
4.8. PheWAS, Gene Expression of Bulk Tissue and Mouse Gene Knockout
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdul-Rahman, F.; Tranchina, D.; Gresham, D. Fluctuating Environments Maintain Genetic Diversity through Neutral Fitness Effects and Balancing Selection. Mol. Biol. Evol. 2021, 38, 4362–4375. [Google Scholar] [CrossRef] [PubMed]
- Wall, E.; Visscher, P.M.; Hospital, F.; Woolliams, J.A. Genomic Contributions in Livestock Gene Introgression Programmes. Genet. Sel. Evol. 2005, 37, 291–313. [Google Scholar] [CrossRef] [PubMed]
- Ai, H.; Fang, X.; Yang, B.; Huang, Z.; Chen, H.; Mao, L.; Zhang, F.; Zhang, L.; Cui, L.; He, W.; et al. Adaptation and Possible Ancient Interspecies Introgression in Pigs Identified by Whole-Genome Sequencing. Nat. Genet. 2015, 47, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Bosse, M.; Megens, H.-J.; Frantz, L.A.F.; Madsen, O.; Larson, G.; Paudel, Y.; Duijvesteijn, N.; Harlizius, B.; Hagemeijer, Y.; Crooijmans, R.P.M.A.; et al. Genomic Analysis Reveals Selection for Asian Genes in European Pigs Following Human-Mediated Introgression. Nat. Commun. 2014, 5, 4392. [Google Scholar] [CrossRef]
- Bosse, M.; Madsen, O.; Megens, H.-J.; Frantz, L.A.F.; Paudel, Y.; Crooijmans, R.P.M.A.; Groenen, M.A.M. Hybrid Origin of European Commercial Pigs Examined by an In-Depth Haplotype Analysis on Chromosome 1. Front. Genet. 2014, 5, 442. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Frantz, L.A.F.; Schraiber, J.G.; Madsen, O.; Megens, H.-J.; Cagan, A.; Bosse, M.; Paudel, Y.; Crooijmans, R.P.M.A.; Larson, G.; Groenen, M.A.M. Evidence of Long-Term Gene Flow and Selection during Domestication from Analyses of Eurasian Wild and Domestic Pig Genomes. Nat. Genet. 2015, 47, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Mary, N.; Iannuccelli, N.; Petit, G.; Bonnet, N.; Pinton, A.; Barasc, H.; Faure, A.; Calgaro, A.; Grosbois, V.; Servin, B.; et al. Genome-Wide Analysis of Hybridization in Wild Boar Populations Reveals Adaptive Introgression from Domestic Pig. Evol. Appl. 2022, 15, 1115–1128. [Google Scholar] [CrossRef]
- Wang, M.-S.; Murray, G.G.R.; Mann, D.; Groves, P.; Vershinina, A.O.; Supple, M.A.; Kapp, J.D.; Corbett-Detig, R.; Crump, S.E.; Stirling, I.; et al. A Polar Bear Paleogenome Reveals Extensive Ancient Gene Flow from Polar Bears into Brown Bears. Nat. Ecol. Evol. 2022, 6, 936–944. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, L.; Duan, D.; Qiao, R.; Li, X.; Li, X.; Han, X. Genomic Analysis Reveals Human-Mediated Introgression From European Commercial Pigs to Henan Indigenous Pigs. Front. Genet. 2021, 12, 705803. [Google Scholar] [CrossRef]
- Ramos-Onsins, S.E.; Burgos-Paz, W.; Manunza, A.; Amills, M. Mining the Pig Genome to Investigate the Domestication Process. Heredity 2014, 113, 471–484. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.; Li, Z.; Lü, P.; Gardner, J.D.; Ye, M.; Wang, J.; Yang, M.; Shao, J.; Wang, W.; et al. Ancient DNA Reveals the Maternal Genetic History of East Asian Domestic Pigs. J. Genet. Genom. 2022, 49, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Zheng, Z.; Gong, M.; Li, D.; Hu, S.; Cai, Y.; Wang, Y.; Nanaei, H.A.; Zhang, N.; Yu, T.; et al. Ancient Genomes Reveal the Genetic Inheritance and Recent Introgression in Chinese Indigenous Pigs. Sci. China Life Sci. 2022, 65, 842–845. [Google Scholar] [CrossRef]
- Ai, H.; Zhang, M.; Yang, B.; Goldberg, A.; Li, W.; Ma, J.; Brandt, D.; Zhang, Z.; Nielsen, R.; Huang, L. Human-Mediated Admixture and Selection Shape the Diversity on the Modern Swine (Sus scrofa) Y Chromosomes. Mol. Biol. Evol. 2021, 38, 5051–5065. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Yu, Y.; Feng, W.; Du, H.; Yu, J.; Kang, H.; Zheng, X.; Wang, Z.; Liu, G.E.; Ernst, C.W.; et al. Evidence of Evolutionary History and Selective Sweeps in the Genome of Meishan Pig Reveals Its Genetic and Phenotypic Characterization. Gigascience 2018, 7, giy058. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, C.; Chen, J.; Bai, Y.; Wang, K.; Wang, Y.; Fang, M. Genome-Wide Analysis Reveals Human-Mediated Introgression from Western Pigs to Indigenous Chinese Breeds. Genes 2020, 11, 275. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, H.; Huang, M.; Tang, J.; Yang, L.; Yu, Z.; Li, D.; Li, G.; Jiang, Y.; Sun, Y.; et al. Whole-Genome SNP Markers Reveal Conservation Status, Signatures of Selection, and Introgression in Chinese Laiwu Pigs. Evol. Appl. 2021, 14, 383–398. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ding, W. Spatial Pattern of the Ecological Environment in Yunnan Province. PLoS ONE 2021, 16, e0248090. [Google Scholar] [CrossRef]
- Hua, Z. The Floras of Southern and Tropical Southeastern Yunnan Have Been Shaped by Divergent Geological Histories. PLoS ONE 2013, 8, e64213. [Google Scholar] [CrossRef]
- National Committee of animal genetic resources. National List of Livestock and Poultry Genetic Resources in China; Ministry of Agriculture and Rural Affairs of the People’s Republic of China: Beijing, China, 2022.
- Quan, J.; Gou, X.; Zhao, S. The genetic diversity of mitochondrial DNA D-loop in Yunnan native pigs. J. Sichuan Agric. Univ. 2015, 33, 422–428. [Google Scholar] [CrossRef]
- Diao, S.; Huang, S.; Chen, Z.; Teng, J.; Ma, Y.; Yuan, X.; Chen, Z.; Zhang, H.; Li, J.; Zhang, Z. Genome-Wide Signatures of Selection Detection in Three South China Indigenous Pigs. Genes 2019, 10, 346. [Google Scholar] [CrossRef]
- Diao, S.; Xu, Z.; Ye, S.; Huang, S.; Teng, J.; Yuan, X.; Chen, Z.; Zhang, H.; Li, J.; Zhang, Z. Exploring the Genetic Features and Signatures of Selection in South China Indigenous Pigs. J. Integr. Agric. 2021, 20, 1359–1371. [Google Scholar] [CrossRef]
- Layos, J.K.N.; Godinez, C.J.P.; Liao, L.M.; Yamamoto, Y.; Masangkay, J.S.; Mannen, H.; Nishibori, M. Origin and Demographic History of Philippine Pigs Inferred from Mitochondrial DNA. Front. Genet. 2021, 12, 823364. [Google Scholar] [CrossRef]
- Marincs, F.; Molnár, J.; Tóth, G.; Stéger, V.; Barta, E. Introgression and Isolation Contributed to the Development of Hungarian Mangalica Pigs from a Particular European Ancient Bloodline. Genet. Sel. Evol. 2013, 45, 22. [Google Scholar] [CrossRef]
- Moest, M.; Van Belleghem, S.M.; James, J.E.; Salazar, C.; Martin, S.H.; Barker, S.L.; Moreira, G.R.P.; Merot, C.; Joron, M.; Nadeau, N.J.; et al. Selective Sweeps on Novel and Introgressed Variation Shape Mimicry Loci in a Butterfly Adaptive Radiation. PLoS. Biol. 2020, 18, e3000597. [Google Scholar] [CrossRef]
- Liu, Y.; Mao, X.; Krause, J.; Fu, Q. Insights into Human History from the First Decade of Ancient Human Genomics. Science 2021, 373, 1479–1484. [Google Scholar] [CrossRef] [PubMed]
- Teng, J.; Gao, Y.; Yin, H.; Bai, Z.; Liu, S.; Zeng, H.; PigGTEx Consortium; Bai, L.; Cai, Z.; Zhao, B.; et al. A Compendium of Genetic Regulatory Effects across Pig Tissues. Nat. Genet. 2024, 56, 112–123. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium The GTEx Consortium Atlas of Genetic Regulatory Effects across Human Tissues. Science 2020, 369, 1318–1330. [CrossRef]
- Dickinson, M.E.; Flenniken, A.M.; Ji, X.; Teboul, L.; Wong, M.D.; White, J.K.; Meehan, T.F.; Weninger, W.J.; Westerberg, H.; Adissu, H.; et al. High-Throughput Discovery of Novel Developmental Phenotypes. Nature 2016, 537, 508–514. [Google Scholar] [CrossRef]
- Zeng, H.; Zhang, W.; Lin, Q.; Gao, Y.; Teng, J.; Xu, Z.; Cai, X.; Zhong, Z.; Wu, J.; Liu, Y.; et al. PigBiobank: A Valuable Resource for Understanding Genetic and Biological Mechanisms of Diverse Complex Traits in Pigs. Nucleic Acids Res. 2024, 52, D980–D989. [Google Scholar] [CrossRef]
- Tripathy, S.; Lytle, K.A.; Stevens, R.D.; Bain, J.R.; Newgard, C.B.; Greenberg, A.S.; Huang, L.-S.; Jump, D.B. Fatty Acid Elongase-5 (Elovl5) Regulates Hepatic Triglyceride Catabolism in Obese C57BL/6J Mice. J. Lipid Res. 2014, 55, 1448–1464. [Google Scholar] [CrossRef]
- Zhao, L.; Li, F.; Liu, T.; Yuan, L.; Zhang, X.; Zhang, D.; Li, X.; Zhang, Y.; Zhao, Y.; Song, Q.; et al. Ovine ELOVL5 and FASN Genes Polymorphisms and Their Correlations with Sheep Tail Fat Deposition. Gene 2022, 807, 145954. [Google Scholar] [CrossRef] [PubMed]
- Hinman, M.N.; Richardson, J.I.; Sockol, R.A.; Aronson, E.D.; Stednitz, S.J.; Murray, K.N.; Berglund, J.A.; Guillemin, K. Zebrafish Mbnl Mutants Model Physical and Molecular Phenotypes of Myotonic Dystrophy. Dis. Model. Mech. 2021, 14, dmm045773. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Liang, C.; Li, Q.; Yan, C.; Wang, C.; Gu, Y.; Zhu, M.; Du, F.; Wang, H.; Dai, J.; et al. Role of ATG10 Expression Quantitative Trait Loci in Non-Small Cell Lung Cancer Survival. Int. J. Cancer 2016, 139, 1564–1573. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Xue, J.; He, Y.; Ma, H.; Jin, G.; Chen, J.; Hu, Z.; Liu, X.; Shen, H. Potentially Functional Polymorphisms in ATG10 Are Associated with Risk of Breast Cancer in a Chinese Population. Gene 2013, 527, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-Y.; Shang, J.; Mao, X.-M.; Fan, R.; Li, H.-Q.; Li, R.-H.; Shen, D.-Y. Diosgenin Exerts Anti-Tumor Effects through Inactivation of cAMP/PKA/CREB Signaling Pathway in Colorectal Cancer. Eur. J. Pharmacol. 2021, 908, 174370. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Liu, J.; Liang, S.; Sun, M.; Duan, J. Low-Dose Exposure of Silica Nanoparticles Induces Neurotoxicity via Neuroactive Ligand-Receptor Interaction Signaling Pathway in Zebrafish Embryos. Int. J. Nanomed. 2020, 15, 4407–4415. [Google Scholar] [CrossRef]
- Grabner, G.F.; Xie, H.; Schweiger, M.; Zechner, R. Lipolysis: Cellular Mechanisms for Lipid Mobilization from Fat Stores. Nat. Metab. 2021, 3, 1445–1465. [Google Scholar] [CrossRef]
- Ge, Q.; Gao, C.; Cai, Y.; Jiao, T.; Quan, J.; Guo, Y.; Zheng, W.; Zhao, S. The Domestication Event of the Tibetan Pig Revealed to Be in the Upstream Region of the Yellow River Based on the mtDNA D-Loop. Asian-Australas. J. Anim. Sci. 2020, 33, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Cho, S.; Caetano-Anolles, K.; Kim, H.; Ryu, Y.-C. Genome-Wide Detection and Characterization of Positive Selection in Korean Native Black Pig from Jeju Island. BMC Genet. 2015, 16, 3. [Google Scholar] [CrossRef]
- Li, M.; Tian, S.; Jin, L.; Zhou, G.; Li, Y.; Zhang, Y.; Wang, T.; Yeung, C.K.; Chen, L.; Ma, J.; et al. Genomic Analyses Identify Distinct Patterns of Selection in Domesticated Pigs and Tibetan Wild Boars. Nat. Genet. 2013, 45, 1431–1438. [Google Scholar] [CrossRef]
- Sanchez, M.; Kannengiesser, C.; Hoang, S.; Potier, L.; Fumeron, F.; Venteclef, N.; Scheen, A.; Gautier, J.-F.; Hadjadj, S.; Marre, M.; et al. Leukocyte Telomere Length, Allelic Variations in Related Genes and Risk of Coronary Heart Disease in People with Long-Standing Type 1 Diabetes. Cardiovasc. Diabetol. 2022, 21, 206. [Google Scholar] [CrossRef] [PubMed]
- Olsson, J.B.; Gugerel, M.B.; Jessen, S.B.; Jørgensen, J.; Gögenur, I.; Hansen, C.; Kirkeby, L.T.; Olsen, J.; Pedersen, O.B.V.; Vestlev, P.M.; et al. Colorectal Cancer-Associated SNP Rs17042479 Is Involved in the Regulation of NAF1 Promoter Activity. PLoS ONE 2022, 17, e0274033. [Google Scholar] [CrossRef] [PubMed]
- Kalbe, C.; Puppe, B. Long-Term Cognitive Enrichment Affects Opioid Receptor Expression in the Amygdala of Domestic Pigs. Genes. Brain Behav. 2010, 9, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, J.; Wang, Y.; Wu, Y.; Fu, J.; Liu, J.-F. Genome-Wide Detection of Selection Signatures in Chinese Indigenous Laiwu Pigs Revealed Candidate Genes Regulating Fat Deposition in Muscle. BMC Genet. 2018, 19, 31. [Google Scholar] [CrossRef] [PubMed]
- Finucane, H.K.; Reshef, Y.A.; Anttila, V.; Slowikowski, K.; Gusev, A.; Byrnes, A.; Gazal, S.; Loh, P.-R.; Lareau, C.; Shoresh, N.; et al. Heritability Enrichment of Specifically Expressed Genes Identifies Disease-Relevant Tissues and Cell Types. Nat. Genet. 2018, 50, 621–629. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Han, D. BWA-MEME: BWA-MEM Emulated with a Machine Learning Approach. Bioinformatics 2022, 38, 2404–2413. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; McCarthy, S.A. BCFtools/Csq: Haplotype-Aware Variant Consequences. Bioinformatics 2017, 33, 2037–2039. [Google Scholar] [CrossRef]
- Fang, M.; Larson, G.; Ribeiro, H.S.; Li, N.; Andersson, L. Contrasting Mode of Evolution at a Coat Color Locus in Wild and Domestic Pigs. PLoS Genet. 2009, 5, e1000341. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kramer, F.; Bayerlová, M.; Klemm, F.; Bleckmann, A.; Beissbarth, T. rBiopaxParser--an R Package to Parse, Modify and Visualize BioPAX Data. Bioinformatics 2013, 29, 520–522. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE Algorithm for Individual Ancestry Estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Patterson, N.; Price, A.L.; Reich, D. Population Structure and Eigenanalysis. PLoS Genet. 2006, 2, e190. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Kao, H.; Dong, S. Population Genetic Structure and Gene Flow of Rare and Endangered Tetraena Mongolica Maxim. Revealed by Reduced Representation Sequencing. BMC Plant Biol. 2020, 20, 391. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Pritchard, J.K. Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef]
- Xu, T.; Wang, Y.; Sun, J.; Chen, C.; Watanabe, H.K.; Chen, J.; Qian, P.-Y.; Qiu, J.-W. Hidden Historical Habitat-Linked Population Divergence and Contemporary Gene Flow of a Deep-Sea Patellogastropod Limpet. Mol. Biol. Evol. 2021, 38, 5640–5654. [Google Scholar] [CrossRef]
- Petr, M.; Vernot, B.; Kelso, J. Admixr-R Package for Reproducible Analyses Using ADMIXTOOLS. Bioinformatics 2019, 35, 3194–3195. [Google Scholar] [CrossRef]
- Garcia-Erill, G.; Jorgensen, C.H.F.; Muwanika, V.B.; Wang, X.; Rasmussen, M.S.; de Jong, Y.A.; Gaubert, P.; Olayemi, A.; Salmona, J.; Butynski, T.M.; et al. Warthog Genomes Resolve an Evolutionary Conundrum and Reveal Introgression of Disease Resistance Genes. Mol. Biol. Evol. 2022, 39, msac134. [Google Scholar] [CrossRef]
- Keller, M.C.; Visscher, P.M.; Goddard, M.E. Quantification of Inbreeding Due to Distant Ancestors and Its Detection Using Dense Single Nucleotide Polymorphism Data. Genetics 2011, 189, 237–249. [Google Scholar] [CrossRef]
- Hu, Z.-L.; Park, C.A.; Reecy, J.M. Bringing the Animal QTLdb and CorrDB into the Future: Meeting New Challenges and Providing Updated Services. Nucleic Acids Res. 2022, 50, D956–D961. [Google Scholar] [CrossRef]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The Structure of Haplotype Blocks in the Human Genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef] [PubMed]
- Vens, M.; Ziegler, A. Estimating Disequilibrium Coefficients. Methods Mol. Biol. 2012, 850, 103–117. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent Prioritization and Exploratory Visualization of Biological Functions for Gene Enrichment Analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef] [PubMed]
- Colwill, K.; Renewable Protein Binder Working Group; Gräslund, S. A Roadmap to Generate Renewable Protein Binders to the Human Proteome. Nat. Methods 2011, 8, 551–558. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A Single-Cell Type Transcriptomics Map of Human Tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A Subcellular Map of the Human Proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Carroll, R.J.; Bastarache, L.; Denny, J.C. R PheWAS: Data Analysis and Plotting Tools for Phenome-Wide Association Studies in the R Environment. Bioinformatics 2014, 30, 2375–2376. [Google Scholar] [CrossRef]
- Groza, T.; Gomez, F.L.; Mashhadi, H.H.; Muñoz-Fuentes, V.; Gunes, O.; Wilson, R.; Cacheiro, P.; Frost, A.; Keskivali-Bond, P.; Vardal, B.; et al. The International Mouse Phenotyping Consortium: Comprehensive Knockout Phenotyping Underpinning the Study of Human Disease. Nucleic Acids Res. 2023, 51, D1038–D1045. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Liu, Y.; Feng, X.; Diao, S.; Zhong, Z.; Li, B.; Teng, J.; Zhang, W.; Zeng, H.; Cai, X.; et al. Integrating Multiple Database Resources to Elucidate the Gene Flow in Southeast Asian Pig Populations. Int. J. Mol. Sci. 2024, 25, 5689. https://doi.org/10.3390/ijms25115689
Li G, Liu Y, Feng X, Diao S, Zhong Z, Li B, Teng J, Zhang W, Zeng H, Cai X, et al. Integrating Multiple Database Resources to Elucidate the Gene Flow in Southeast Asian Pig Populations. International Journal of Molecular Sciences. 2024; 25(11):5689. https://doi.org/10.3390/ijms25115689
Chicago/Turabian StyleLi, Guangzhen, Yuqiang Liu, Xueyan Feng, Shuqi Diao, Zhanming Zhong, Bolang Li, Jinyan Teng, Wenjing Zhang, Haonan Zeng, Xiaodian Cai, and et al. 2024. "Integrating Multiple Database Resources to Elucidate the Gene Flow in Southeast Asian Pig Populations" International Journal of Molecular Sciences 25, no. 11: 5689. https://doi.org/10.3390/ijms25115689
APA StyleLi, G., Liu, Y., Feng, X., Diao, S., Zhong, Z., Li, B., Teng, J., Zhang, W., Zeng, H., Cai, X., Gao, Y., Liu, X., Yuan, X., Li, J., & Zhang, Z. (2024). Integrating Multiple Database Resources to Elucidate the Gene Flow in Southeast Asian Pig Populations. International Journal of Molecular Sciences, 25(11), 5689. https://doi.org/10.3390/ijms25115689







