EGFR and PI3K Signalling Pathways as Promising Targets on Circulating Tumour Cells from Patients with Metastatic Gastric Adenocarcinoma
Abstract
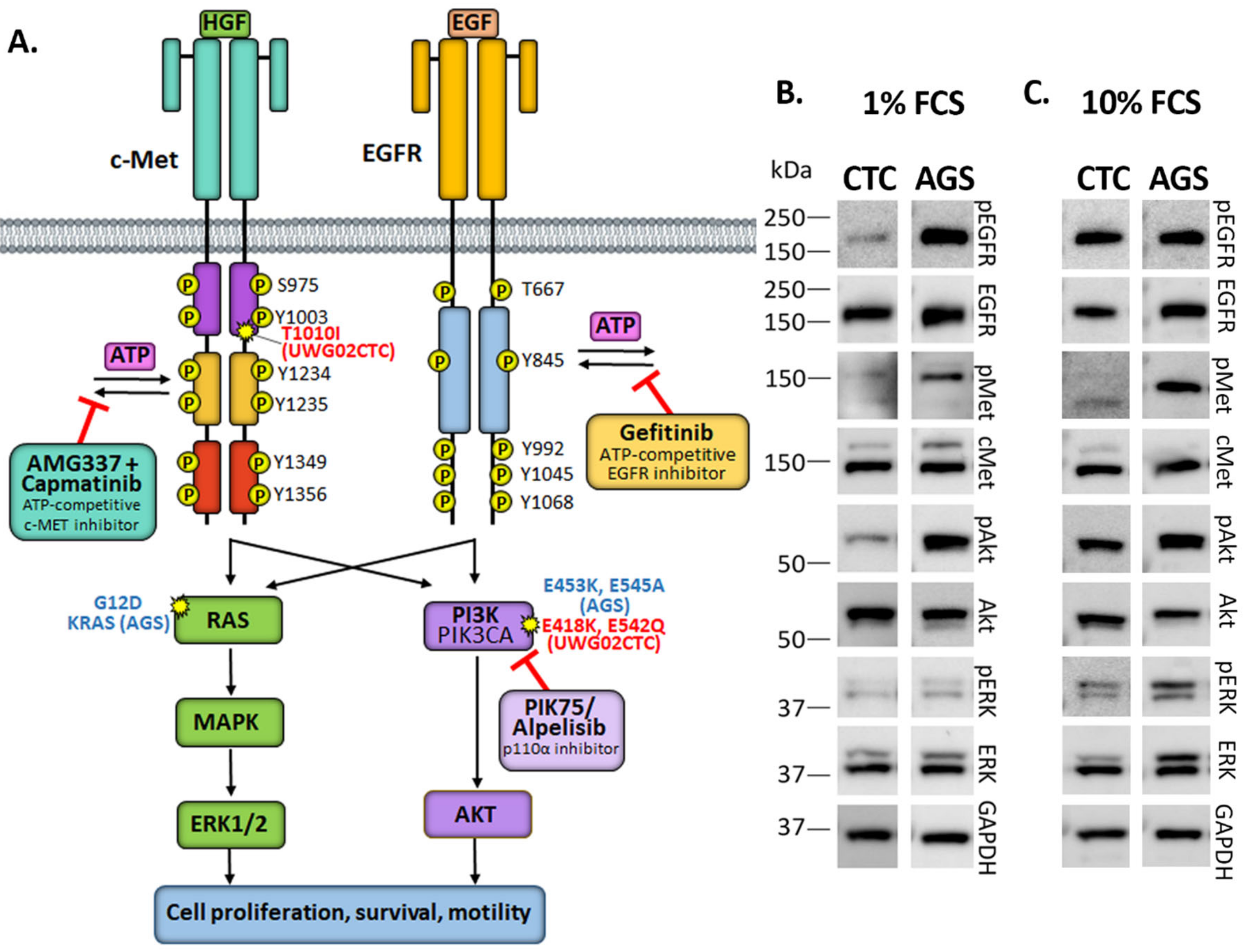
1. Introduction
2. Results
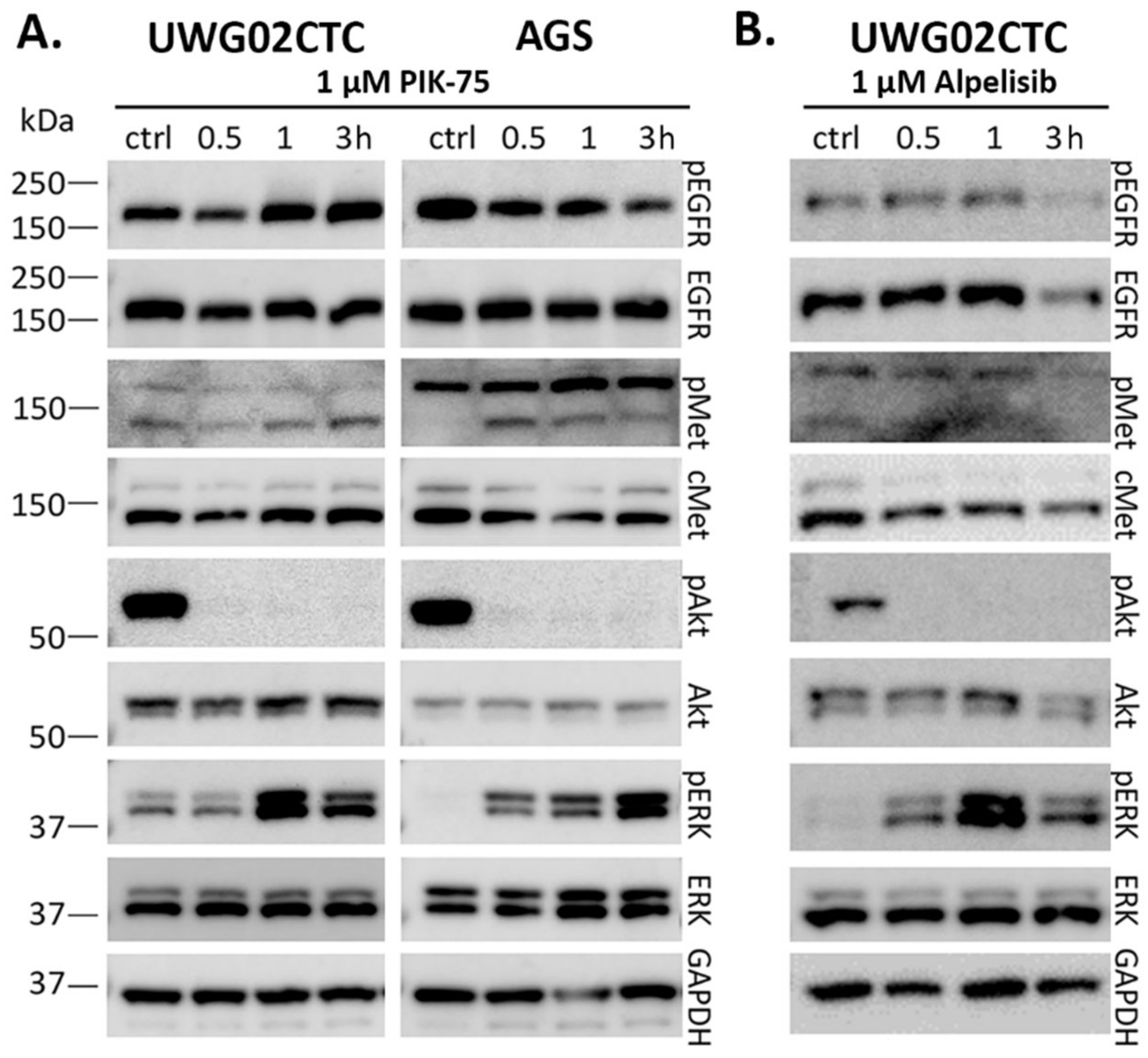
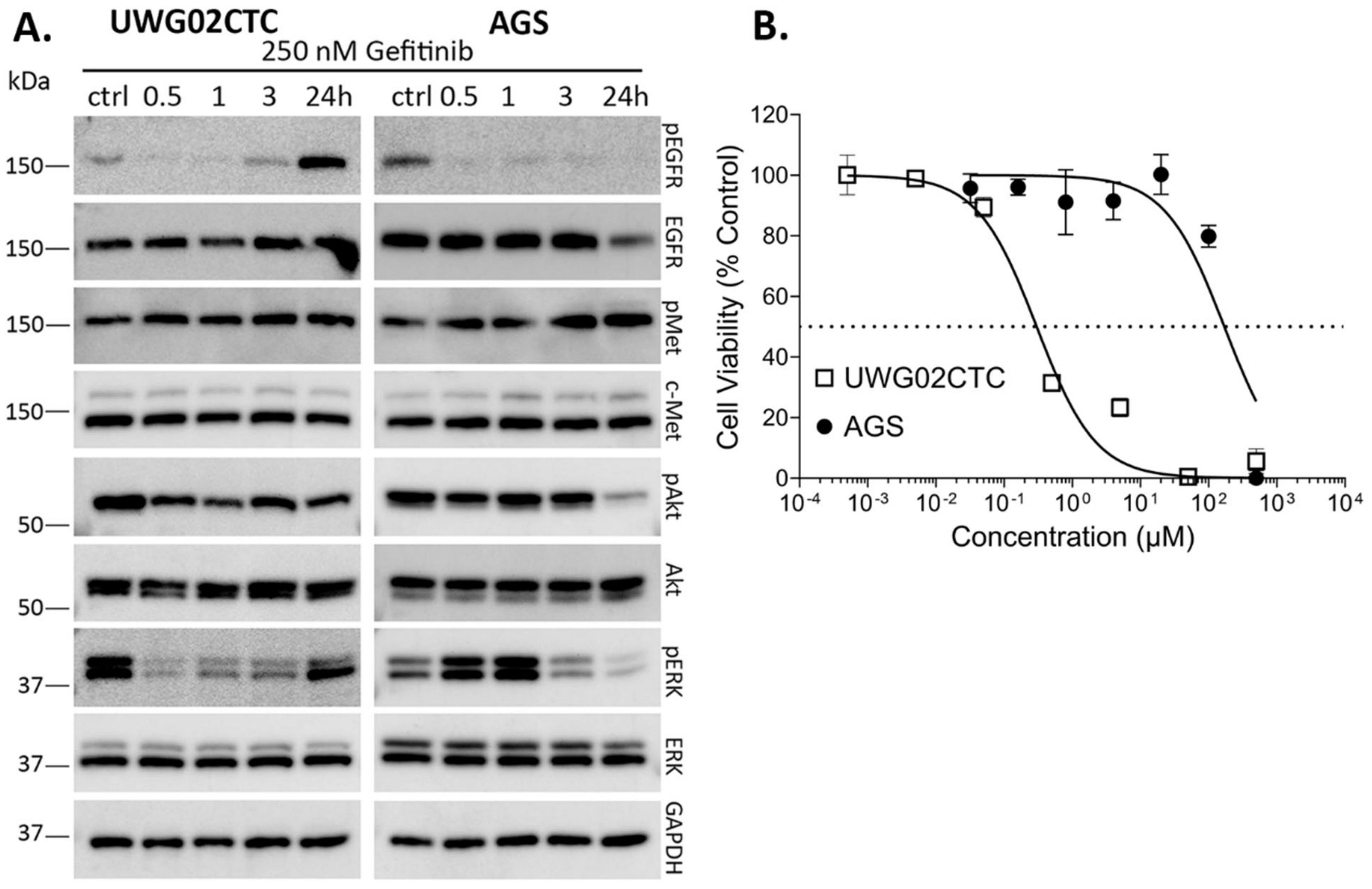
2.1. Monotherapy Effects
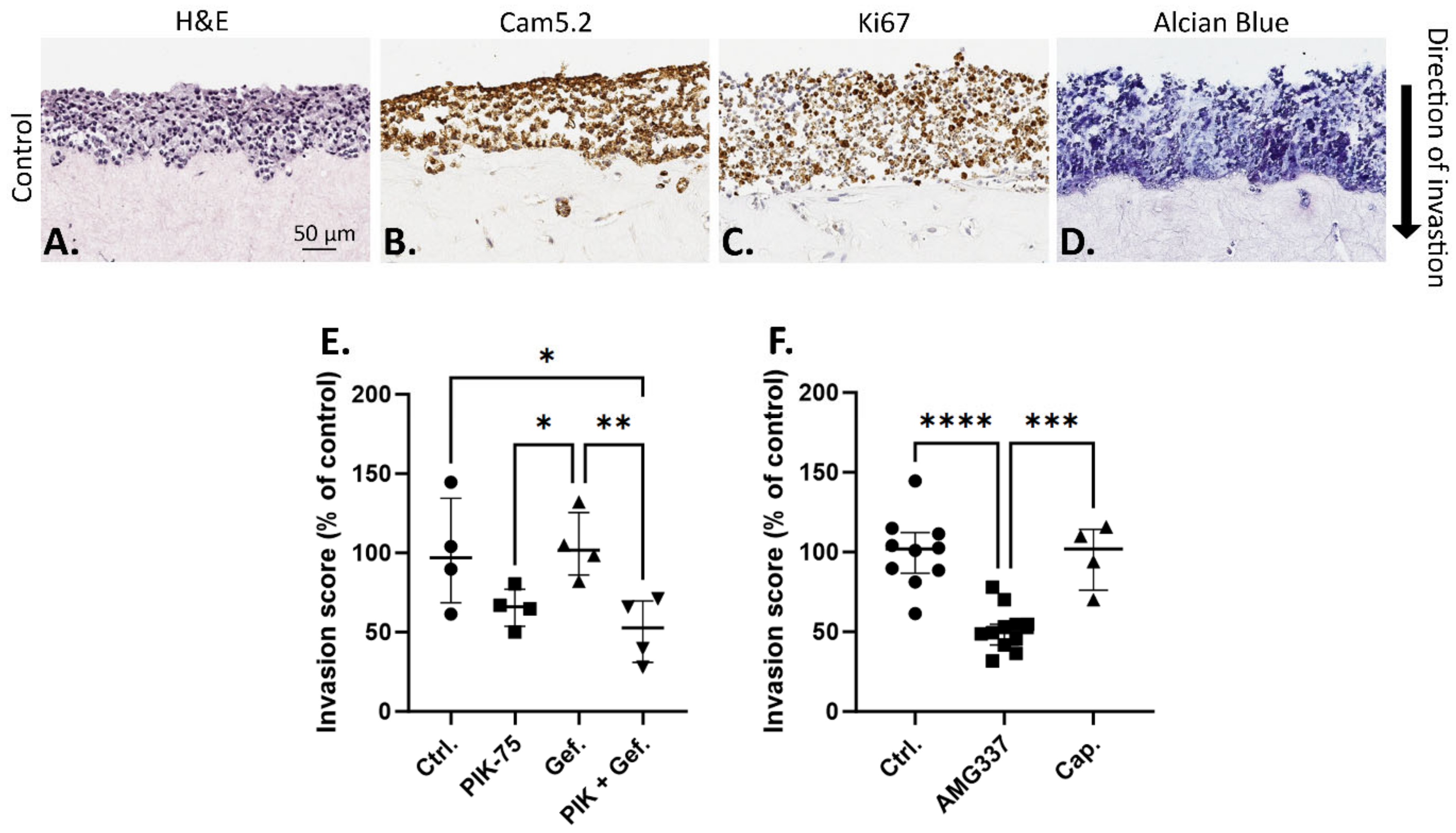
2.2. Effect of Drugs on UWG02CTC Invasion in 3D Organotypic Culture
2.3. Drug Responsiveness in 2D versus 3D Culture Models
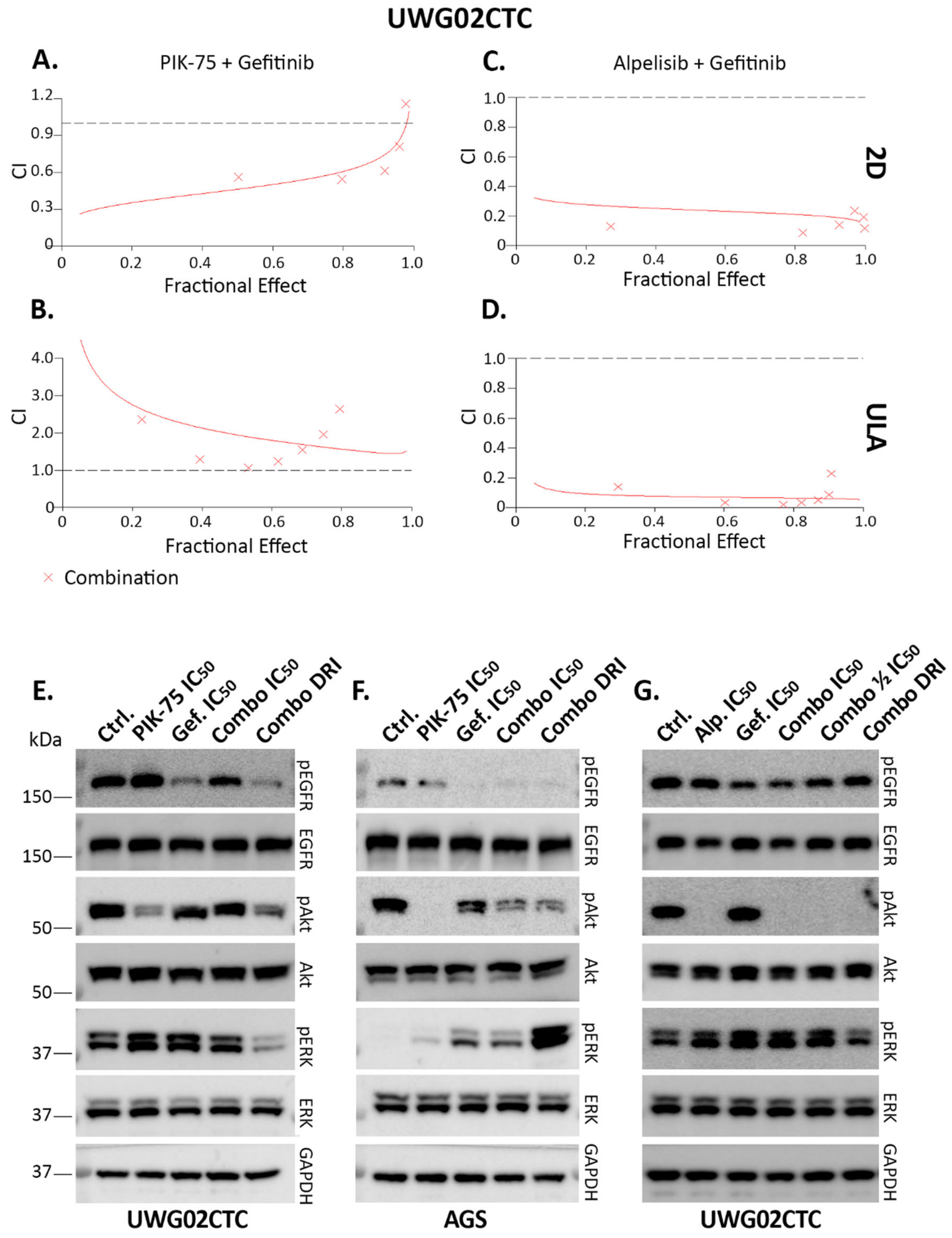
2.4. Combination Drug Analyses
3. Discussion
Summary
4. Materials and Methods
4.1. Cell Culture
4.2. Western Blotting
4.3. Cell Viability Assays
4.4. Organotypic Invasion Assays
4.5. Collagen-Embedded Spheroids
4.6. RASTRUM Bioprinting
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.M.; Chiu, N.C.; Li, A.F.; Liu, C.A.; Chou, Y.H.; Chiou, Y.Y. Unusual gastric tumors and tumor-like lesions: Radiological with pathological correlation and literature review. World J. Gastroenterol. 2017, 23, 2493–2504. [Google Scholar] [CrossRef] [PubMed]
- Lordick, F.; Carneiro, F.; Cascinu, S.; Fleitas, T.; Haustermans, K.; Piessen, G.; Vogel, A.; Smyth, E.C.; on behalf of theESMO Guidelines Committee. Gastric cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2022, 33, 1005–1020. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.J.; Gao, J.B. Molecular mechanisms of chemoresistance in gastric cancer. World J. Gastrointest. Oncol. 2016, 8, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Rha, S.Y.; Oh, D.; Yañez, P.; Bai, Y.X.; Ryu, M.H.; Lee, J.Y.; Rivera, F.; Alves, G.V.; Garrido, M.; Shiu, K.K.; et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for HER2-negative advanced gastric cancer (KEYNOTE-859): A multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 2023, 24, 1181–1195. [Google Scholar] [CrossRef] [PubMed]
- Teng, L.S.; Lu, J. cMET as a potential therapeutic target in gastric cancer. Int. J. Mol. Med. 2013, 32, 1247–1254. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Ito, F. Receptor tyrosine kinases and targeted cancer therapeutics. Biol. Pharm. Bull. 2011, 34, 1774–1780. [Google Scholar] [CrossRef] [PubMed]
- Pottier, C.; Fresnais, M.; Gilon, M.; Jerusalem, G.; Longuespee, R.; Sounni, N.E. Tyrosine Kinase Inhibitors in Cancer: Breakthrough and Challenges of Targeted Therapy. Cancers 2020, 12, 731. [Google Scholar] [CrossRef] [PubMed]
- El Darsa, H.; El Sayed, R.; Abdel-Rahman, O. MET Inhibitors for the Treatment of Gastric Cancer: What’s Their Potential? J. Exp. Pharmacol. 2020, 12, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Janjigian, Y.Y.; Tang, L.H.; Coit, D.G.; Kelsen, D.P.; Francone, T.D.; Weiser, M.R.; Jhanwar, S.C.; Shah, M.A. MET expression and amplification in patients with localized gastric cancer. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1021–1027. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, H.; Okamoto, I. MET-targeted therapy for gastric cancer: The importance of a biomarker-based strategy. Gastric Cancer 2016, 19, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.C.; Vlachogiannis, G.; Hedayat, S.; Harbery, A.; Hulkki-Wilson, S.; Salati, M.; Kouvelakis, K.; Fernandez-Mateos, J.; Cresswell, G.D.; Fontana, E. EGFR amplification and outcome in a randomised phase III trial of chemotherapy alone or chemotherapy plus panitumumab for advanced gastro-oesophageal cancers. Gut 2021, 70, 1632–1641. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Sasaki, A.; Yukami, H.; Jogo, T.; Kawazoe, A.; Kuboki, Y.; Taniguchi, H.; Yamashita, R.; Kuwata, T.; Ozawa, M. Emergence of concurrent multiple EGFR mutations and MET amplification in a patient with EGFR-amplified advanced gastric cancer treated with cetuximab. JCO Precis. Oncol. 2020, 4, PO.20.00263. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.M.; Janjigian, Y.Y. The genomics and therapeutics of HER2-positive gastric cancer-from trastuzumab and beyond. J. Gastrointest. Oncol. 2016, 7, 750–762. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Gravalos, C.; Jimeno, A. HER2 in gastric cancer: A new prognostic factor and a novel therapeutic target. Ann. Oncol. 2008, 19, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Prins, M.J.; Ruurda, J.P.; van Diest, P.J.; van Hillegersberg, R.; Ten Kate, F.J. The significance of the HER-2 status in esophageal adenocarcinoma for survival: An immunohistochemical and an in situ hybridization study. Ann. Oncol. 2013, 24, 1290–1297. [Google Scholar] [CrossRef]
- Bradley, C.A.; Salto-Tellez, M.; Laurent-Puig, P.; Bardelli, A.; Rolfo, C.; Tabernero, J.; Khawaja, H.A.; Lawler, M.; Johnston, P.G.; Van Schaeybroeck, S.; et al. Targeting c-MET in gastrointestinal tumours: Rationale, opportunities and challenges. Nat. Rev. Clin. Oncol. 2017, 14, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Kneissl, J.; Grabher-Meier, V.; Heindl, S.; Hasenauer, J.; Maier, D.; Mattes, J.; Winter, P.; Luber, B. Evaluation of epidermal growth factor receptor signaling effects in gastric cancer cell lines by detailed motility-focused phenotypic characterization linked with molecular analysis. BMC Cancer 2017, 17, 845. [Google Scholar] [CrossRef]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, T.; Yashiro, M. Biomarkers of gastric cancer: Current topics and future perspective. World J. Gastroenterol. 2018, 24, 2818. [Google Scholar] [CrossRef] [PubMed]
- Brungs, D.; Minaei, E.; Piper, A.K.; Perry, J.; Splitt, A.; Carolan, M.; Ryan, S.; Wu, X.J.; Corde, S.; Tehei, M.; et al. Establishment of novel long-term cultures from EpCAM positive and negative circulating tumour cells from patients with metastatic gastroesophageal cancer. Sci. Rep. 2020, 10, 539. [Google Scholar] [CrossRef]
- Kim, J.-W.; Lee, H.S.; Nam, K.H.; Ahn, S.; Kim, J.W.; Ahn, S.-H.; Park, D.J.; Kim, H.-H.; Lee, K.-W. PIK3CA mutations are associated with increased tumor aggressiveness and Akt activation in gastric cancer. Oncotarget 2017, 8, 90948. [Google Scholar] [CrossRef] [PubMed]
- Ito, C.; Nishizuka, S.S.; Ishida, K.; Uesugi, N.; Sugai, T.; Tamura, G.; Koeda, K.; Sasaki, A. Analysis of PIK3CA mutations and PI3K pathway proteins in advanced gastric cancer. J. Surg. Res. 2017, 212, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.M.; Xu, G.; Ma, W.; Li, Y.; Luo, W.; Xiao, Y.; Liu, Y.; Zhang, Z. Significant function and research progress of biomarkers in gastric cancer. Oncol. Lett. 2020, 19, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Juric, D.; Rodon, J.; Tabernero, J.; Janku, F.; Burris, H.A.; Schellens, J.H.M.; Middleton, M.R.; Berlin, J.; Schuler, M.; Gil-Martin, M.; et al. Phosphatidylinositol 3-Kinase alpha-Selective Inhibition With Alpelisib (BYL719) in PIK3CA-Altered Solid Tumors: Results From the First-in-Human Study. J. Clin. Oncol. 2018, 36, 1291–1299. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Hu, Y. Targeting PI3K/Akt/mTOR cascade: The medicinal potential, updated research highlights and challenges ahead. Curr. Med. Chem. 2013, 20, 2991–3010. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Wang, T.; Zang, R.; Zang, Y.; Feng, Y.; Yan, S.; Geng, C.; Zhu, N.; Wang, Q. Dual PI3K/mTOR inhibitor PF-04979064 regulates tumor growth in gastric cancer and enhances drug sensitivity of gastric cancer cells to 5-FU. Biomed. Pharmacother. 2024, 170, 116086. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Han, J.; Lin, K.; Wang, J.; Li, G.; Li, X.; Gao, Y. PTEN/AKT and Wnt/beta-catenin signaling pathways regulate the proliferation of Lgr5+ cells in liver cancer. Biochem. Biophys. Res. Commun. 2023, 683, 149117. [Google Scholar] [CrossRef] [PubMed]
- Adelstein, D.J.; Rodriguez, C.P.; Rybicki, L.A.; Ives, D.I.; Rice, T.W. A phase II trial of gefitinib for recurrent or metastatic cancer of the esophagus or gastroesophageal junction. Investig. New Drugs 2012, 30, 1684–1689. [Google Scholar] [CrossRef] [PubMed]
- Kazandjian, D.; Blumenthal, G.M.; Yuan, W.; He, K.; Keegan, P.; Pazdur, R. FDA Approval of Gefitinib for the Treatment of Patients with Metastatic EGFR Mutation-Positive Non-Small Cell Lung Cancer. Clin. Cancer Res. 2016, 22, 1307–1312. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Yoshida, T.; Masuda, K.; Matsumoto, Y.; Shinno, Y.; Okuma, Y.; Goto, Y.; Horinouchi, H.; Yamamoto, N.; Ohe, Y. Comparison of clinical outcomes of osimertinib and first-generation EGFR-tyrosine kinase inhibitors (TKIs) in TKI-untreated EGFR-mutated non-small-cell lung cancer with leptomeningeal metastases. ESMO Open 2023, 8, 101594. [Google Scholar] [CrossRef]
- Guo, A.; Villen, J.; Kornhauser, J.; Lee, K.A.; Stokes, M.P.; Rikova, K.; Possemato, A.; Nardone, J.; Innocenti, G.; Wetzel, R.; et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc. Natl. Acad. Sci. USA 2008, 105, 692–697. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Q.; Yang, G.; Marando, C.; Koblish, H.K.; Hall, L.M.; Fridman, J.S.; Behshad, E.; Wynn, R.; Li, Y.; et al. A novel kinase inhibitor, INCB28060, blocks c-MET-dependent signaling, neoplastic activities, and cross-talk with EGFR and HER-3. Clin. Cancer Res. 2011, 17, 7127–7138. [Google Scholar] [CrossRef] [PubMed]
- Puri, N.; Salgia, R. Synergism of EGFR and c-Met pathways, cross-talk and inhibition, in non-small cell lung cancer. J. Carcinog. 2008, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L.; et al. Improved Overall Survival in Melanoma with Combined Dabrafenib and Trametinib. N. Engl. J. Med. 2015, 372, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabieres, C. Circulating tumour cells in cancer patients: Challenges and perspectives. Trends Mol. Med. 2010, 16, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Smit, D.J.; Pantel, K.; Jucker, M. Circulating tumor cells as a promising target for individualized drug susceptibility tests in cancer therapy. Biochem. Pharmacol. 2021, 188, 114589. [Google Scholar] [CrossRef] [PubMed]
- Neklason, D.W.; Done, M.W.; Sargent, N.R.; Schwartz, A.G.; Anton-Culver, H.; Griffin, C.A.; Ahnen, D.J.; Schildkraut, J.M.; Tomlinson, G.E.; Strong, L.C.; et al. Activating mutation in MET oncogene in familial colorectal cancer. Bmc Cancer 2011, 11, 424. [Google Scholar] [CrossRef] [PubMed]
- Chandarlapaty, S.; Sawai, A.; Scaltriti, M.; Rodrik-Outmezguine, V.; Grbovic-Huezo, O.; Serra, V.; Majumder, P.K.; Baselga, J.; Rosen, N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 2011, 19, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Serra, V.; Scaltriti, M.; Prudkin, L.; Eichhorn, P.J.; Ibrahim, Y.H.; Chandarlapaty, S.; Markman, B.; Rodriguez, O.; Guzman, M.; Rodriguez, S.; et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene 2011, 30, 2547–2557. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.Y.; Ma, W.L.; Lu, Y.S. Role of Alpelisib in the Treatment of PIK3CA-Mutated Breast Cancer: Patient Selection and Clinical Perspectives. Ther. Clin. Risk Manag. 2021, 17, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.J.; Kim, J.W.; Sung, J.H.; Suh, K.J.; Lee, J.Y.; Kim, S.H.; Lee, J.O.; Kim, J.W.; Kim, Y.J.; Kim, J.H.; et al. PI3K-targeting strategy using alpelisib to enhance the antitumor effect of paclitaxel in human gastric cancer. Sci. Rep. 2020, 10, 12308. [Google Scholar] [CrossRef] [PubMed]
- Tria, S.M.; Burge, M.E.; Whitehall, V.L.J. The Therapeutic Landscape for KRAS-Mutated Colorectal Cancers. Cancers 2023, 15, 2375. [Google Scholar] [CrossRef] [PubMed]
- Hughes, P.E.; Rex, K.; Caenepeel, S.; Yang, Y.; Zhang, Y.; Broome, M.A.; Kha, H.T.; Burgess, T.L.; Amore, B.; Kaplan-Lefko, P.J.; et al. In Vitro and In Vivo Activity of AMG 337, a Potent and Selective MET Kinase Inhibitor, in MET-Dependent Cancer Models. Mol. Cancer Ther. 2016, 15, 1568–1579. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, R. The research progress of c-Met inhibitors in clinical trials. Canc Cell Res. 2018, 20, 524–528. [Google Scholar]
- Du, Z.; Caenepeel, S.; Shen, Y.; Rex, K.; Zhang, Y.; He, Y.; Tang, E.T.; Wang, O.; Zhong, W.; Zhou, H.; et al. Preclinical Evaluation of AMG 337, a Highly Selective Small Molecule MET Inhibitor, in Hepatocellular Carcinoma. Mol. Cancer Ther. 2016, 15, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xia, M.; Jin, K.; Wang, S.; Wei, H.; Fan, C.; Wu, Y.; Li, X.; Li, X.; Li, G.; et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol. Cancer 2018, 17, 45. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef]
- Hervieu, A.; Kermorgant, S. The Role of PI3K in Met Driven Cancer: A Recap. Front. Mol. Biosci. 2018, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Timpson, P.; McGhee, E.J.; Erami, Z.; Nobis, M.; Quinn, J.A.; Edward, M.; Anderson, K.I. Organotypic collagen I assay: A malleable platform to assess cell behaviour in a 3-dimensional context. J. Vis. Exp. 2011, e3089. [Google Scholar] [CrossRef] [PubMed]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells 2019, 11, 1065–1083. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.R.; Erler, J.T. Remodeling and homeostasis of the extracellular matrix: Implications for fibrotic diseases and cancer. Dis. Model. Mech. 2011, 4, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Sa, J.K.; Hong, J.Y.; Lee, I.K.; Kim, J.S.; Sim, M.H.; Kim, H.J.; An, J.Y.; Sohn, T.S.; Lee, J.H.; Bae, J.M.; et al. Comprehensive pharmacogenomic characterization of gastric cancer. Genome Med. 2020, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.Q.; Chan, A.S.; Yokomori, R.; Huang, X.Z.; Theardy, M.S.; Yeoh, A.E.J.; Tan, S.H.; Sanda, T. Targeting dual oncogenic machineries driven by TAL1 and PI3K-AKT pathways in T-cell acute lymphoblastic leukemia. Haematologica 2023, 108, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Maron, S.B.; Xu, J.; Janjigian, Y.Y. Targeting EGFR in Esophagogastric Cancer. Front. Oncol. 2020, 10, 553876. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, A.V.; Marie, P.K.; Bitner, L.; Syed, M.; Woods, M.; Manyam, G.; Kwong, L.N.; Johnson, B.; Morris, V.; Jones, P.; et al. Targeting RAS Mutant Colorectal Cancer with Dual Inhibition of MEK and CDK4/6. Cancer Res. 2022, 82, 3335–3344. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Hong, R.; Cao, Y.; Wu, Q.; Wang, Y.; Chen, J.; Li, J.; Zhang, W.; Zhan, Q. Combined Wee1 and EGFR inhibition reveals synergistic antitumor effect in esophageal squamous cell carcinoma. Carcinogenesis 2023, 44, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Song, Z.; Ru, Y.; Zhang, J.; Luo, L.; Yang, W.; Wu, H.; Jin, H.; Bao, X.; Wei, D.; et al. Small-molecule nanoprodrug with high drug loading and EGFR, PI3K/AKT dual-inhibiting properties for bladder cancer treatment. Exploration 2023, 3, 20220141. [Google Scholar] [CrossRef]
- Brisson, R.J.; Kochanny, S.; Arshad, S.; Dekker, A.; DeSouza, J.A.; Saloura, V.; Vokes, E.E.; Seiwert, T.Y. A pilot study of the pan-class I PI3K inhibitor buparlisib in combination with cetuximab in patients with recurrent or metastatic head and neck cancer. Head. Neck 2019, 41, 3842–3849. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xiao, Z.; Lei, C.; Ma, C.; Ding, L.; Tang, Q.; He, Y.; Chen, Y.; Chang, X.; Zhu, Y.; et al. BYL719 reverses gefitinib-resistance induced by PI3K/AKT activation in non-small cell lung cancer cells. Bmc Cancer 2023, 23, 732. [Google Scholar] [CrossRef] [PubMed]
- Alzeeb, G.; Metges, J.-P.; Corcos, L.; Jossic-Corcos, L. Three-dimensional culture systems in gastric cancer research. Cancers 2020, 12, 2800. [Google Scholar] [CrossRef] [PubMed]
- Fontoura, J.C.; Viezzer, C.; Dos Santos, F.G.; Ligabue, R.A.; Weinlich, R.; Puga, R.D.; Antonow, D.; Severino, P.; Bonorino, C. Comparison of 2D and 3D cell culture models for cell growth, gene expression and drug resistance. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 107, 110264. [Google Scholar] [CrossRef] [PubMed]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling physiological events in 2D vs. 3D cell culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kleeff, J.; Giese, N.; Buchler, M.W.; Korc, M.; Friess, H. Gefitinib (‘Iressa’, ZD1839), a selective epidermal growth factor receptor tyrosine kinase inhibitor, inhibits pancreatic cancer cell growth, invasion, and colony formation. Int. J. Oncol. 2004, 25, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Guanghui, L.; Xiefu, Z. Knockdown of EGFR inhibits growth and invasion of gastric cancer cells. Cancer Gene Ther. 2014, 21, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Yoshida, S.; Muroi, E.; Yoshida, N.; Kawamura, M.; Kouchi, Z.; Nakamura, Y.; Sakai, R.; Fukami, K. Phosphoinositide 3-kinase signaling pathway mediated by p110alpha regulates invadopodia formation. J. Cell Biol. 2011, 193, 1275–1288. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.C.; Kijima, T.; Maulik, G.; Fox, E.A.; Sattler, M.; Griffin, J.D.; Johnson, B.E.; Salgia, R. c-MET mutational analysis in small cell lung cancer: Novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003, 63, 6272–6281. [Google Scholar]
- Liu, S.Y.; Meric-Bernstam, F.; Parinyanitikul, N.; Wang, B.L.; Eterovic, A.K.; Zheng, X.F.; Gagea, M.; Chavez-MacGregor, M.; Ueno, N.T.; Lei, X.D.; et al. Functional consequence of the MET-T1010I polymorphism in breast cancer. Oncotarget 2015, 6, 2604–2614. [Google Scholar] [CrossRef]
- Wolf, J.; Seto, T.; Han, J.Y.; Reguart, N.; Garon, E.B.; Groen, H.J.M.; Tan, D.S.W.; Hida, T.; de Jonge, M.; Orlov, S.V.; et al. Capmatinib in MET Exon 14-Mutated or MET-Amplified Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 944–957. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Park, S.Y.; Lee, Y.; Lim, K.Y.; Park, M.; Lee, G.K.; Han, J.Y. The Clinical Impact of Capmatinib in the Treatment of Advanced Non-Small Cell Lung Cancer with MET Exon 14 Skipping Mutation or Gene Amplification. Cancer Res. Treat. 2021, 53, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Harris, N.L.E.; Vennin, C.; Conway, J.R.W.; Vine, K.L.; Pinese, M.; Cowley, M.J.; Shearer, R.F.; Lucas, M.C.; Herrmann, D.; Allam, A.H.; et al. SerpinB2 regulates stromal remodelling and local invasion in pancreatic cancer. Oncogene 2017, 36, 4288–4298. [Google Scholar] [CrossRef] [PubMed]
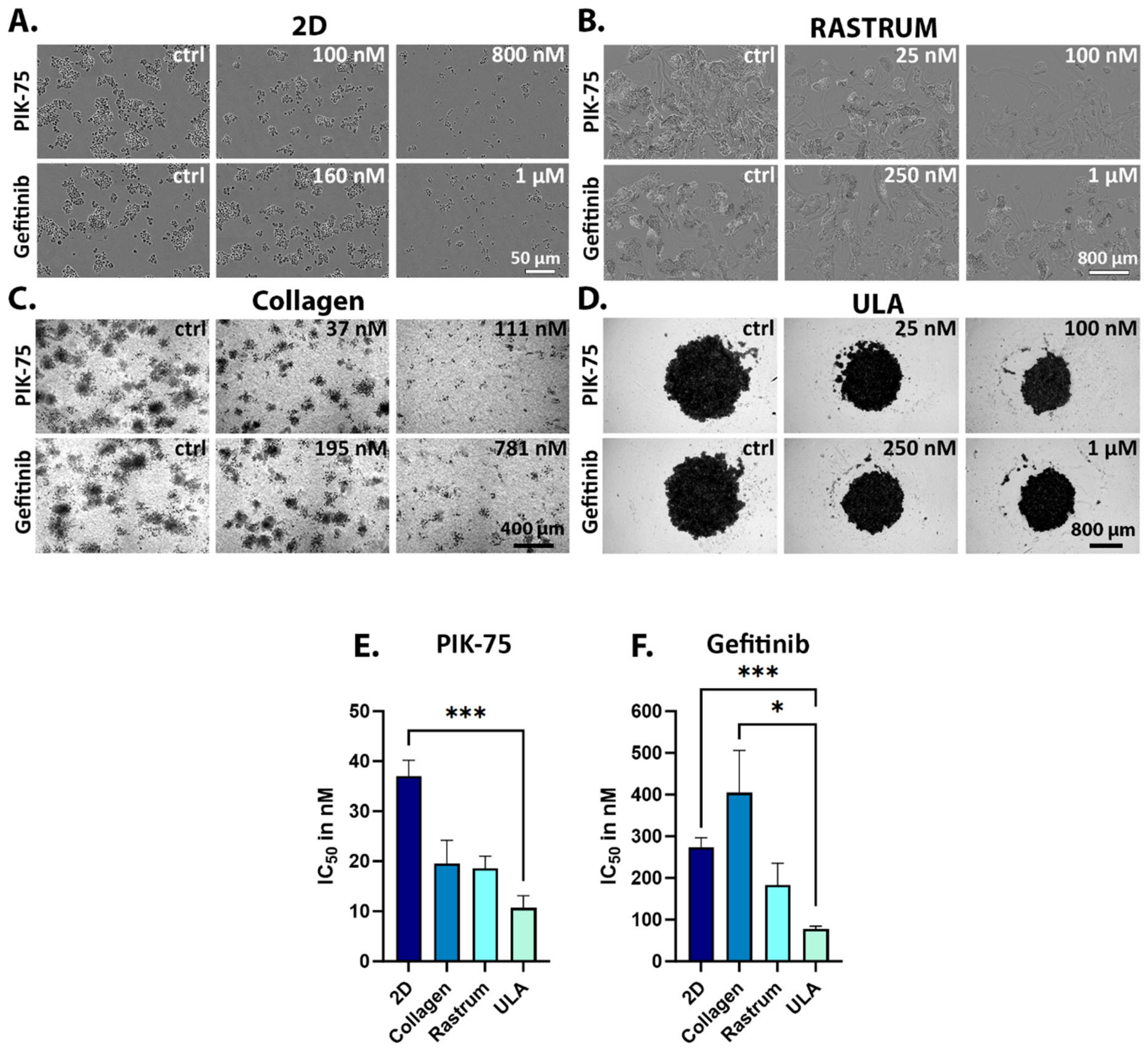
| Drug IC50 (Mean ± SEM) | |||||
|---|---|---|---|---|---|
| Conditions | |||||
| Cell Line | Drug | 2D | ULA | Collagen | Rastrum |
| UWG02CTC | PIK-75 | 37.0 ± 11.1 nM (n = 12) | 10.7 ± 3.4 nM (n = 2) 1 | 19.5 ± 6.6 nM (n = 2) | 18.6 ± 4.2 nM (n = 3) |
| Alpelisib | 7.05 ± 3.7 µM (n = 9) | 3.7 ± 1.0 µM (n = 3) | ND | ND | |
| Gefitinib | 273.0 ± 56.6 nM (n = 6) | 77.2 ± 9.9 nM (n = 2) 1 | 404.6 ± 202.3 nM (n = 4) 2 | 182.8 ± 74.3 nM (n = 2) | |
| AGS | PIK-75 | 42.4 ± 12.7 nM (n = 9) | 30.4 ± 15.2 nM (n = 2) | ||
| Gefitinib | 24.7 ± 15.6 µM (n = 4) | 19.47 ± 5.4 µM (n = 4) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piper, A.-K.; Penney, C.; Holliday, J.; Tincknell, G.; Ma, Y.; Napaki, S.; Pantel, K.; Brungs, D.; Ranson, M. EGFR and PI3K Signalling Pathways as Promising Targets on Circulating Tumour Cells from Patients with Metastatic Gastric Adenocarcinoma. Int. J. Mol. Sci. 2024, 25, 5565. https://doi.org/10.3390/ijms25105565
Piper A-K, Penney C, Holliday J, Tincknell G, Ma Y, Napaki S, Pantel K, Brungs D, Ranson M. EGFR and PI3K Signalling Pathways as Promising Targets on Circulating Tumour Cells from Patients with Metastatic Gastric Adenocarcinoma. International Journal of Molecular Sciences. 2024; 25(10):5565. https://doi.org/10.3390/ijms25105565
Chicago/Turabian StylePiper, Ann-Katrin, Chelsea Penney, Jacqueline Holliday, Gary Tincknell, Yafeng Ma, Sarbar Napaki, Klaus Pantel, Daniel Brungs, and Marie Ranson. 2024. "EGFR and PI3K Signalling Pathways as Promising Targets on Circulating Tumour Cells from Patients with Metastatic Gastric Adenocarcinoma" International Journal of Molecular Sciences 25, no. 10: 5565. https://doi.org/10.3390/ijms25105565
APA StylePiper, A.-K., Penney, C., Holliday, J., Tincknell, G., Ma, Y., Napaki, S., Pantel, K., Brungs, D., & Ranson, M. (2024). EGFR and PI3K Signalling Pathways as Promising Targets on Circulating Tumour Cells from Patients with Metastatic Gastric Adenocarcinoma. International Journal of Molecular Sciences, 25(10), 5565. https://doi.org/10.3390/ijms25105565






