The Epigenomic Features and Potential Functions of PEG- and PDS-Favorable DNA G-Quadruplexes in Rice
Abstract
1. Introduction
2. Results
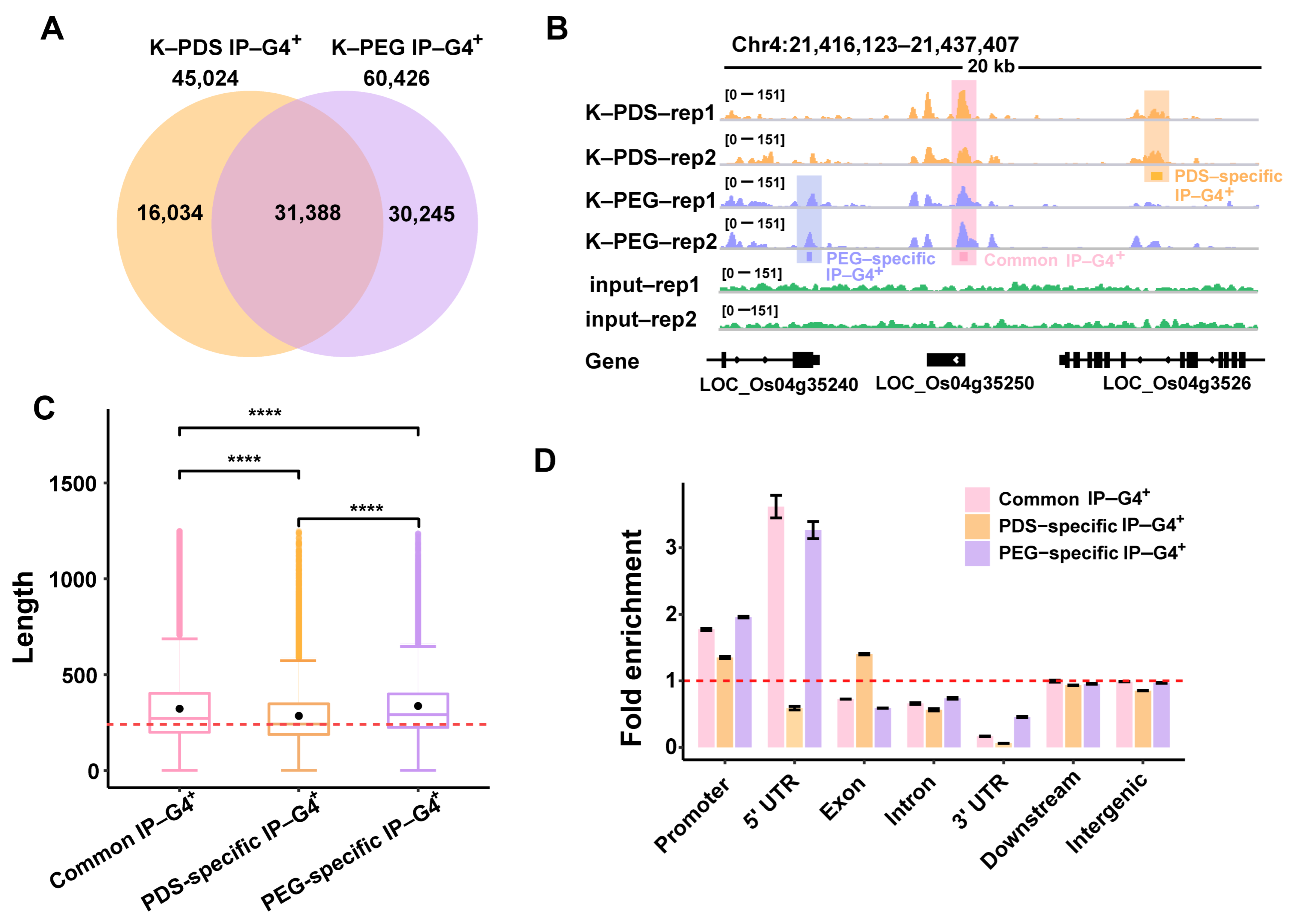
2.1. Identification of PDS/PEG-Specific IP-G4s+
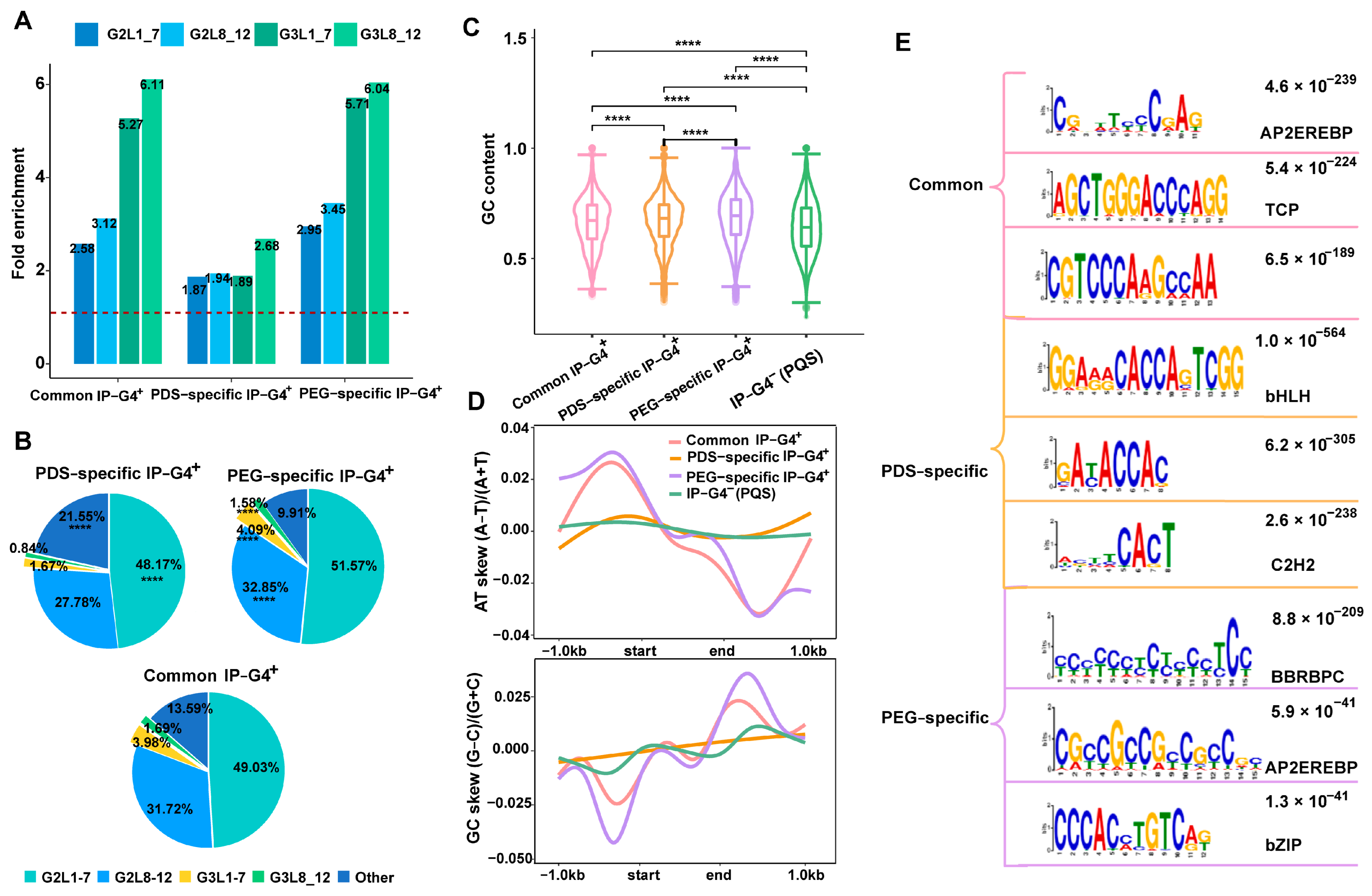
2.2. Genomic Features of Each Subtype of IP-G4s+
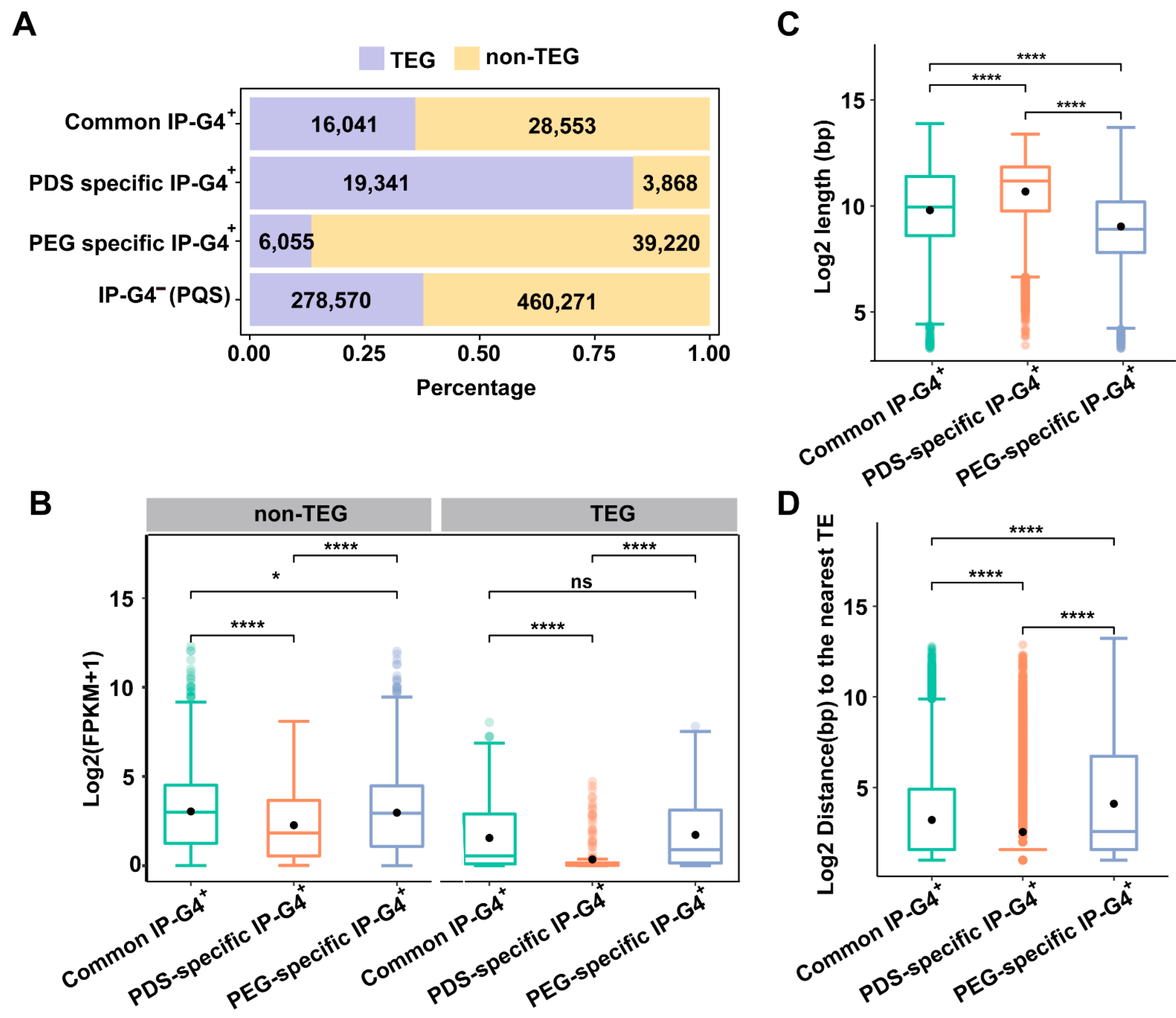
2.3. Distinct Associations of PDS/PEG-Specific IP-G4s+ with TEs or TEGs
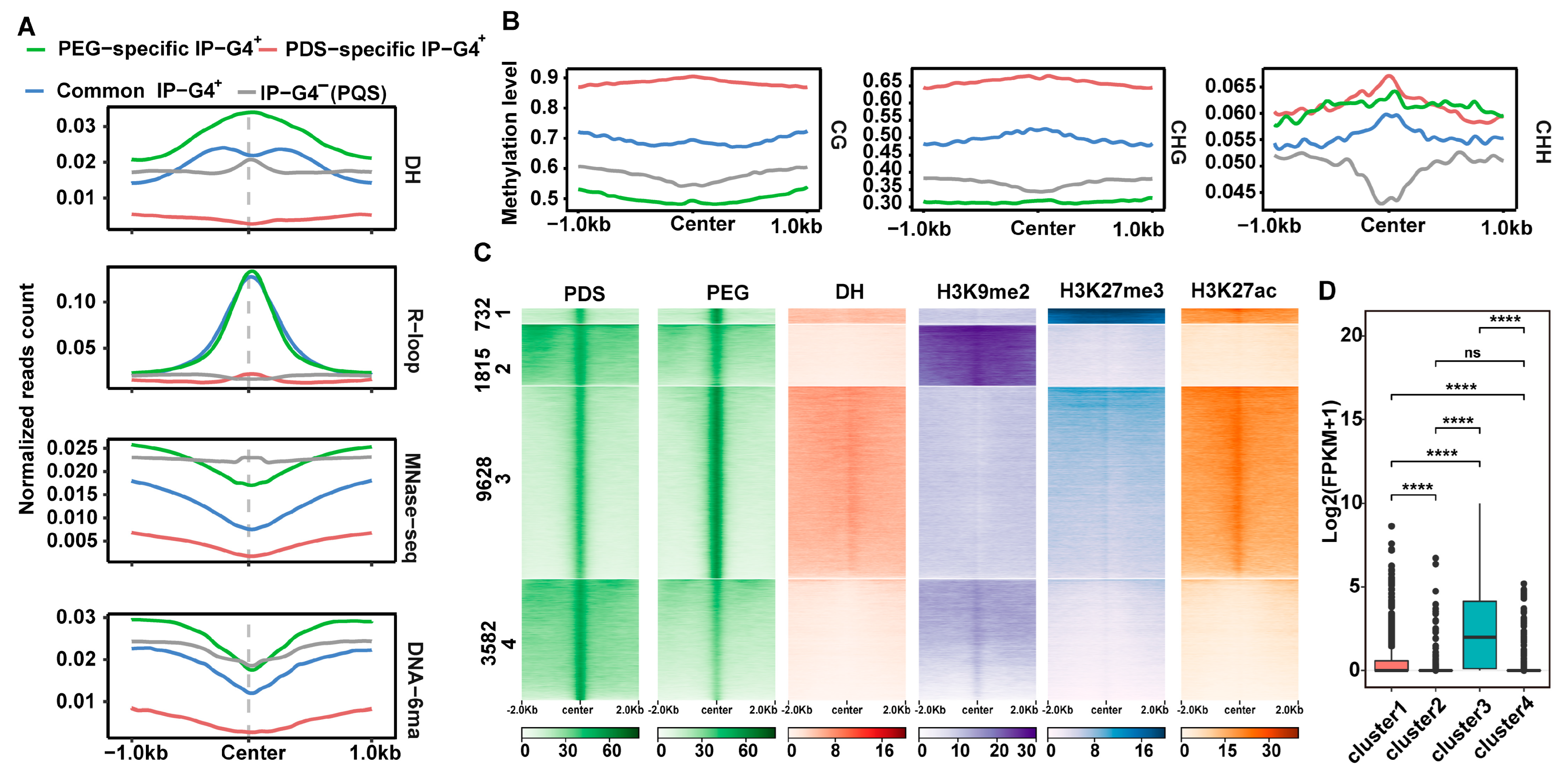
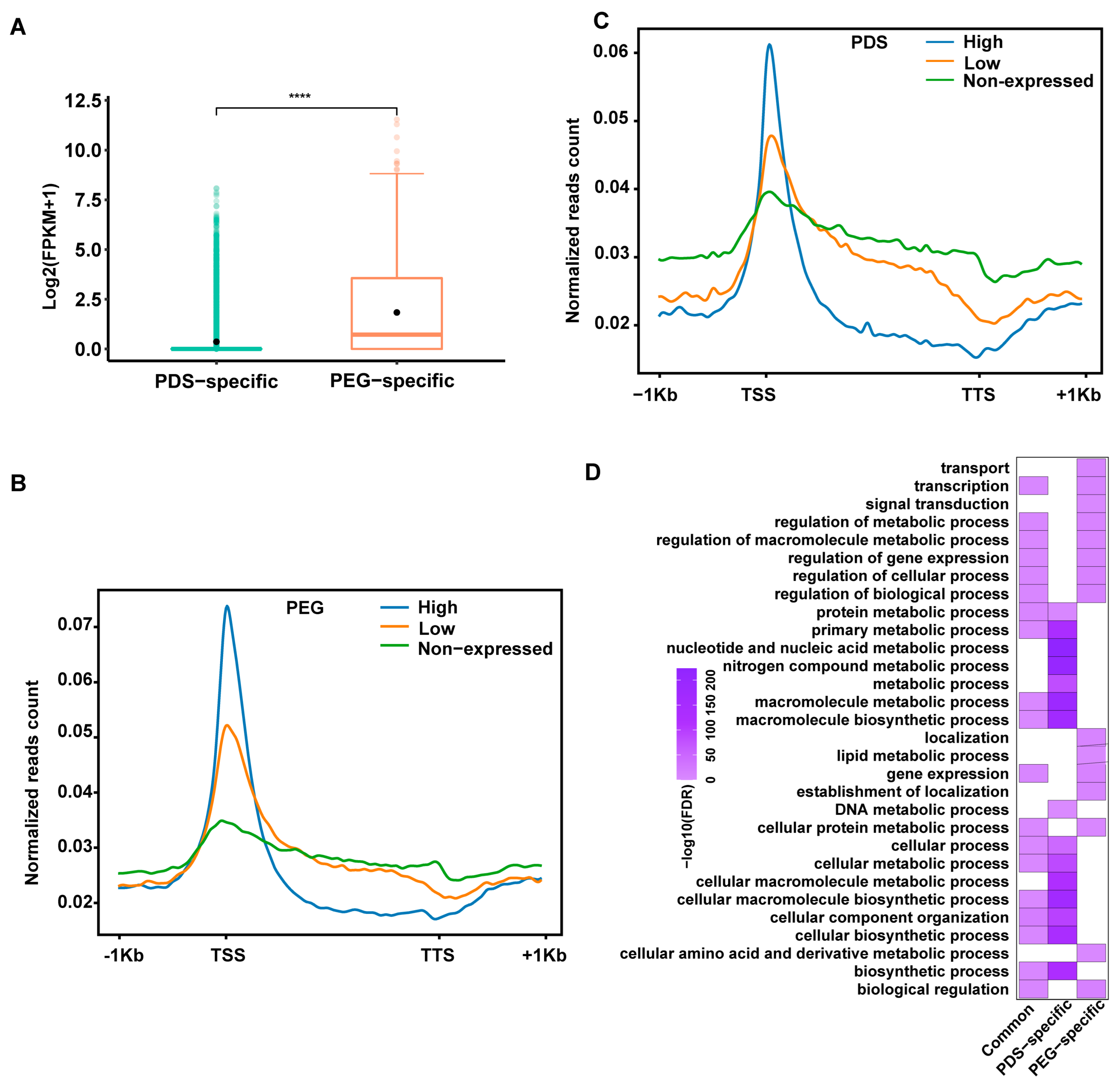
2.4. Epigenomic Features of PEG/PDS-Specific and Common IP-G4s+
2.5. Relationships of PEG/PDS-Specific IP-G4s+ with the Expression Levels of Overlapping Genes
3. Discussion
4. Materials and Methods
4.1. Growth of Rice Seedlings
4.2. BG4-DNA-IP-Seq
4.3. Analyses of BG4-DNA-IP-Seq Data
4.4. Analyses of GC Content and GC/AT Skews
4.5. Public Data Analyses
4.6. In Silico Prediction of Motifs
4.7. Gene Ontology Enrichment Analyses
4.8. Normalization of Read Counts
4.9. In Silico Identification of PQFSs
4.10. Circular Dichroism (CD) Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef]
- Bang, I. Untersuchungen über die Guanylsre. Biochem. Z. 1910, 26, 293–311. [Google Scholar]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Nakken, S.; Rognes, T.; Hovig, E. The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 2009, 37, 5749–5756. [Google Scholar] [CrossRef] [PubMed]
- Eddy, J.; Maizels, N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006, 34, 3887–3896. [Google Scholar] [CrossRef] [PubMed]
- Griffin, B.D.; Bass, H.W. Review: Plant G-quadruplex (G4) motifs in DNA and RNA; abundant, intriguing sequences of unknown function. Plant Sci. 2018, 269, 143–147. [Google Scholar] [CrossRef]
- Feng, Y.; Tao, S.; Zhang, P.; Sperti, F.R.; Liu, G.; Cheng, X.; Zhang, T.; Yu, H.; Wang, X.E.; Chen, C.; et al. Epigenomic features of DNA G-quadruplexes and their roles in regulating rice gene transcription. Plant Physiol. 2022, 188, 1632–1648. [Google Scholar] [CrossRef]
- Lexa, M.; Kejnovský, E.; Steflová, P.; Konvalinová, H.; Vorlícková, M.; Vyskot, B. Quadruplex-forming sequences occupy discrete regions inside plant LTR retrotransposons. Nucleic Acids Res. 2014, 42, 968–978. [Google Scholar] [CrossRef]
- Kang, S.G.; Henderson, E. Identification of non-telomeric G4-DNA binding proteins in human, E. coli, yeast, and Arabidopsis. Mol. Cells 2002, 14, 404–410. [Google Scholar] [CrossRef]
- Capra, J.A.; Paeschke, K.; Singh, M.; Zakian, V.A. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 2010, 6, e1000861. [Google Scholar] [CrossRef]
- Hershman, S.G.; Chen, Q.; Lee, J.Y.; Kozak, M.L.; Yue, P.; Wang, L.S.; Johnson, F.B. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008, 36, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [PubMed]
- Todd, A.K.; Johnston, M.; Neidle, S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef] [PubMed]
- Sparks, M.A.; Singh, S.P.; Burgers, P.M.; Galletto, R. Complementary roles of Pif1 helicase and single stranded DNA binding proteins in stimulating DNA replication through G-quadruplexes. Nucleic Acids Res. 2019, 47, 8595–8605. [Google Scholar] [CrossRef] [PubMed]
- Gowan, S.M.; Harrison, J.R.; Patterson, L.; Valenti, M.; Read, M.A.; Neidle, S.; Kelland, L.R. A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol. Pharmacol. 2002, 61, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Incles, C.M.; Schultes, C.M.; Kempski, H.; Koehler, H.; Kelland, L.R.; Neidle, S. A G-quadruplex telomere targeting agent produces p16-associated senescence and chromosomal fusions in human prostate cancer cells. Mol. Cancer Ther. 2004, 3, 1201–1206. [Google Scholar] [CrossRef]
- Hänsel-Hertsch, R.; Simeone, A.; Shea, A.; Hui, W.W.I.; Zyner, K.G.; Marsico, G.; Rueda, O.M.; Bruna, A.; Martin, A.; Zhang, X.; et al. Landscape of G-quadruplex DNA structural regions in breast cancer. Nat. Genet. 2020, 52, 878–883. [Google Scholar] [CrossRef]
- Bagga, J.S.; D’Antonio, L.A. Role of conserved cis-regulatory elements in the post-transcriptional regulation of the human MECP2 gene involved in autism. Hum. Genom. 2013, 7, 19. [Google Scholar] [CrossRef][Green Version]
- Mao, S.Q.; Ghanbarian, A.T.; Spiegel, J.; Martinez Cuesta, S.; Beraldi, D.; Di Antonio, M.; Marsico, G.; Hansel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. DNA G-quadruplex structures mold the DNA methylome. Nat. Struct. Mol. Biol. 2018, 25, 951–957. [Google Scholar] [CrossRef]
- Wong, L.H.; McGhie, J.D.; Sim, M.; Anderson, M.A.; Ahn, S.; Hannan, R.D.; George, A.J.; Morgan, K.A.; Mann, J.R.; Choo, K.H. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010, 20, 351–360. [Google Scholar] [CrossRef]
- Varizhuk, A.; Isaakova, E.; Pozmogova, G. DNA G-Quadruplexes (G4s) Modulate Epigenetic (Re)Programming and Chromatin Remodeling: Transient Genomic G4s Assist in the Establishment and Maintenance of Epigenetic Marks, While Persistent G4s May Erase Epigenetic Marks. BioEssays 2019, 41, e1900091. [Google Scholar] [CrossRef] [PubMed]
- Sahayasheela, V.J.; Yu, Z.; Hidaka, T.; Pandian, G.N.; Sugiyama, H. Mitochondria and G-quadruplex evolution: An intertwined relationship. Trends Genet. 2023, 39, 15–30. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, F.; Gong, H.; Cai, C. Double G-quadruplexes in a copper nanoparticle based fluorescent probe for determination of HIV genes. Mikrochim. Acta 2018, 186, 30. [Google Scholar] [CrossRef] [PubMed]
- Moruno-Manchon, J.F.; Koellhoffer, E.C.; Gopakumar, J.; Hambarde, S.; Kim, N.; McCullough, L.D.; Tsvetkov, A.S. The G-quadruplex DNA stabilizing drug pyridostatin promotes DNA damage and downregulates transcription of Brca1 in neurons. Aging 2017, 9, 1957–1970. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Aggarwal, J.; Thakkar, B. Genome-wide discovery of G-quadruplex forming sequences and their functional relevance in plants. Sci. Rep. 2016, 6, 28211. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Zhu, J.; Ding, Y.; Burrows, C.J. Location dependence of the transcriptional response of a potential G-quadruplex in gene promoters under oxidative stress. Nucleic Acids Res. 2019, 47, 5049–5060. [Google Scholar] [CrossRef]
- Pecinka, P.; Bohalova, N.; Volna, A.; Kundratova, K.; Brazda, V.; Bartas, M. Analysis of G-Quadruplex-Forming Sequences in Drought Stress-Responsive Genes, and Synthesis Genes of Phenolic Compounds in Arabidopsis thaliana. Life 2023, 13, 199. [Google Scholar] [CrossRef]
- Wu, G.; Chen, L.; Liu, W.; Yang, D. Molecular Recognition of the Hybrid-Type G-Quadruplexes in Human Telomeres. Molecules 2019, 24, 1578. [Google Scholar] [CrossRef]
- Hansel-Hertsch, R.; Spiegel, J.; Marsico, G.; Tannahill, D.; Balasubramanian, S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2018, 13, 551–564. [Google Scholar] [CrossRef]
- Liu, H.Y.; Zhao, Q.; Zhang, T.P.; Wu, Y.; Xiong, Y.X.; Wang, S.K.; Ge, Y.L.; He, J.H.; Lv, P.; Ou, T.M.; et al. Conformation Selective Antibody Enables Genome Profiling and Leads to Discovery of Parallel G-Quadruplex in Human Telomeres. Cell Chem. Biol. 2016, 23, 1261–1270. [Google Scholar] [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Antonio, M.D.; Balasubramanian, S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.W.; Zhang, J.Y.; He, Y.D.; Gong, J.Y.; Wen, C.J.; Chen, J.N.; Hao, Y.H.; Zhao, Y.; Tan, Z. Detection of genomic G-quadruplexes in living cells using a small artificial protein. Nucleic Acids Res. 2020, 48, 11706–11720. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Shao, R.; Kwong Yung, P.Y.; Elsasser, S.J. Genome-wide mapping of G-quadruplex structures with CUT&Tag. Nucleic Acids Res. 2022, 50, e13. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; He, Z.; Luo, Z.; Sperti, F.R.; Valverde, I.E.; Zhang, W.; Monchaud, D. Side-by-side comparison of G-quadruplex (G4) capture efficiency of the antibody BG4 versus the small-molecule ligands TASQs. iScience 2023, 26, 106846. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Gilbert, D.E.; Feigon, J. Multistranded DNA structures. Curr. Opin. Struct. Biol. 1999, 9, 305–314. [Google Scholar] [CrossRef]
- Buscaglia, R.; Miller, M.C.; Dean, W.L.; Gray, R.D.; Lane, A.N.; Trent, J.O.; Chaires, J.B. Polyethylene glycol binding alters human telomere G-quadruplex structure by conformational selection. Nucleic Acids Res. 2013, 41, 7934–7946. [Google Scholar] [CrossRef]
- Kan, Z.Y.; Lin, Y.; Wang, F.; Zhuang, X.Y.; Zhao, Y.; Pang, D.W.; Hao, Y.H.; Tan, Z. G-quadruplex formation in human telomeric (TTAGGG)4 sequence with complementary strand in close vicinity under molecularly crowded condition. Nucleic Acids Res. 2007, 35, 3646–3653. [Google Scholar] [CrossRef]
- Miyoshi, D.; Fujimoto, T.; Sugimoto, N. Molecular crowding and hydration regulating of G-quadruplex formation. Top. Curr. Chem. 2013, 330, 87–110. [Google Scholar] [CrossRef]
- Miyoshi, D.; Nakao, A.; Sugimoto, N. Molecular crowding regulates the structural switch of the DNA G-quadruplex. Biochemistry 2002, 41, 15017–15024. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.W.; Chen, Z.; Hao, Y.H.; Tan, Z. Molecular crowding creates an essential environment for the formation of stable G-quadruplexes in long double-stranded DNA. Nucleic Acids Res. 2010, 38, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Abou Assi, H.; Garavís, M.; González, C.; Damha, M.J. i-Motif DNA: Structural features and significance to cell biology. Nucleic Acids Res. 2018, 46, 8038–8056. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Y.; Ma, T.Z.; Zeng, Y.L.; Liu, W.; Mao, Z.W. Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes. J. Am. Chem. Soc. 2022, 144, 11878–11887. [Google Scholar] [CrossRef] [PubMed]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Di Antonio, M.; Ponjavic, A.; Radzevičius, A.; Ranasinghe, R.T.; Catalano, M.; Zhang, X.; Shen, J.; Needham, L.M.; Lee, S.F.; Klenerman, D.; et al. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat. Chem. 2020, 12, 832–837. [Google Scholar] [CrossRef]
- Beniaminov, A.; Chashchina, G.; Shchyolkina, A.; Kaluzhny, D. Oxidative probing of the G4 DNA structure induced in double-stranded DNA by molecular crowding or pyridostatin. Biochimie 2021, 191, 33–36. [Google Scholar] [CrossRef]
- Ge, F.; Wang, Y.; Li, H.; Zhang, R.; Wang, X.; Li, Q.; Liang, Z.; Yang, L. Plant-GQ: An Integrative Database of G-Quadruplex in Plant. J. Comput. Biol. 2019, 26, 1013–1019. [Google Scholar] [CrossRef]
- Takahashi, H.; Nakagawa, A.; Kojima, S.; Takahashi, A.; Cha, B.Y.; Woo, J.T.; Nagai, K.; Machida, Y.; Machida, C. Discovery of novel rules for G-quadruplex-forming sequences in plants by using bioinformatics methods. J. Biosci. Bioeng. 2012, 114, 570–575. [Google Scholar] [CrossRef]
- Andorf, C.M.; Kopylov, M.; Dobbs, D.; Koch, K.E.; Stroupe, M.E.; Lawrence, C.J.; Bass, H.W. G-quadruplex (G4) motifs in the maize (Zea mays L.) genome are enriched at specific locations in thousands of genes coupled to energy status, hypoxia, low sugar, and nutrient deprivation. J. Genet. Genom. 2014, 41, 627–647. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, M.; Zhang, Q.; Zhu, G.F.; Li, F.F.; Du, L.F. Genomic distribution and possible functional roles of putative G-quadruplex motifs in two subspecies of Oryza sativa. Comput. Biol. Chem. 2015, 56, 122–130. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Sen, T.Z. Genome-Wide Discovery of G-Quadruplexes in Wheat: Distribution and Putative Functional Roles. G3 2020, 10, 2021–2032. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Budak, H.; Sen, T.Z. Genome-wide discovery of G-quadruplexes in barley. Sci. Rep. 2021, 11, 7876. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Luo, Z.; Huang, R.; Yang, X.; Cheng, X.; Zhang, W. Epigenomic Features and Potential Functions of K+ and Na+ Favorable DNA G-Quadruplexes in Rice. Int. J. Mol. Sci. 2022, 23, 8404. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Hemansi, N.; Kim, N.; Tuteja, N.; Yadav, P. G Quadruplex in Plants: A Ubiquitous Regulatory Element and Its Biological Relevance. Front. Plant Sci. 2017, 8, 1163. [Google Scholar] [CrossRef] [PubMed]
- Mullen, M.A.; Olson, K.J.; Dallaire, P.; Major, F.; Assmann, S.M.; Bevilacqua, P.C. RNA G-Quadruplexes in the model plant species Arabidopsis thaliana: Prevalence and possible functional roles. Nucleic Acids Res. 2010, 38, 8149–8163. [Google Scholar] [CrossRef]
- Volna, A.; Bartas, M.; Nezval, J.; Pech, R.; Pecinka, P.; Spunda, V.; Cerven, J. Beyond the Primary Structure of Nucleic Acids: Potential Roles of Epigenetics and Noncanonical Structures in the Regulations of Plant Growth and Stress Responses. Methods Mol. Biol. 2023, 2642, 331–361. [Google Scholar] [CrossRef]
- Bai, D.; Shan, S.W.; Zhang, X.; Li, Y.; Xie, J.; Wu, W.Q. Comprehensive insights into the structures and dynamics of plant telomeric G-quadruplexes. Int. J. Biol. Macromol. 2023, 231, 123281. [Google Scholar] [CrossRef]
- De Rache, A.; Mergny, J.L. Assessment of selectivity of G-quadruplex ligands via an optimised FRET melting assay. Biochimie 2015, 115, 194–202. [Google Scholar] [CrossRef]
- Fujimoto, A.; Fujita, M.; Hasegawa, T.; Wong, J.H.; Maejima, K.; Oku-Sasaki, A.; Nakano, K.; Shiraishi, Y.; Miyano, S.; Yamamoto, G.; et al. Comprehensive analysis of indels in whole-genome microsatellite regions and microsatellite instability across 21 cancer types. Genome Res. 2020, 30, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Chen, Z.; Luo, F.; Liao, M.; Wei, S.; Yang, Z.; Yang, J. RicetissueTFDB: A Genome-Wide Identification of Tissue-Specific Transcription Factors in Rice. Plant Genome 2019, 12, 170081. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP Transcription Factors in Plant Salt Stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef]
- Sahu, A.; Singh, R.; Verma, P.K. Plant BBR/BPC transcription factors: Unlocking multilayered regulation in development, stress and immunity. Planta 2023, 258, 31. [Google Scholar] [CrossRef]
- Fang, Y.; Chen, L.; Lin, K.; Feng, Y.; Zhang, P.; Pan, X.; Sanders, J.; Wu, Y.; Wang, X.E.; Su, Z.; et al. Characterization of functional relationships of R-loops with gene transcription and epigenetic modifications in rice. Genome Res. 2019, 29, 1287–1297. [Google Scholar] [CrossRef]
- Sannohe, Y.; Sugiyama, H. Overview of formation of G-quadruplex structures. Curr. Protoc. Nucleic Acid. Chem. 2010, 40, 17.2.1–17.2.17. [Google Scholar] [CrossRef]
- Nishio, M.; Tsukakoshi, K.; Ikebukuro, K. G-quadruplex: Flexible conformational changes by cations, pH, crowding and its applications to biosensing. Biosens. Bioelectron. 2021, 178, 113030. [Google Scholar] [CrossRef]
- Tang, C.F.; Shafer, R.H. Engineering the quadruplex fold: Nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J. Am. Chem. Soc. 2006, 128, 5966–5973. [Google Scholar] [CrossRef]
- Rachwal, P.A.; Brown, T.; Fox, K.R. Effect of G-tract length on the topology and stability of intramolecular DNA quadruplexes. Biochemistry 2007, 46, 3036–3044. [Google Scholar] [CrossRef] [PubMed]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- Risitano, A.; Fox, K.R. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004, 32, 2598–2606. [Google Scholar] [CrossRef] [PubMed]
- Rachwal, P.A.; Findlow, I.S.; Werner, J.M.; Brown, T.; Fox, K.R. Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Res. 2007, 35, 4214–4222. [Google Scholar] [CrossRef] [PubMed]
- Tippana, R.; Xiao, W.; Myong, S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014, 42, 8106–8114. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Nair, D.R.; Maiti, S. Effect of flanking bases on quadruplex stability and Watson-Crick duplex competition. FEBS J. 2009, 276, 3628–3640. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Sharma, S.; Chowdhury, S. Non-duplex G-Quadruplex Structures Emerge as Mediators of Epigenetic Modifications. Trends Genet. 2019, 35, 129–144. [Google Scholar] [CrossRef]
- Mekmaysy, C.S.; Petraccone, L.; Garbett, N.C.; Ragazzon, P.A.; Gray, R.; Trent, J.O.; Chaires, J.B. Effect of O6-methylguanine on the stability of G-quadruplex DNA. J. Am. Chem. Soc. 2008, 130, 6710–6711. [Google Scholar] [CrossRef]
- Gros, J.; Rosu, F.; Amrane, S.; De Cian, A.; Gabelica, V.; Lacroix, L.; Mergny, J.L. Guanines are a quartet’s best friend: Impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007, 35, 3064–3075. [Google Scholar] [CrossRef]
- Virgilio, A.; Esposito, V.; Randazzo, A.; Mayol, L.; Galeone, A. 8-methyl-2′-deoxyguanosine incorporation into parallel DNA quadruplex structures. Nucleic Acids Res. 2005, 33, 6188–6195. [Google Scholar] [CrossRef]
- Esposito, V.; Randazzo, A.; Piccialli, G.; Petraccone, L.; Giancola, C.; Mayol, L. Effects of an 8-bromodeoxyguanosine incorporation on the parallel quadruplex structure [d(TGGGT)]4. Org. Biomol. Chem. 2004, 2, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, A.G.; Polavarapu, P.L. Quadruplex structure of polyriboinosinic acid: Dependence on alkali metal ion concentration, pH and temperature. J. Phys. Chem. B 2008, 112, 2255–2260. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Yu, Z.; Zhou, W.; Li, Y.; Fan, L.; Li, X. Investigation of Na+ and K+ Competitively Binding with a G-Quadruplex and Discovery of a Stable K+-Na+-Quadruplex. J. Phys. Chem. B 2019, 123, 5405–5411. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, D.; Mirihana Arachchilage, G.; Basu, S. Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Renciuk, D.; Kejnovska, I.; Skolakova, P.; Bednarova, K.; Motlova, J.; Vorlickova, M. Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Res. 2009, 37, 6625–6634. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Kan, Z.Y.; Wang, Q.; Yao, Y.; Liu, J.; Hao, Y.H.; Tan, Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007, 129, 11185–11191. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Mohamed, H.I.; Deng, J.; Umer, M.; Anwar, N.; Chen, J.; Wu, Q.; Wang, Z.; He, Y. Effects of Molecular Crowding on the Structure, Stability, and Interaction with Ligands of G-quadruplexes. ACS Omega 2023, 8, 14342–14348. [Google Scholar] [CrossRef]
- Yan, Y.Y.; Tan, J.H.; Lu, Y.J.; Yan, S.C.; Wong, K.Y.; Li, D.; Gu, L.Q.; Huang, Z.S. G-Quadruplex conformational change driven by pH variation with potential application as a nanoswitch. Biochim. Biophys. Acta 2013, 1830, 4935–4942. [Google Scholar] [CrossRef]
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008, 36, 5482–5515. [Google Scholar] [CrossRef]
- Kumar, N.; Basundra, R.; Maiti, S. Elevated polyamines induce c-MYC overexpression by perturbing quadruplex-WC duplex equilibrium. Nucleic Acids Res. 2009, 37, 3321–3331. [Google Scholar] [CrossRef]
- Yin, F.; Liu, J.; Peng, X. Triethylene tetraamine: A novel telomerase inhibitor. Bioorg. Med. Chem. Lett. 2003, 13, 3923–3926. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.; Miller, K.M.; Forment, J.V.; Bradshaw, C.R.; Nikan, M.; Britton, S.; Oelschlaegel, T.; Xhemalce, B.; Balasubramanian, S.; Jackson, S.P. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Salvati, E.; Scarsella, M.; Porru, M.; Rizzo, A.; Iachettini, S.; Tentori, L.; Graziani, G.; D’Incalci, M.; Stevens, M.F.; Orlandi, A.; et al. PARP1 is activated at telomeres upon G4 stabilization: Possible target for telomere-based therapy. Oncogene 2010, 29, 6280–6293. [Google Scholar] [CrossRef] [PubMed]
- Ohnmacht, S.A.; Marchetti, C.; Gunaratnam, M.; Besser, R.J.; Haider, S.M.; Di Vita, G.; Lowe, H.L.; Mellinas-Gomez, M.; Diocou, S.; Robson, M.; et al. A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer. Sci. Rep. 2015, 5, 11385. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Deng, J.; Anwar, N.; Umer, M.; Chen, J.; Wu, Q.; Dong, X.; Xu, H.; He, Y.; Wang, Z. Molecular crowding promotes the aggregation of parallel structured G-quadruplexes. Int. J. Biol. Macromol. 2023, 240, 124442. [Google Scholar] [CrossRef]
- Miyoshi, D.; Karimata, H.; Sugimoto, N. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 2006, 128, 7957–7963. [Google Scholar] [CrossRef]
- Trajkovski, M.; Endoh, T.; Tateishi-Karimata, H.; Ohyama, T.; Tanaka, S.; Plavec, J.; Sugimoto, N. Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations. Nucleic Acids Res. 2018, 46, 4301–4315. [Google Scholar] [CrossRef]
- Knowles, D.B.; LaCroix, A.S.; Deines, N.F.; Shkel, I.; Record, M.T., Jr. Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability. Proc. Natl. Acad. Sci. USA 2011, 108, 12699–12704. [Google Scholar] [CrossRef]
- Kan, Z.Y.; Yao, Y.; Wang, P.; Li, X.H.; Hao, Y.H.; Tan, Z. Molecular crowding induces telomere G-quadruplex formation under salt-deficient conditions and enhances its competition with duplex formation. Angew. Chem. Int. Ed. Engl. 2006, 45, 1629–1632. [Google Scholar] [CrossRef]
- Petraccone, L.; Malafronte, A.; Amato, J.; Giancola, C. G-quadruplexes from human telomeric DNA: How many conformations in PEG containing solutions? J. Phys. Chem. B 2012, 116, 2294–2305. [Google Scholar] [CrossRef]
- Han, J.H.; Kim, J.H.; Kim, S.K.; Jang, Y.J. Conformational change of a G-quadruplex under molecular crowding conditions. J. Biomol. Struct. Dyn. 2020, 38, 2575–2581. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Sanders, D.A.; Di Antonio, M.; Matsis, S.; Riou, J.F.; Rodriguez, R.; Balasubramanian, S. Pyridostatin analogues promote telomere dysfunction and long-term growth inhibition in human cancer cells. Org. Biomol. Chem. 2012, 10, 6537–6546. [Google Scholar] [CrossRef] [PubMed]
- Di Fonzo, S.; Amato, J.; D’Aria, F.; Caterino, M.; D’Amico, F.; Gessini, A.; Brady, J.W.; Cesaro, A.; Pagano, B.; Giancola, C. Ligand binding to G-quadruplex DNA: New insights from ultraviolet resonance Raman spectroscopy. Phys. Chem. Chem. Phys. PCCP 2020, 22, 8128–8140. [Google Scholar] [CrossRef] [PubMed]
- Lejault, P.; Moruno-Manchon, J.F.; Vemu, S.M.; Honarpisheh, P.; Zhu, L.; Kim, N.; Urayama, A.; Monchaud, D.; McCullough, L.D.; Tsvetkov, A.S. Regulation of autophagy by DNA G-quadruplexes. Autophagy 2020, 16, 2252–2259. [Google Scholar] [CrossRef] [PubMed]
- Cree, S.L.; Fredericks, R.; Miller, A.; Pearce, F.G.; Filichev, V.; Fee, C.; Kennedy, M.A. DNA G-quadruplexes show strong interaction with DNA methyltransferases in vitro. FEBS Lett. 2016, 590, 2870–2883. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Hou, J.Q.; Xiang, H.D.; Yan, Y.Y.; Gu, Y.C.; Tan, J.H.; Li, D.; Gu, L.Q.; Ou, T.M.; Huang, Z.S. Stabilization of G-quadruplex DNA by C-5-methyl-cytosine in bcl-2 promoter: Implications for epigenetic regulation. Biochem. Biophys. Res. Commun. 2013, 433, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Hardin, C.C.; Corregan, M.; Brown, B.A., 2nd; Frederick, L.N. Cytosine-cytosine+ base pairing stabilizes DNA quadruplexes and cytosine methylation greatly enhances the effect. Biochemistry 1993, 32, 5870–5880. [Google Scholar] [CrossRef]
- Oyoshi, T.; Masuzawa, T. Modulation of histone modifications and G-quadruplex structures by G-quadruplex-binding proteins. Biochem. Biophys. Res. Commun. 2020, 531, 39–44. [Google Scholar] [CrossRef]
- Abuín, J.M.; Pichel, J.C.; Pena, T.F.; Amigo, J. BigBWA: Approaching the Burrows–Wheeler aligner to Big Data technologies. Bioinformatics 2015, 31, 4003–4005. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Boyle, A.P.; Davis, S.; Shulha, H.P.; Meltzer, P.; Margulies, E.H.; Weng, Z.; Furey, T.S.; Crawford, G.E. High-resolution mapping and characterization of open chromatin across the genome. Cell 2008, 132, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wang, C.; Liu, H.; Zhou, Q.; Liu, Q.; Guo, Y.; Peng, T.; Song, J.; Zhang, J.; Chen, L.; et al. Identification and analysis of adenine N6-methylation sites in the rice genome. Nat. Plants 2018, 4, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, W.; Jiang, J. Genome-Wide Nucleosome Occupancy and Positioning and Their Impact on Gene Expression and Evolution in Plants. Plant Physiol. 2015, 168, 1406–1416. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, R.; Feng, Y.; Gao, Z.; Ahmed, A.; Zhang, W. The Epigenomic Features and Potential Functions of PEG- and PDS-Favorable DNA G-Quadruplexes in Rice. Int. J. Mol. Sci. 2024, 25, 634. https://doi.org/10.3390/ijms25010634
Huang R, Feng Y, Gao Z, Ahmed A, Zhang W. The Epigenomic Features and Potential Functions of PEG- and PDS-Favorable DNA G-Quadruplexes in Rice. International Journal of Molecular Sciences. 2024; 25(1):634. https://doi.org/10.3390/ijms25010634
Chicago/Turabian StyleHuang, Ranran, Yilong Feng, Zhicheng Gao, Asgar Ahmed, and Wenli Zhang. 2024. "The Epigenomic Features and Potential Functions of PEG- and PDS-Favorable DNA G-Quadruplexes in Rice" International Journal of Molecular Sciences 25, no. 1: 634. https://doi.org/10.3390/ijms25010634
APA StyleHuang, R., Feng, Y., Gao, Z., Ahmed, A., & Zhang, W. (2024). The Epigenomic Features and Potential Functions of PEG- and PDS-Favorable DNA G-Quadruplexes in Rice. International Journal of Molecular Sciences, 25(1), 634. https://doi.org/10.3390/ijms25010634






