In Vivo and In Silico Studies of the Hepatoprotective Activity of Tert-Butylhydroquinone
Abstract
:1. Introduction
2. Results
2.1. Mortality and Survival
2.2. Body and Liver Weights
2.3. Macroscopic Findings of Livers
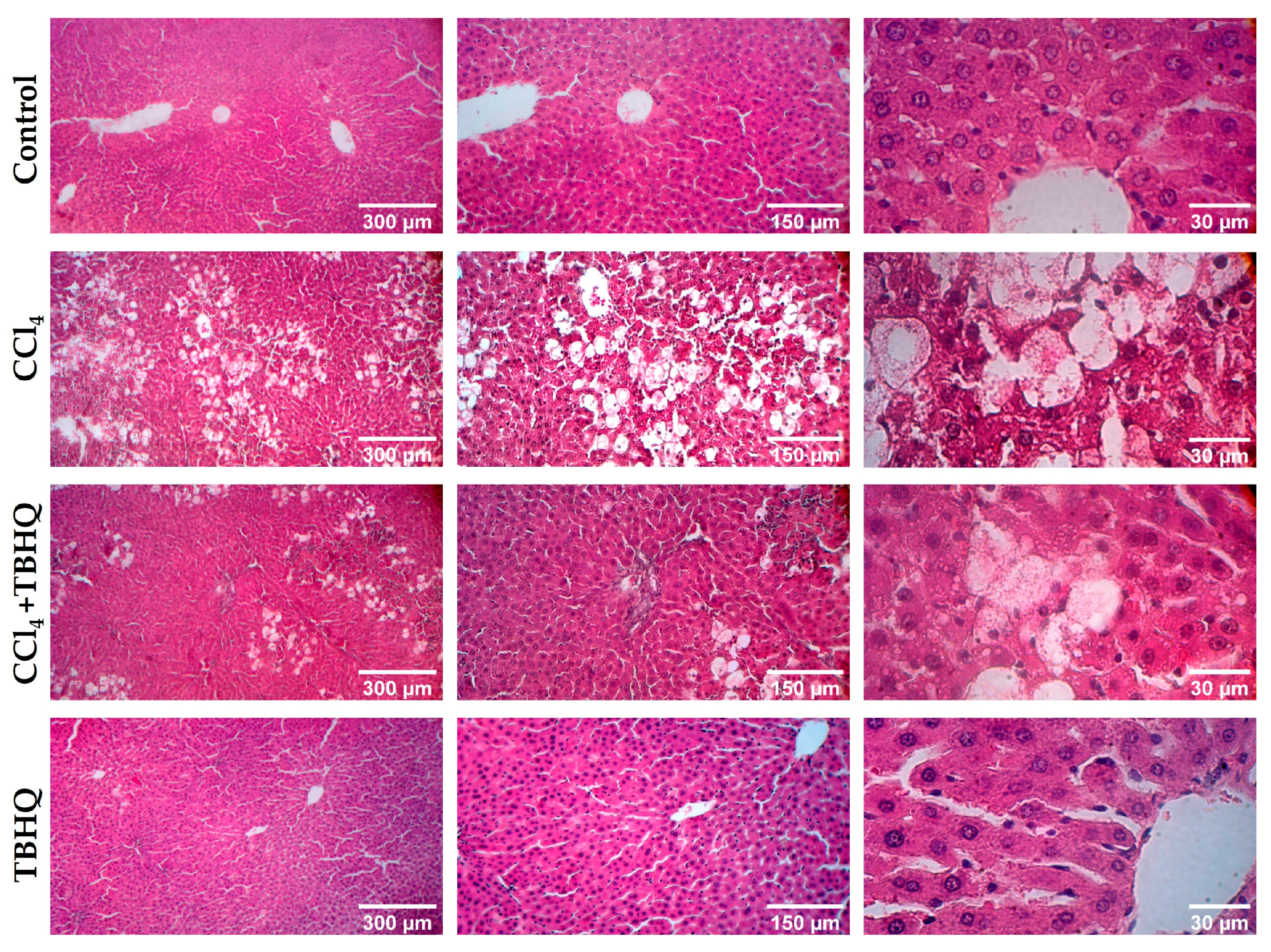
2.4. Microscopic Evaluation of Liver Damage Induced by a Sublethal Dose of CCl4
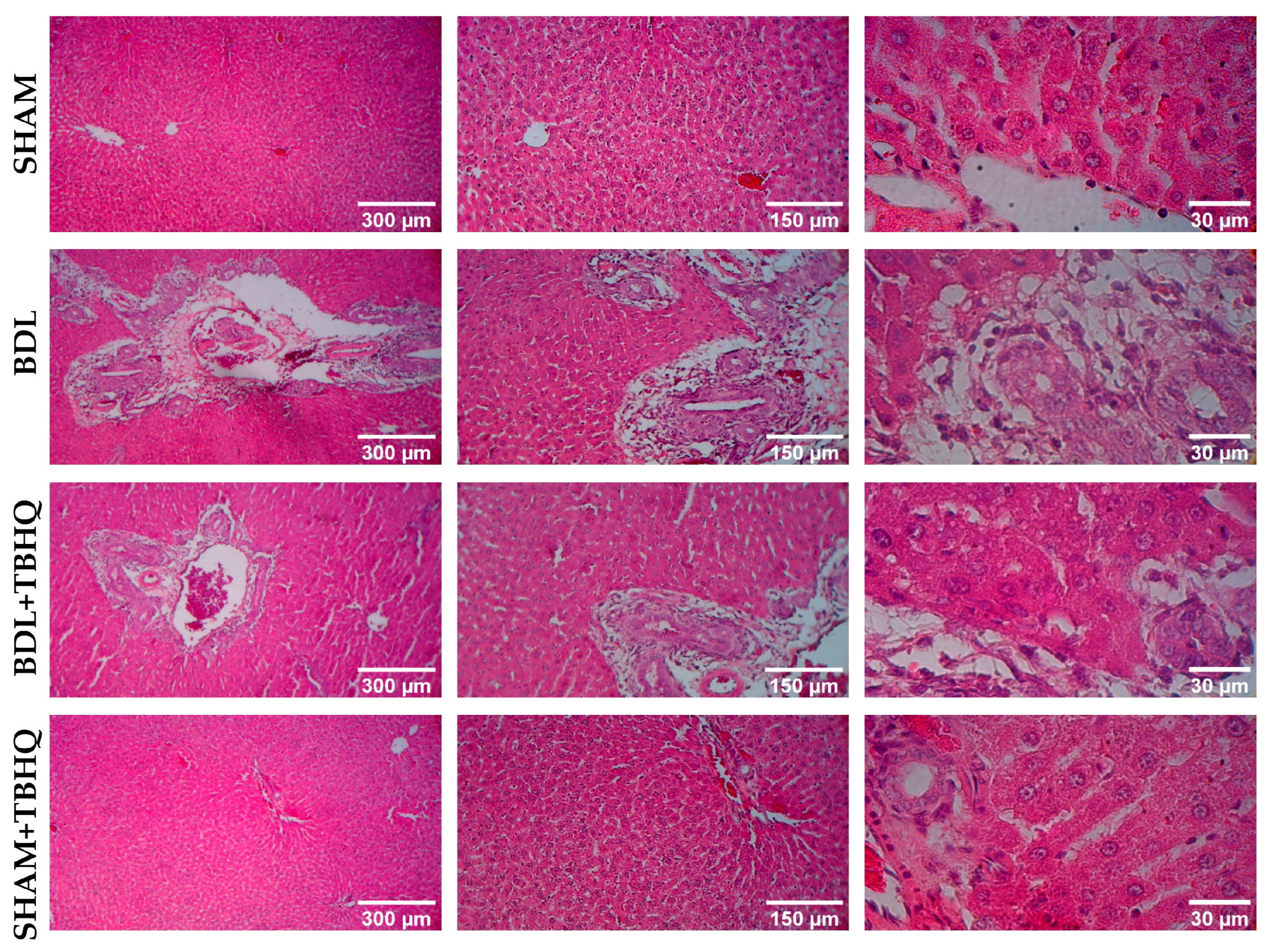
2.5. Microscopic Evaluation of Liver Damage Induced by BDL
2.6. Serum Biochemical Markers of Liver Damage
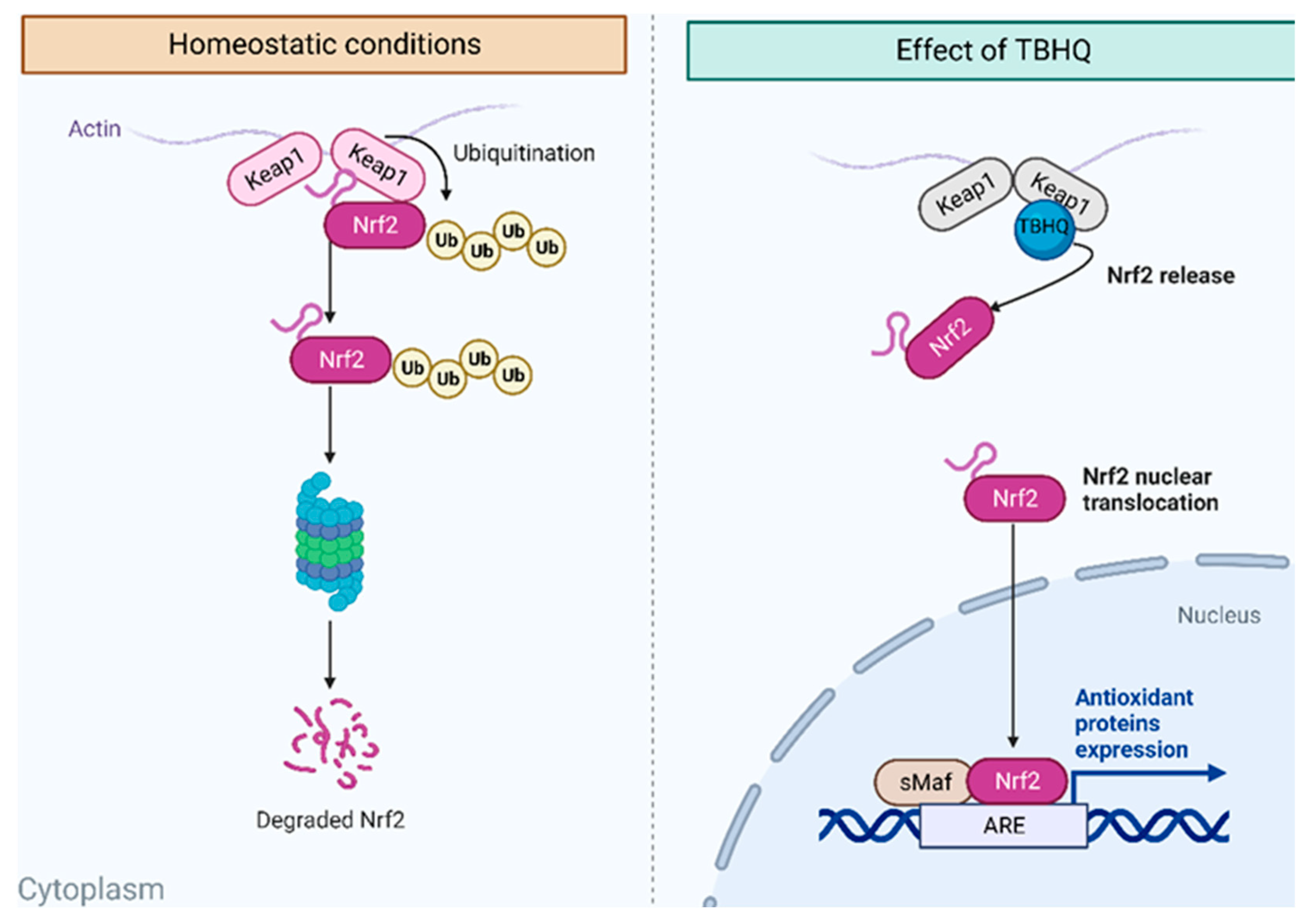
2.7. Identification of Potential TBHQ Protein Targets in the Liver
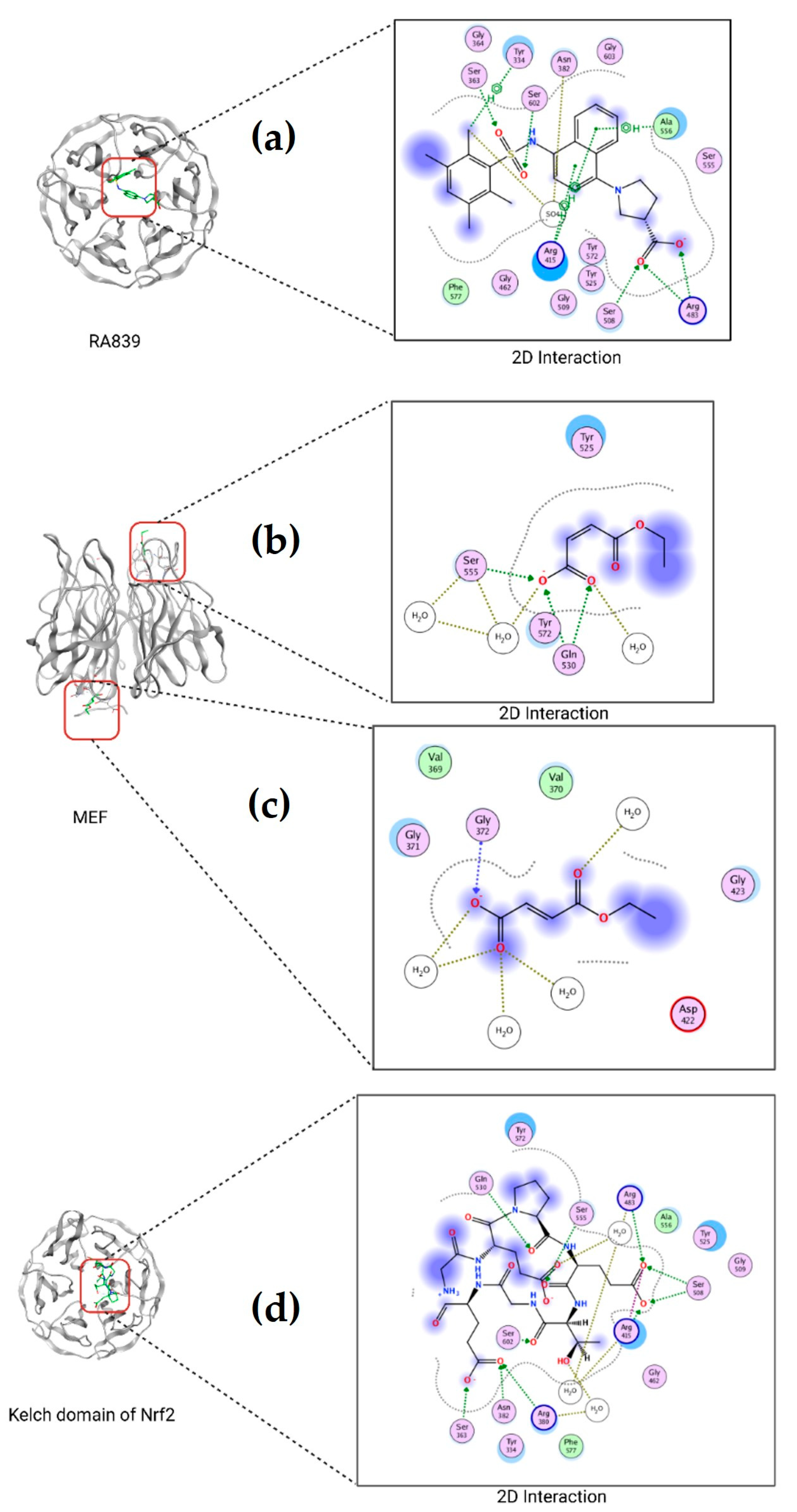
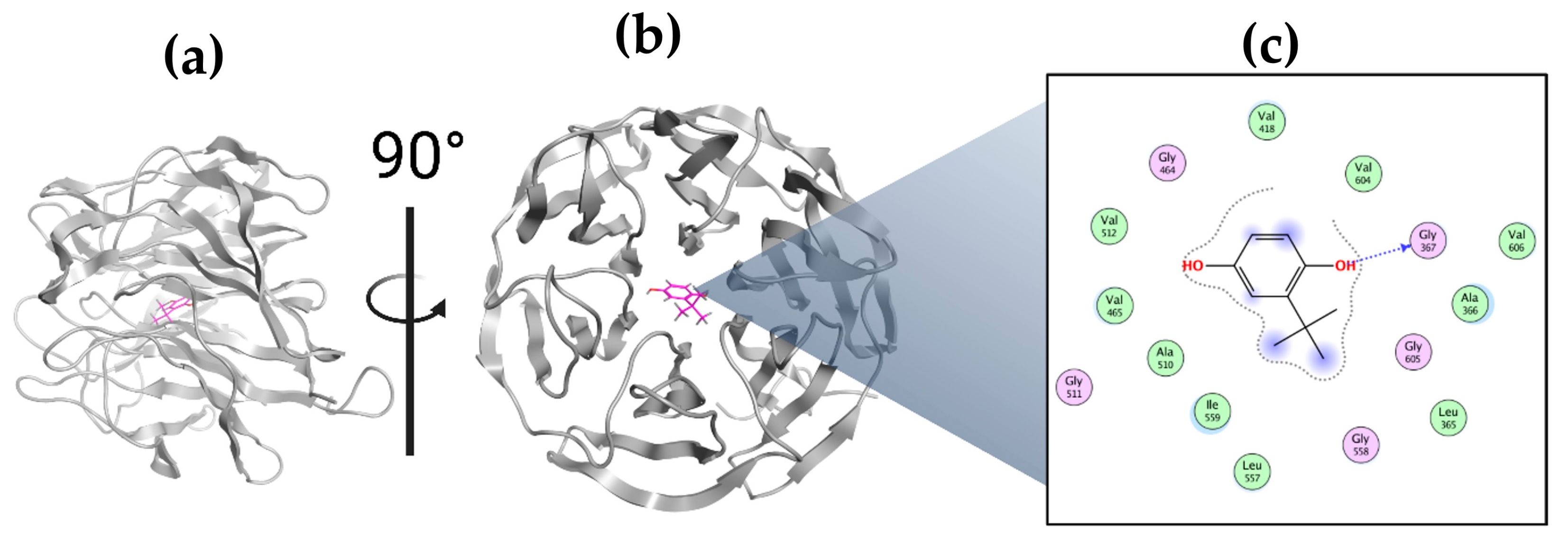
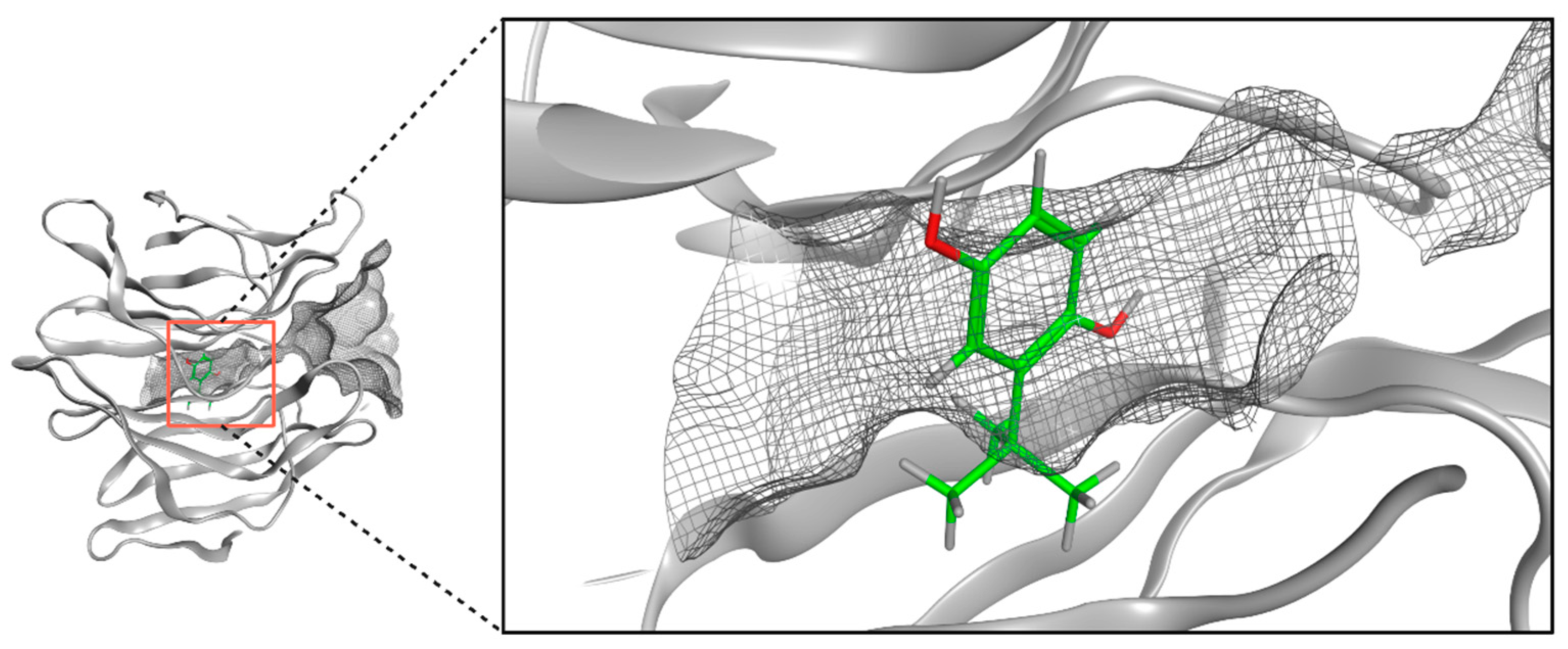
2.8. Molecular Docking of TBHQ and Keap1
3. Discussion
4. Materials and Methods
4.1. Animals
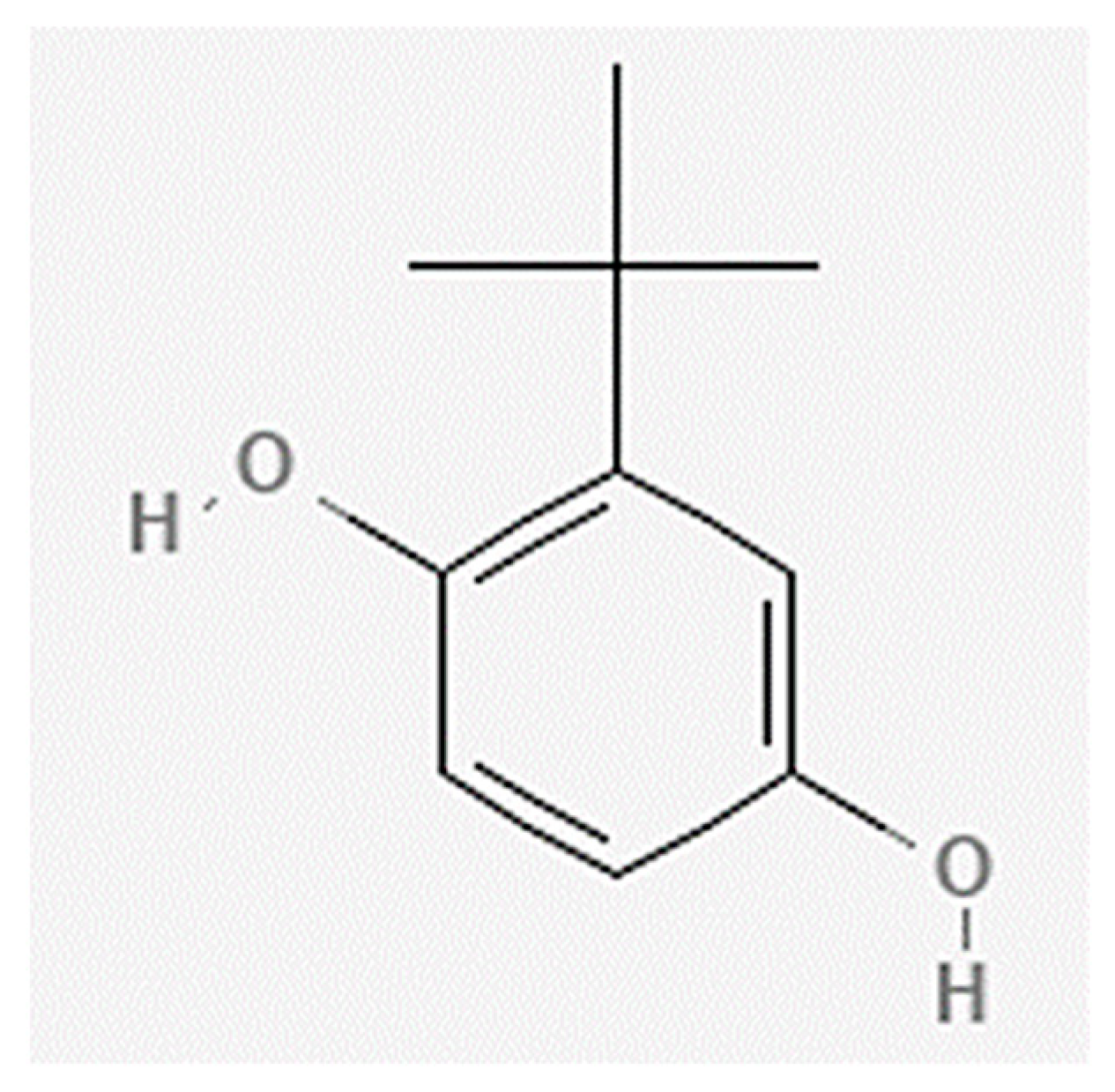
4.2. Preparation of TBHQ
4.3. Experimental Protocol of the CCl4 Model
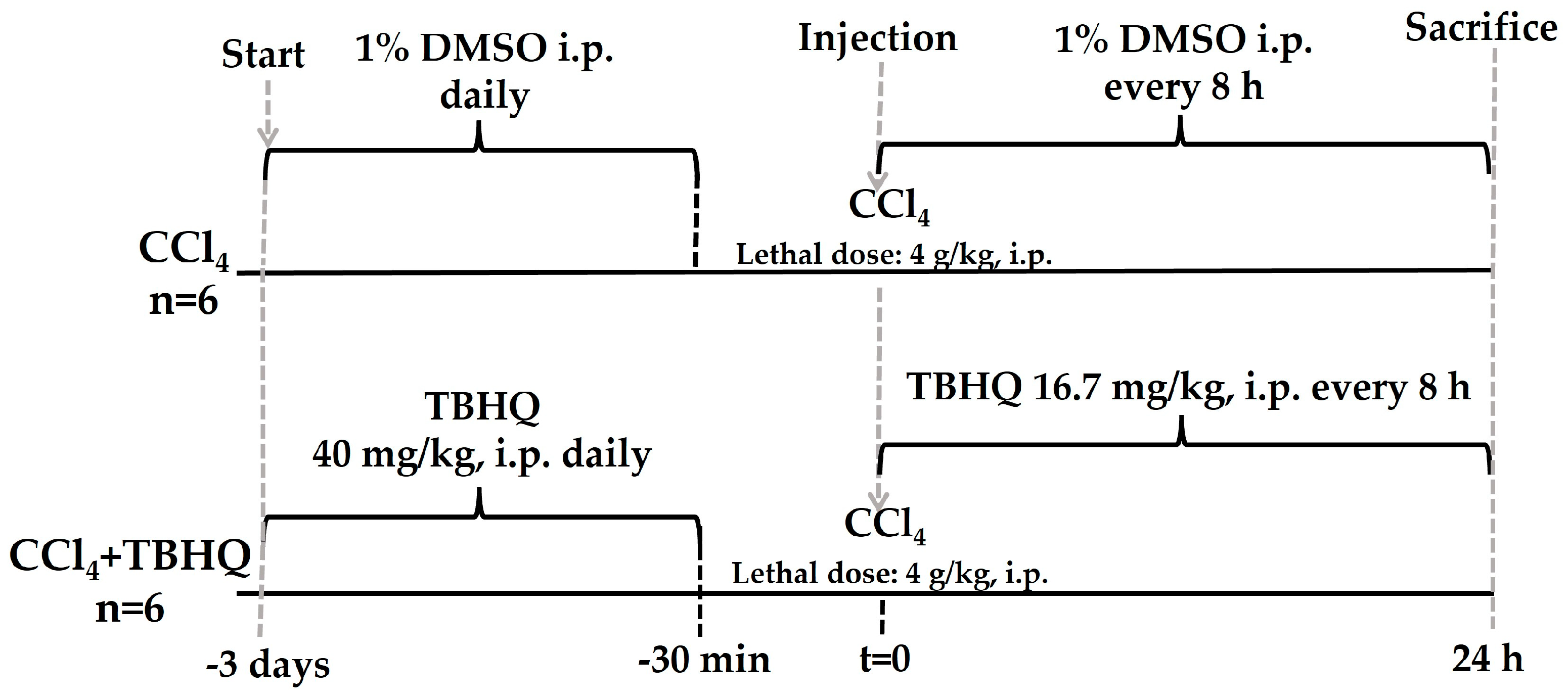
4.3.1. Administration of TBHQ to Rats Intoxicated with CCl4
4.3.2. Mortality Assessment during the Lethal Dose of CCl4
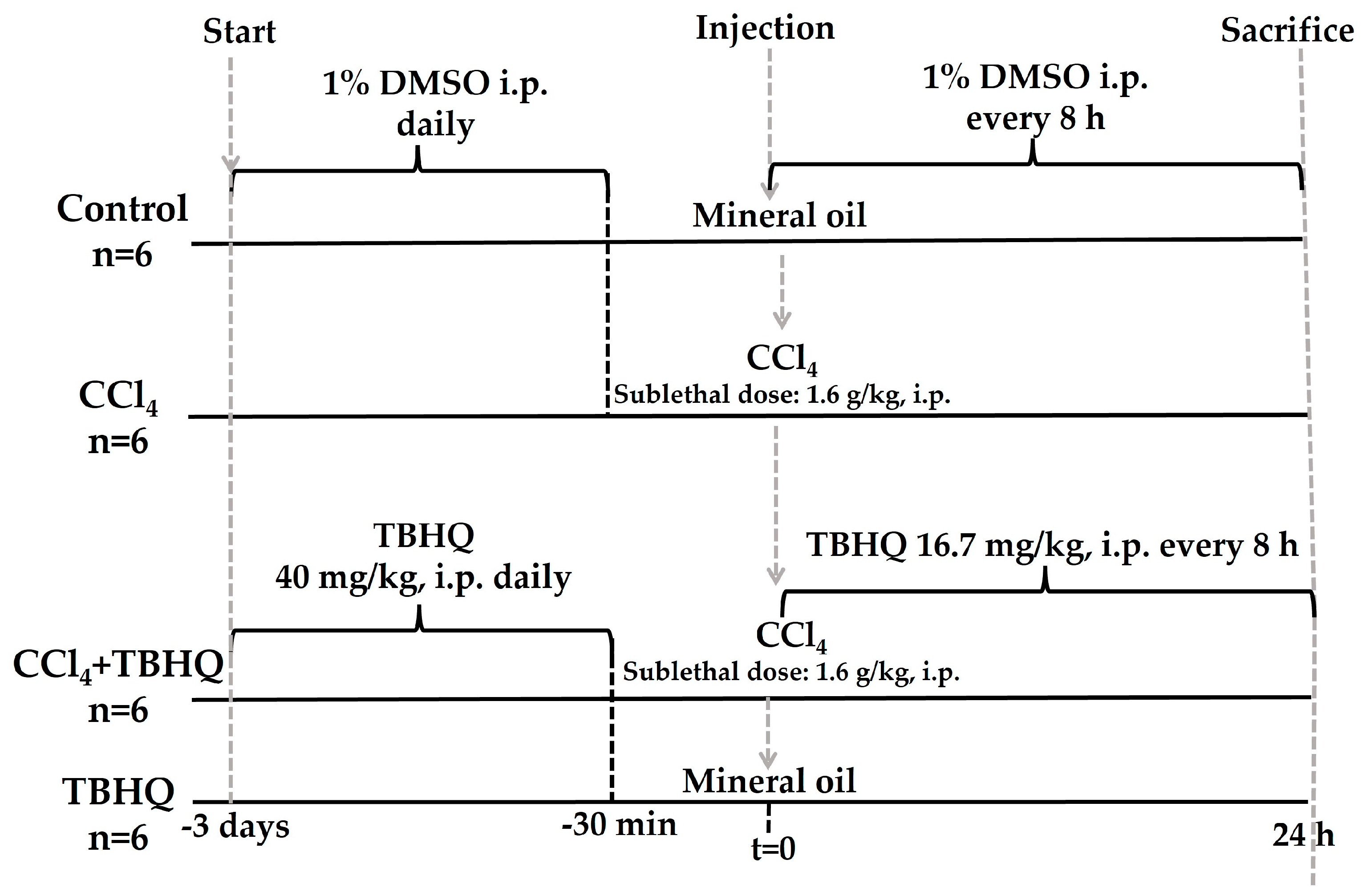
4.3.3. Hepatoprotective Activity of TBHQ during the Sublethal Dose of CCl4
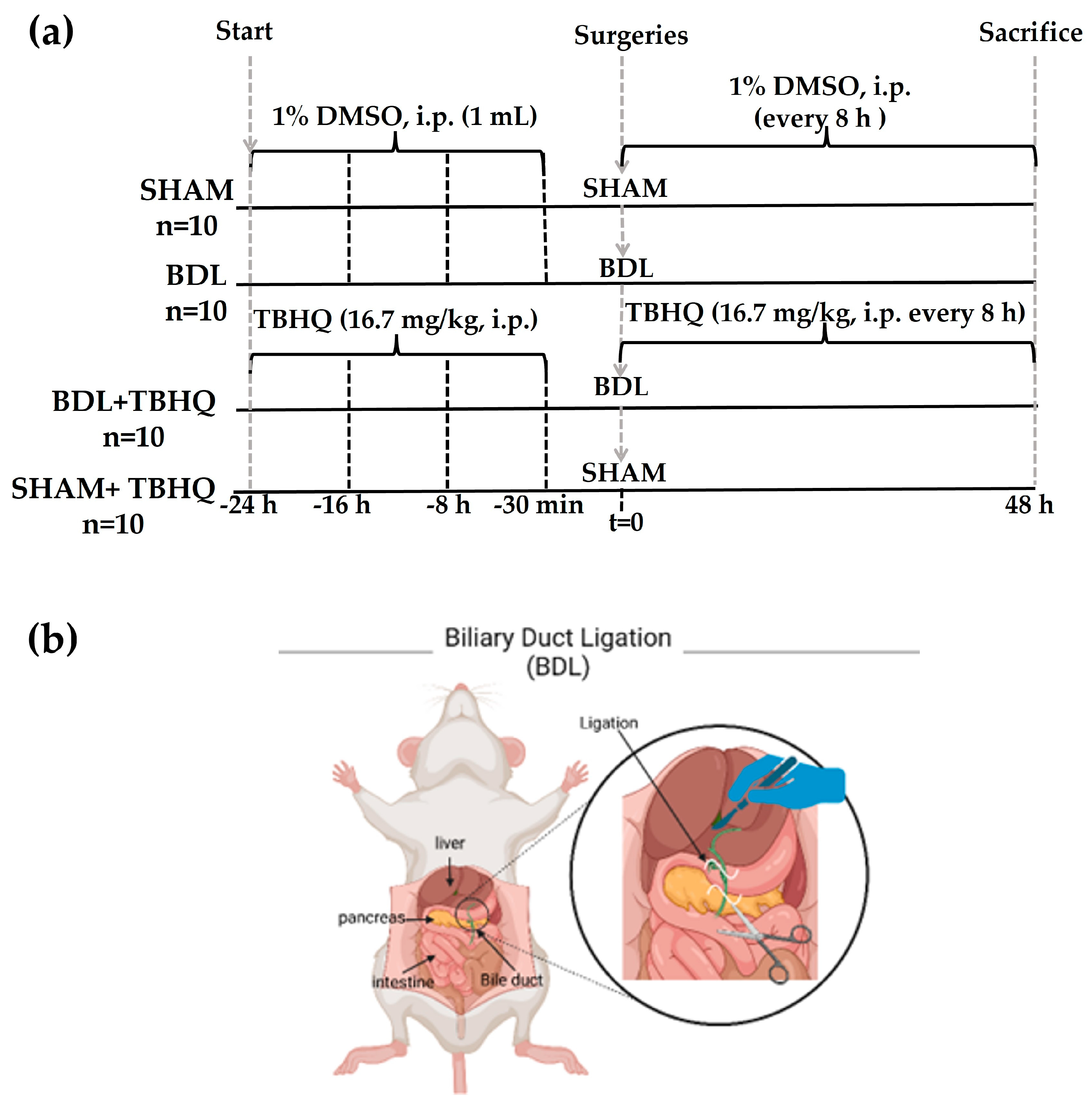
4.4. Experimental Protocol of the BDL Model
4.5. Sacrifice of Experimental Animals
4.6. H&E Staining Procedure
4.7. Photographic Images of H&E Staining
4.8. Evaluation of Serum Biochemical Markers of Liver Damage
4.9. Statistical Analysis
4.10. Identification of Potential Targets for TBHQ Protein in the Liver
4.11. Molecular Docking of TBHQ with Keap1
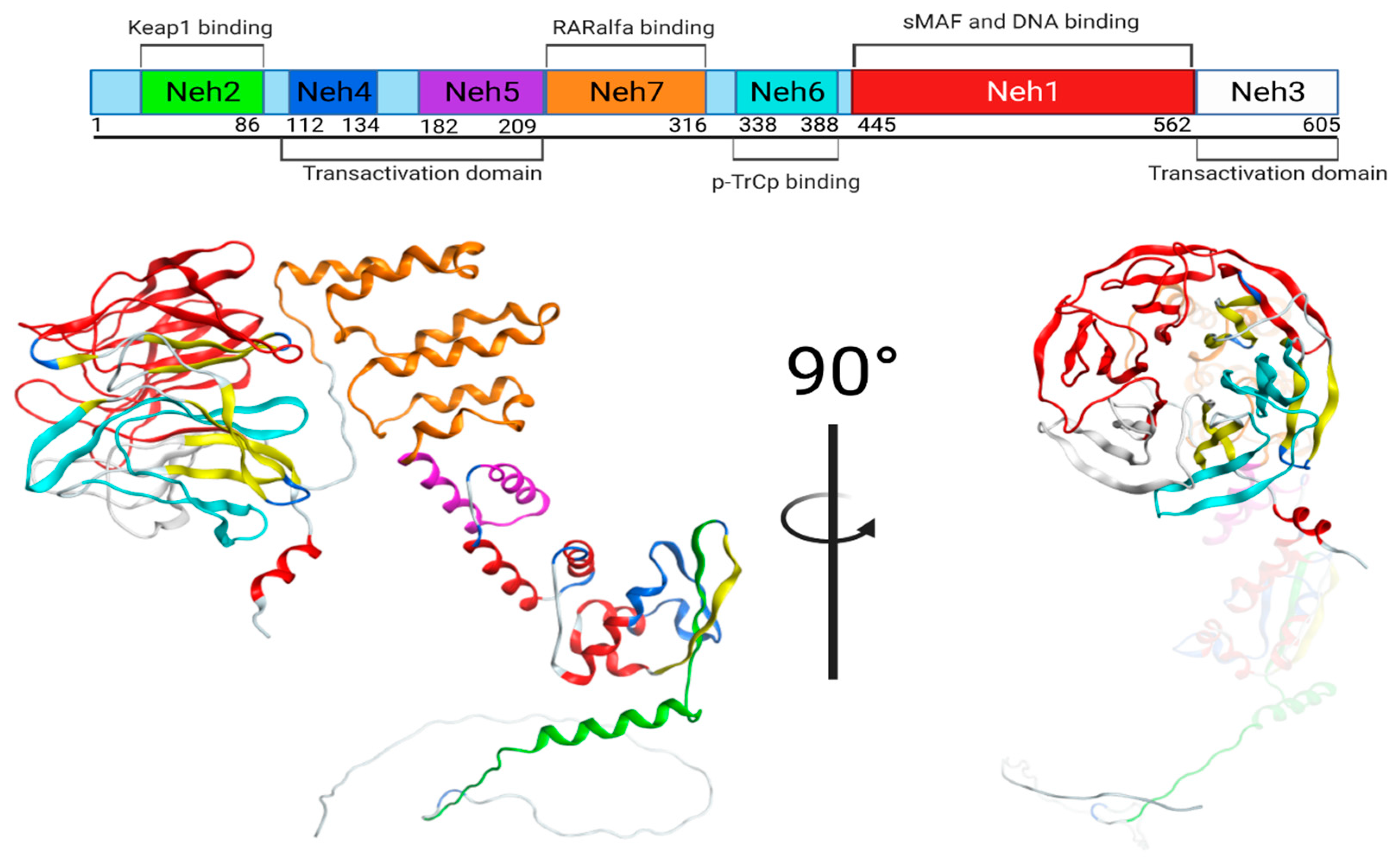
4.11.1. Keap1/Nrf2 Protein
4.11.2. Molecular Docking
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, C.; Ziani, S.; Nelson, L.J.; Ávila, M.A.; Nevzorova, Y.A.; Cubero, F.J. Fibrotic Events in the Progression of Cholestatic Liver Disease. Cells 2021, 10, 1107. [Google Scholar] [CrossRef] [PubMed]
- Shojaie, L.; Iorga, A.; Dara, L. Cell Death in Liver Diseases: A Review. Int. J. Mol. Sci. 2020, 21, 9682. [Google Scholar] [CrossRef] [PubMed]
- Ben-Moshe, S.; Itzkovitz, S. Spatial heterogeneity in the mammalian liver. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 395–410. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.; Regev, A.; Lindor, K.; Avigan, M.I.; Dimick-Santos, L.; Treem, W.; Marcinak, J.F.; Lewis, J.H.; Anania, F.A.; Seekins, D.; et al. Consensus guidelines: Best practices for detection, assessment and management of suspected acute drug-induced liver injury occurring during clinical trials in adults with chronic cholestatic liver disease. Aliment. Pharmacol. Ther. 2020, 51, 90–109. [Google Scholar] [CrossRef] [PubMed]
- Meirelles Júnior, R.F.; Salvalaggio, P.; Rezende, M.B.; Evangelista, A.S.; Guardia, B.D.; Matielo, C.E.; Neves, D.B.; Pandullo, F.L.; Felga, G.E.; Alves, J.A.; et al. Liver transplantation: History, outcomes and perspectives. Einstein 2015, 13, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Ezhilarasan, D. Oxidative stress is bane in chronic liver diseases: Clinical and experimental perspective. Arab. J. Gastroenterol. 2018, 19, 56–64. [Google Scholar] [CrossRef]
- PubChem. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 2004. PubChem Compound Summary for CID 16043, Tert-Butylhydroquinone. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/16043 (accessed on 6 October 2023).
- Farhoosh, R. Initiation and propagation kinetics of inhibited lipid peroxidation. Sci. Rep. 2021, 11, 6864. [Google Scholar] [CrossRef]
- Nna, V.U.; Ujah, G.A.; Suleiman, J.B.; Mohamed, M.; Nwokocha, C.; Akpan, T.J.; Ekuma, H.C.; Fubara, V.V.; Kekung-Asu, C.B.; Osim, E.E. Tert-butylhydroquinone preserve testicular steroidogenesis and spermatogenesis in cisplatin-intoxicated rats by targeting oxidative stress, inflammation and apoptosis. Toxicology 2020, 441, 152528. [Google Scholar] [CrossRef]
- Rahman, Z.; Dwivedi, D.K.; Jena, G.B. Ethanol-induced gastric ulcer in rats and intervention of tert-butylhydroquinone: Involvement of Nrf2/HO-1 signalling pathway. Hum. Exp. Toxicol. 2020, 39, 547–562. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, X.; Shao, X.; Wang, H.; Liu, X.; Ke, X.; Xiong, C.; Wei, L.; Zou, H. tert-Butylhydroquinone Treatment Alleviates Contrast-Induced Nephropathy in Rats by Activating the Nrf2/Sirt3/SOD2 Signaling Pathway. Oxid. Med. Cell. Longev. 2019, 2019, 4657651. [Google Scholar] [CrossRef] [PubMed]
- Gharavi, N.; Haggarty, S.; El-Kadi, A.O. Chemoprotective and carcinogenic effects of tert-butylhydroquinone and its metabolites. Curr. Drug Metab. 2007, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Silva-Islas, C.A.; Maldonado, P.D. Canonical and non-canonical mechanisms of Nrf2 activation. Pharmacol. Res. 2018, 134, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zheng, Q.; Chen, Z. The Nrf2 Pathway in Liver Diseases. Front. Cell Dev. Biol. 2022, 10, 826204. [Google Scholar] [CrossRef] [PubMed]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Nowak, A.J.; Relja, B. The Impact of Acute or Chronic Alcohol Intake on the NF-κB Signaling Pathway in Alcohol-Related Liver Disease. Int. J. Mol. Sci. 2020, 21, 9407. [Google Scholar] [CrossRef]
- Bai, J.; Yu, X.J.; Liu, K.L.; Wang, F.F.; Li, H.B.; Shi, X.L.; Zhang, Y.; Huo, C.J.; Li, X.; Gao, H.L.; et al. Tert-butylhydroquinone attenuates oxidative stress and inflammation in hypothalamic paraventricular nucleus in high salt-induced hypertension. Toxicol. Lett. 2017, 281, 1–9. [Google Scholar] [CrossRef]
- Gong, D.J.; Wang, L.; Yang, Y.Y.; Zhang, J.J.; Liu, X.H. Diabetes aggravates renal ischemia and reperfusion injury in rats by exacerbating oxidative stress, inflammation, and apoptosis. Ren. Fail. 2019, 41, 750–761. [Google Scholar] [CrossRef]
- Veskemaa, L.; Graw, J.A.; Pickerodt, P.A.; Taher, M.; Boemke, W.; González-López, A.; Francis, R.C.E. Tert-butylhydroquinone augments Nrf2-dependent resilience against oxidative stress and improves survival of ventilator-induced lung injury in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 2021, 320, L17–L28. [Google Scholar] [CrossRef]
- Nimiya, Y.; Wang, W.; Du, Z.; Sukamtoh, E.; Zhu, J.; Decker, E.; Zhang, G. Redox modulation of curcumin stability: Redox active antioxidants increase chemical stability of curcumin. Mol. Nutr. Food Res. 2016, 60, 487–494. [Google Scholar] [CrossRef]
- Yueh, M.F.; Tukey, R.H. Nrf2-Keap1 signaling pathway regulates human UGT1A1 expression in vitro and in transgenic UGT1 mice. J. Biol. Chem. 2007, 282, 8749–8758. [Google Scholar] [CrossRef] [PubMed]
- Muriel, P. Some Experimental Models of Liver Damage. In Hepatotoxicity; Sahu, S.C., Ed.; John Wiles & Sons, Ltd.: Hoboken, NJ, USA, 2008; pp. 119–137. [Google Scholar] [CrossRef]
- Tag, C.G.; Sauer-Lehnen, S.; Weiskirchen, S.; Borkham-Kamphorst, E.; Tolba, R.H.; Tacke, F.; Weiskirchen, R. Bile duct ligation in mice: Induction of inflammatory liver injury and fibrosis by obstructive cholestasis. J. Vis. Exp. 2015, 96, 52438. [Google Scholar] [CrossRef]
- Kloek, J.J.; Levi, M.; Heger, M.; van der Loos, C.M.; Gouma, D.J.; van Gulik, T.M. Cholestasis enhances liver ischemia/reperfusion-induced coagulation activation in rats. Hepatol. Res. 2010, 40, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Van Campenhout, S.; Van Vlierberghe, H.; Devisscher, L. Common Bile Duct Ligation as Model for Secondary Biliary Cirrhosis. Methods Mol. Biol. 2019, 1981, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, J.M.; Lee, E.G. A quantitative assessment of the structural changes the rat’s liver following obstruction of the common bile duct. Br. J. Exp. Pathol. 1976, 57, 85–94. [Google Scholar] [PubMed]
- Al Amin, A.S.M.; Menezes, R.G. Carbon Tetrachloride Toxicity. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK562180/ (accessed on 1 March 2023).
- Lackner, C.; Gogg-Kamerer, M.; Zatloukal, K.; Stumptner, C.; Brunt, E.M.; Denk, H. Ballooned hepatocytes in steatohepatitis: The value of keratin immunohistochemistry for diagnosis. J. Hepatol. 2008, 48, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Platt, D.S.; Cockrill, B.L. Liver enlargement and hepatoxicity: An investigation into the effects of several agents on rat liver enzyme activities. Biochem. Pharmacol. 1967, 16, 2257–2270. [Google Scholar] [CrossRef]
- Ahmed, S.M.; Luo, L.; Namani, A.; Wang, X.J.; Tang, X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim. Biophys. 2017, 1863, 585–597. [Google Scholar] [CrossRef]
- Li, R.; Zhang, P.; Li, C.; Yang, W.; Yin, Y.; Tao, K. Tert-butylhydroquinone mitigates Carbon Tetrachloride induced Hepatic Injury in mice. Int. J. Med. Sci. 2020, 17, 2095–2103. [Google Scholar] [CrossRef]
- Zeng, X.P.; Li, X.J.; Zhang, Q.Y.; Liu, Q.W.; Li, L.; Xiong, Y.; He, C.X.; Wang, Y.F.; Ye, Q.F. Tert-Butylhydroquinone Protects Liver Against Ischemia/Reperfusion Injury in Rats Through Nrf2-Activating Anti-Oxidative Activity. Transplant. Proc. 2017, 49, 366–372. [Google Scholar] [CrossRef]
- Khezerlou, A.; Akhlaghi, A.P.; Alizadeh, A.M.; Dehghan, P.; Maleki, P. Alarming impact of the excessive use of tert-butylhydroquinone in food products: A narrative review. Toxicol. Rep. 2022, 9, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Zhang, S.; Chan, J.Y.; Zhang, D.D. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol. Cell. Biol. 2007, 27, 6334–6349. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Li, H.; Liu, Q.; Liu, F.; Tang, L.; Li, C.; Yuan, Y.; Zhan, Y.; Xu, W.; Li, W.; et al. Nuclear factor p65 interacts with Keap1 to repress the Nrf2-ARE pathway. Cell. Signal. 2011, 23, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Wei, H.; Song, Y.; Chen, M.; Fan, Z.; Qiu, R.; Zhu, W.; Xu, W.; Wang, F. NF-κB and Keap1 Interaction Represses Nrf2-Mediated Antioxidant Response in Rabbit Hemorrhagic Disease Virus Infection. J. Virol. 2020, 94, e00016-20. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, T.; Perkins, N.D.; Wilson, C.L. NFKB1: A suppressor of inflammation, ageing and cancer. FEBS J. 2016, 283, 1812–1822. [Google Scholar] [CrossRef]
- Kim, S.M.; Choi, J.E.; Hur, W.; Kim, J.H.; Hong, S.W.; Lee, E.B.; Lee, J.H.; Li, T.Z.; Sung, P.S.; Yoon, S.K. RAR-Related Orphan Receptor Gamma (ROR-γ) Mediates Epithelial-Mesenchymal Transition Of Hepatocytes During Hepatic Fibrosis. J. Cell. Biochem. 2017, 118, 2026–2036. [Google Scholar] [CrossRef]
- Maia, L.; Vala, A.; Mira, L. NADH oxidase activity of rat liver xanthine dehydrogenase and xanthine oxidase-contribution for damage mechanisms. Free Radic. Res. 2005, 39, 979–986. [Google Scholar] [CrossRef]
- Ma, Y.; Lu, L.; Tan, K.; Li, Z.; Guo, T.; Wu, Y.; Wu, W.; Zheng, L.; Fan, F.; Mo, J.; et al. Reduced peroxisome proliferator-activated receptor-α and bile acid nuclear receptor NR1H4/FXR may affect the hepatic immune microenvironment of biliary atresia. Front. Immunol. 2022, 13, 875593. [Google Scholar] [CrossRef]
- Gomez, C.B.; de la Cruz, S.H.; Medina-Terol, G.J.; Beltran-Ornelas, J.H.; Sánchez-López, A.; Silva-Velasco, D.L.; Centurión, D. Chronic administration of NaHS and L-Cysteine restores cardiovascular changes induced by high-fat diet in rats. Eur J Pharmacol. 2019, 863, 172707. [Google Scholar] [CrossRef]
- Norma Oficial Mexicana NOM-062-ZOO-1999: Especificaciones Técnicas para la Producción, Cuidado y Uso de los Animales de Laboratorio. 1999. Available online: https://www.fmvz.unam.mx/fmvz/principal/archivos/062ZOO.PDF (accessed on 27 February 2023).
- Tezuka, M.; Momiyama, K.; Edano, T.; Okada, S. Protective effect of chromium(III) on acute lethal toxicity of carbon tetrachloride in rats and mice. J. Inorg. Biochem. 1991, 42, 1–8. [Google Scholar] [CrossRef]
- Bakdemir, M.; Çetin, E. Hepatoprotective effects of ethyl pyruvate against carbon tetrachloride-induced oxidative stress, biochemical and histological alterations in rats. Arch. Physiol. Biochem. 2021, 127, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; Kongo, M.; Sasaki, E.; Nishida, K.; Ishiguro, I. Therapeutic effect of melatonin on carbon tetrachloride-induced acute liver injury in rats. J. Pineal Res. 2000, 28, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Georgiev, P.; Jochum, W.; Heinrich, S.; Jang, J.H.; Nocito, A.; Dahm, F.; Clavien, P.A. Characterization of time-related changes after experimental bile duct ligation. Br. J. Surg. 2008, 95, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Aragón, P.; Noguera, P.; Bañuls, M.J.; Puchades, R.; Maquieira, Á.; González-Martínez, M.Á. Modulating receptor-ligand binding in biorecognition by setting surface wettability. Anal. Bioanal. Chem. 2018, 410, 5723–5730. [Google Scholar] [CrossRef] [PubMed]
- Reitman, S.; Frankel, S. A colorimetric method for the determination of serum glutamic oxalacetic and glutamic pyruvic transaminases. Am. J. Clin. Pathol. 1957, 28, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Glossmann, H.; Neville, D.M. gamma-Glutamyltransferase in kidney brush border membranes. FEBS Lett. 1972, 19, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Bergmeyer, H.U.; Grassl, M.; Walter, H.E. Methods of Enzymatic Analysis, 3rd ed.; Bergmeyer, H.U.M., Ed.; Verlag Chemie: Deerfield Beach, FL, USA, 1983; Volume 2, pp. 269–270. [Google Scholar]
- Davies, M.; Nowotka, M.; Papadatos, G.; Dedman, N.; Gaulton, A.; Atkinson, F.; Bellis, L.; Overington, J.P. ChEMBL web services: Streamlining access to drug discovery data and utilities. Nucleic Acids Res. 2015, 43, W612–W620. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Liu, X.; Ouyang, S.; Yu, B.; Liu, Y.; Huang, K.; Gong, J.; Zheng, S.; Li, Z.; Li, H.; Jiang, H. PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010, 38, W609–W614. [Google Scholar] [CrossRef]
- Wang, X.; Pan, C.; Gong, J.; Liu, X.; Li, H. Enhancing the Enrichment of Pharmacophore-Based Target Prediction for the Polypharmacological Profiles of Drugs. J. Chem. Inf. Model. 2016, 56, 1175–1183. [Google Scholar] [CrossRef]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef]
- Sheils, T.K.; Mathias, S.L.; Kelleher, K.J.; Siramshetty, V.B.; Nguyen, D.T.; Bologa, C.G.; Jensen, L.J.; Vidović, D.; Koleti, A.; Schürer, S.C.; et al. TCRD and Pharos 2021: Mining the human proteome for disease biology. Nucleic Acids Res. 2021, 49, D1334–D1346. [Google Scholar] [CrossRef] [PubMed]
- Reymond, J.L. PPB Polypharmacology Browser for Target Prediction in ChEMBL Browser. PPB. 13 July 2022. Available online: http://gdbtools.unibe.ch:8080/PPB/contact.html (accessed on 4 February 2023).
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Nickel, J.; Gohlke, B.O.; Erehman, J.; Banerjee, P.; Rong, W.W.; Goede, A.; Dunkel, M.; Preissner, R. SuperPred: Update on drug classification and target prediction. Nucleic Acids Res. 2014, 42, W26–W31. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.J.; Dong, J.; Che, Y.J.; Zhu, M.F.; Wen, M.; Wang, N.N.; Wang, S.; Lu, A.P.; Cao, D.S. TargetNet: A web service for predicting potential drug-target interaction profiling via multi-target SAR models. J. Comput. Aided. Mol. Des. 2016, 30, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Cheminfo.org. OPENBABEL–Chemical File Format Converter. Cheminfo.org. 5 July 2022. Available online: http://www.cheminfo.org/Chemistry/Cheminformatics/FormatConverter/index.html# (accessed on 18 May 2023).
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Lancet Lab at the Weizmann Institute of Science. GeneCards–The Human Gene Database. GeneCardsSuite. 14 July 2022. Available online: https://www.genecards.org/ (accessed on 18 May 2023).
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases, 1st ed.; Abugessaisa, I., Kasukawa, T., Eds.; Springer: Singapore, 2021; pp. 27–56. [Google Scholar]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 10 June 2023).
- Winkel, A.F.; Engel, C.K.; Margerie, D.; Kannt, A.; Szillat, H.; Glombik, H.; Kallus, C.; Ruf, S.; Güssregen, S.; Riedel, J.; et al. Characterization of RA839, a Noncovalent Small Molecule Binder to Keap1 and Selective Activator of Nrf2 Signaling. J. Biol. Chem. 2015, 290, 28446–28455. [Google Scholar] [CrossRef]
- Unni, S.; Deshmukh, P.; Krishnappa, G.; Kommu, P.; Padmanabhan, B. Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1. FEBS J. 2021, 288, 1599–1613. [Google Scholar] [CrossRef]
- Ortet, P.C.; Muellers, S.N.; Viarengo-Baker, L.A.; Streu, K.; Szymczyna, B.R.; Beeler, A.B.; Allen, K.N.; Whitty, A. Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning. J. Am. Chem. Soc. 2021, 143, 3779–3793. [Google Scholar] [CrossRef]
- Edelsbrunner, H. Weighted Alpha Shapes; Technical Paper of the Department of Computer Science of the University of Illinois at Urbana-Champaign; Urbana, Illinois 61810. Available online: https://pub.ista.ac.at/~edels/Papers/1992-R-08-WeightedAlphaShapes.pdf (accessed on 27 June 2023).
- Del Carpio, C.A.; Takahashi, Y.; Sasaki, S. A new approach to the automatic identification of candidates for ligand receptor sites in proteins: (I). Search for pocket regions. J. Mol. Graph. 1993, 11, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Edelsbrunner, H.; Facello, M.; Fu, P.; Liang, J. Measuring Proteins and Voids in Proteins. In Proceedings of the 28th Hawaii International Conference on Systems Science, Wailea, HI, USA, 3–6 January 1995; Volume 5, pp. 256–264. [Google Scholar] [CrossRef]
- Goodford, P.J. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 1985, 28, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Hendlich, M.; Rippmann, F.; Barnickel, G. LIGSITE: Automatic and efficient detection of potential small molecule-binding sites in proteins. J. Mol. Graph. Model. 1997, 15, 359–389. [Google Scholar] [CrossRef] [PubMed]
- Le Guilloux, V.; Schmidtke, P.; Tuffery, P. Fpocket: An open source platform for ligand pocket detection. BMC Bioinform. 2009, 10, 168. [Google Scholar] [CrossRef]
- Miranker, A.; Karplus, M. Functionality maps of binding sites: A multiple copy simultaneous search method. Proteins 1991, 11, 29–34. [Google Scholar] [CrossRef]
- Soga, S.; Shirai, H.; Kobori, M.; Hirayama, N. Use of amino acid composition to predict ligand-binding sites. J. Chem. Inf. Model. 2007, 47, 400–406. [Google Scholar] [CrossRef]
- Volkamer, A.; Griewel, A.; Grombacher, T.; Rarey, M. Analyzing the topology of active sites: On the prediction of pockets and subpockets. J. Chem. Inf. Model. 2010, 50, 2041–2052. [Google Scholar] [CrossRef]

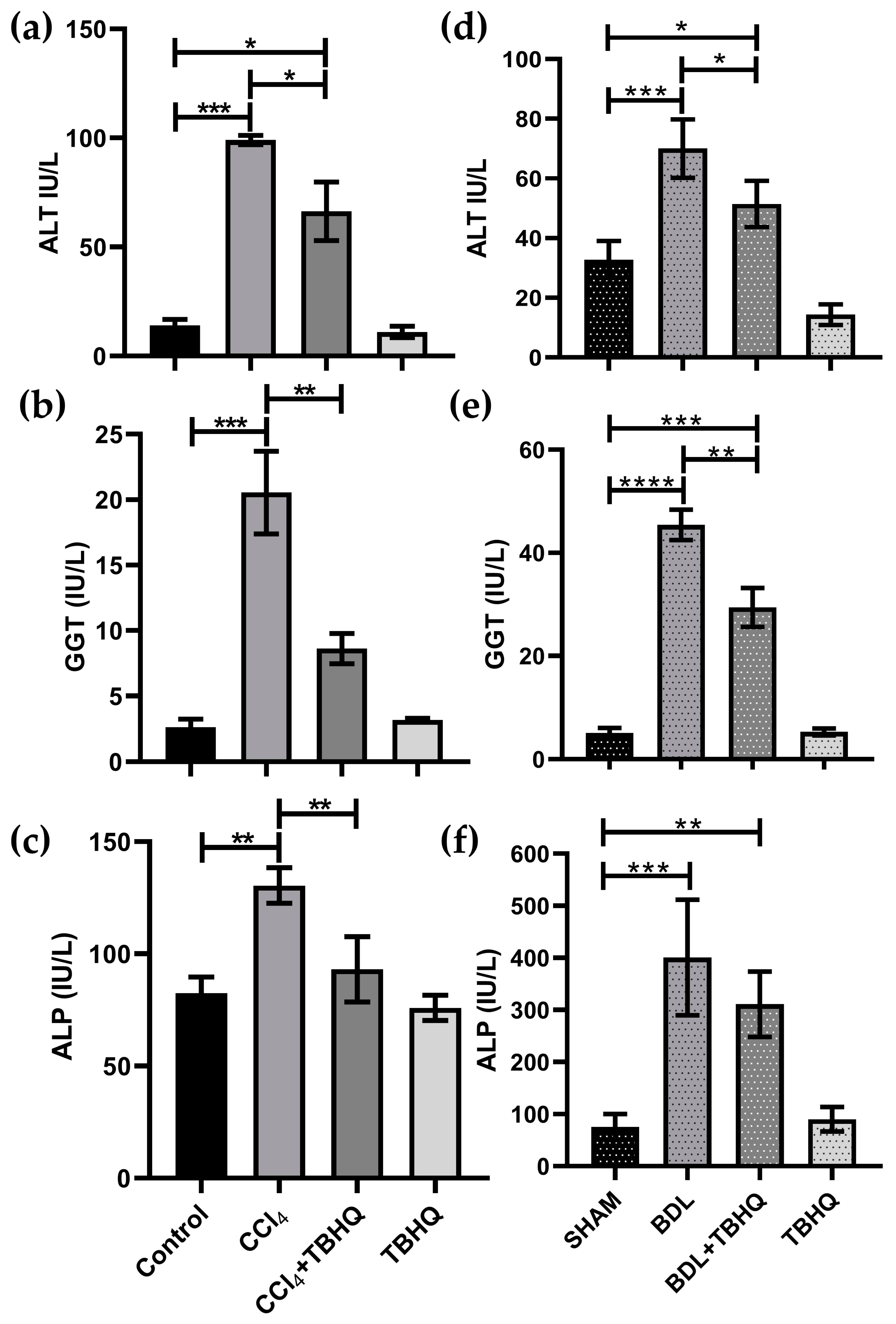

| Liver Damage Model | Mortality | ||
|---|---|---|---|
| (n) | % | Survival % | |
| Lethal dose of CCl4 | |||
| CCl4 (4 g/kg, i.p.) | 6/6 | 100 | 0 |
| CCl4 (4 g/kg, i.p.) + TBHQ | 3/6 | 50 | 50 |
| Sublethal dose of CCl4 | |||
| CCl4 (1.6 g/kg, i.p.) | 0/6 | 0 | 100 |
| CCl4 (1.6 g/kg, i.p.) + TBHQ | 0/6 | 0 | 100 |
| BDL experimental model | |||
| BDL | 5/10 | 50 | 50 |
| BDL+TBHQ | 5/10 | 20 | 80 |
| Group | Body Weight (g) | Liver Weight (g) | Liver/Body Weight |
|---|---|---|---|
| Control | 348.3 ± 4.70 | 11.5 ± 0.65 | 0.033 ± 0.00169 |
| CCl4 | 350.8 ± 12.98 | 16.2 ± 0.67 a | 0.046 ± 0.00194 ac |
| CCl4+TBHQ | 302.9 ± 8.26 ab | 15.21 ± 0.76 a | 0.050 ± 0.00178 ac |
| TBHQ | 370.3 ± 9.53 | 13.92 ± 0.37 | 0.038 ± 0.00010 |
| SHAM | 411.6 ± 3.32 | 16.23 ± 0.061 | 0.039 ± 0.00179 |
| BDL | 373.38 ± 16.55 | 15.36 ± 0.76 | 0.040 ± 0.00323 |
| BDL+TBHQ | 366.9 ± 11.62 | 14.48 ± 0.13 | 0.039 ± 0.00143 |
| SHAM+TBHQ | 335.93 ± 12.43 | 14.6 ± 0.49 | 0.043 ± 0.00056 |
| Target Protein Name | Protein ID | Target Location and/or Function (Human Protein Atlas) | Target Search Platform |
|---|---|---|---|
| Nuclear factor erythroid 2-related factor | NFE2L2 (Nrf2) | Antioxidant Liver—Metabolism | P |
| Nuclear factor-kappa B p105 subunit | NFKB1 | NF-KB subunit | SP |
| Transcription factor p65 | RELA | NF-KB subunit | TN |
| Acetyl-CoA carboxylase 2 | ACACB | Cell type enhanced—Hepatocytes | SP |
| Estrogen receptor | ESR1 | Cell type enhanced—Hepatocytes | PM, S, TN |
| Endoplasmic reticulum-associated amyloid beta-peptide-binding protein | HSD17B10 | Cell type enhanced—Hepatocytes | P, SP |
| Dihydropteridine reductase | QDPR | Cell type enhanced—Hepatocytes | S |
| Nuclear receptor ROR gamma | RORC | Cell type enhanced—Hepatocytes | P |
| Excitatory amino acid transporter 3 | SLC1A1 | Cell type enhanced—Hepatocytes | SP |
| Transthyretin | TTR | Cell type enriched—Hepatocytes | PM, SP |
| Tyrosine-protein kinase receptor RET | NRTN | Cell type enhanced—Hepatocytes | SP |
| Oxysterols receptor LXR-alpha | NR1H3 | Cell type enhanced—Hepatocytes | TN |
| Glyceraldehyde-3-phosphate dehydrogenase, liver * | GAPDH | Tissue enhanced—Liver | ST |
| Hydroxysteroid 17-beta-dehydrogenase 3 | HSD17B3 | Tissue enhanced—Liver | TN |
| Pyruvate dehydrogenase kinase 4 | PDK4 | Cell type enriched—Liver–Hepatocytes | S |
| Aryl hydrocarbon receptor | AHR | Liver—Metabolism | TN |
| Transient receptor potential cation channel subfamily V member 1 | TRPV1 | Liver—Metabolism | TN |
| Aldo-keto reductase family 1 member C3 | AKR1C3 | Liver—Lipid metabolism | PM |
| Monoamine oxidase A | MAOA | Liver—Lipid metabolism | SP, TN |
| Androgen receptor | AR | Liver—Metabolism Cell type enhanced—Hepatocytes | PM, TN |
| Coagulation factor X | F10 | Liver—Metabolism Cell type enhanced—Hepatocytes | PM |
| Monoamine oxidase B | MAOB | Liver—Metabolism Cell type enhanced—Hepatocytes | TN |
| Bile acid receptor | NR1H4 | Liver—Metabolism Cell type enhanced—Hepatocytes | S |
| Cholinergic receptor nicotinic alpha 4 subunit (neuronal acetylcholine receptor) | CHRNA4 | Liver—Metabolism Cell type enhanced—Hepatocytes | SP |
| Cocaine esterase | CES2 | Liver—Lipid metabolism Cell type enhanced—Hepatocytes | TN |
| Nuclear receptor subfamily 1 group I member 2 (pregnane X receptor) | NR1I2 | Liver—Lipid metabolism Cell type enhanced—Hepatocytes | SP, PM |
| Estrogen sulfotransferase | SULT1E1 | Liver—Lipid metabolism Cell type enhanced—Hepatocytes | PM |
| Xanthine dehydrogenase | XDH | Liver—Lipid metabolism Cell type enhanced—Hepatocytes | Ph, SP, TN |
| Serum albumin | ALB | Tissue enriched—Liver Liver—Hemostasis Cell type enriched—Hepatocytes | PM, ST |
| Complement factor B | CFB | Tissue enriched—Liver Liver—Hemostasis Cell type enhanced—Hepatocytes | PM |
| Prothrombin | F2 | Liver—Hemostasis Cell type enriched—Hepatocytes | PM |
| Hydroxysteroid 11-beta dehydrogenase 1 | HSD11B1 | Liver—Hemostasis and lipid metabolism Cell type enriched—Hepatocytes | SP, TN |
| Butyrylcholinesterase | BCHE | Liver—Hemostasis and lipid metabolism Cell type enhanced—Hepatocytes | Ch |
| Carbonic anhydrase 5A, mitochondrial | CA5A | Liver—Hemostasis and lipid metabolism Cell type enriched—Hepatocytes | TN |
| Liver carboxylesterase 1 * | CES1 | Liver—Hemostasis and lipid metabolism Cell type enriched—Hepatocytes | TN |
| Cytochrome P450 1A2 | CYP1A2 | Liver—Hemostasis and lipid metabolism Cell type enriched—Hepatocytes | TN |
| Cytochrome P450 2C9 | CYP2C9 | Liver—Hemostasis and lipid metabolism Cell type enriched—Hepatocytes | P, TN |
| IgG receptor FcRn large subunit p51 | FCGRT | Liver—Lipid metabolism Cell type enhanced—Kupffer cells | SP |
| Arachidonate 5-lipoxygenase | ALOX5 | Cell type enhanced—Kupffer cells | SP, ST, TN |
| C-C chemokine receptor type 2 | CCR2 | Cell type enhanced—Kupffer cells | SP |
| Tyrosine-protein kinase FGR | FGR | Cell type enhanced—Kupffer cells | SP |
| Formyl peptide receptor 1 | FPR1 | Cell type enhanced—Kupffer cells | SP |
| Lipoxin A4 receptor | FPR2 | Cell type enhanced—Kupffer cells | SP |
| G-protein coupled bile acid receptor 1 | GPBAR1 | Cell type enhanced—Kupffer cells | SP |
| Tyrosine-protein kinase SYK | SYK | Cell type enhanced—Kupffer cells | PM |
| Hydroxysteroid17-beta-dehydrogenase 2 | HSD17B2 | Liver—ER transport pathway Cell type enhanced—Hepatocytes | S |
| Endoplasmic reticulum aminopeptidase 1 | ERAP1 | Liver—ER transport pathway | SP |
| Tissue factor pathway inhibitor | TFPI | Tissue enhanced liver Liver—ER transport pathway | SP |
| Tyrosine-protein kinase YES | YES1 | Liver—ER transport pathway | SP |
| Site | Size | Hyd | Side | Residues |
|---|---|---|---|---|
| Approximately the highest propensity for ligand binding | 192 alpha spheres (with the size of a carbon atom sphere) | 42 hydrophobic contact atoms in the receptor | 64 side-chain contact atoms in the receptor | TYR334 SER363 GLY364 LEU365 ALA366 GLY367 CYS368 ASN382 ASN414 ARG415 ILE416 GLY417 VAL418 GLY419 VAL420 GLY462 VAL463 GLY464 VAL465 ALA466 VAL467 SER508 GLY509 ALA510 GLY511 VAL512 CYS513 VAL514 TYR525 GLN530 SER555 ALA556 LEU557 GLY558 ILE559 THR560 VAL561 TYR572 PHE577 SER602 GLY603 VAL604 GLY605 VAL606 ALA607 VAL608 |
| Ligand | Nef2/Nrf2 | MEF | RA839 | TBHQ |
|---|---|---|---|---|
| Energy (kcal/mol) | −10.4673 | −5.2186 | −6.9461 | −5.5491 |
| Domain | Name | Residues/Amino Acids |
|---|---|---|
| Neh 1 | sMAF and DNA binding | 435–562 |
| Neh 2 | Keap1 binding | 50–86 |
| Neh 3 | Transactivation domain | 563–605 |
| Neh 4 | Transactivation domain | 112–134 |
| Neh 5 | Transactivation domain | 182–209 |
| Neh 6 | p-TrCp binding | 338–388 |
| Neh 7 | RARalfa binding | 210–316 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aldaba-Muruato, L.R.; Sánchez-Barbosa, S.; Rodríguez-Purata, V.H.; Cabrera-Cruz, G.; Rosales-Domínguez, E.; Martínez-Valentín, D.; Alarcón-López, Y.A.; Aguirre-Vidal, P.; Hernández-Serda, M.A.; Cárdenas-Granados, L.A.; et al. In Vivo and In Silico Studies of the Hepatoprotective Activity of Tert-Butylhydroquinone. Int. J. Mol. Sci. 2024, 25, 475. https://doi.org/10.3390/ijms25010475
Aldaba-Muruato LR, Sánchez-Barbosa S, Rodríguez-Purata VH, Cabrera-Cruz G, Rosales-Domínguez E, Martínez-Valentín D, Alarcón-López YA, Aguirre-Vidal P, Hernández-Serda MA, Cárdenas-Granados LA, et al. In Vivo and In Silico Studies of the Hepatoprotective Activity of Tert-Butylhydroquinone. International Journal of Molecular Sciences. 2024; 25(1):475. https://doi.org/10.3390/ijms25010475
Chicago/Turabian StyleAldaba-Muruato, Liseth Rubi, Sandra Sánchez-Barbosa, Víctor Hugo Rodríguez-Purata, Georgina Cabrera-Cruz, Estefany Rosales-Domínguez, Daniela Martínez-Valentín, Yoshio Aldo Alarcón-López, Pablo Aguirre-Vidal, Manuel Alejandro Hernández-Serda, Luis Alfonso Cárdenas-Granados, and et al. 2024. "In Vivo and In Silico Studies of the Hepatoprotective Activity of Tert-Butylhydroquinone" International Journal of Molecular Sciences 25, no. 1: 475. https://doi.org/10.3390/ijms25010475
APA StyleAldaba-Muruato, L. R., Sánchez-Barbosa, S., Rodríguez-Purata, V. H., Cabrera-Cruz, G., Rosales-Domínguez, E., Martínez-Valentín, D., Alarcón-López, Y. A., Aguirre-Vidal, P., Hernández-Serda, M. A., Cárdenas-Granados, L. A., Vázquez-Valadez, V. H., Angeles, E., & Macías-Pérez, J. R. (2024). In Vivo and In Silico Studies of the Hepatoprotective Activity of Tert-Butylhydroquinone. International Journal of Molecular Sciences, 25(1), 475. https://doi.org/10.3390/ijms25010475








