Abstract
The Helios protein (encoded by the IKZF2 gene) is a member of the Ikaros transcription family and it has recently been proposed as a promising biomarker for systemic lupus erythematosus (SLE) disease progression in both mouse models and patients. Helios is beginning to be studied extensively for its influence on the T regulatory (Treg) compartment, both CD4+ Tregs and KIR+/Ly49+ CD8+ Tregs, with alterations to the number and function of these cells correlated to the autoimmune phenomenon. This review analyzes the most recent research on Helios expression in relation to the main immune cell populations and its role in SLE immune homeostasis, specifically focusing on the interaction between T cells and tolerogenic dendritic cells (tolDCs). This information could be potentially useful in the design of new therapies, with a particular focus on transfer therapies using immunosuppressive cells. Finally, we will discuss the possibility of using nanotechnology for magnetic targeting to overcome some of the obstacles related to these therapeutic approaches.
1. General Considerations on the Immune System
The immune system of complex organisms like mammals is a coordinated network of innate and adaptive cells, molecules, tissues, and organs that has evolved to fight against the foreign pathogens causing infectious disease. In parallel, the immune system also has the means to protect host self-antigens, part of a dynamic homeostatic equilibrium aimed at preventing any potential side effects due to an overzealous immunogenic response.
Unlike innate cells, one of the essential features of adaptive cells is their ability to recognize a wide range of different antigens (endogenous and exogenous). Adaptive immunity is driven by cellular (helper or CD4+ T cells, and cytotoxic or CD8+ T cells) and humoral immunity (via B cells). From a functional and perhaps simplified perspective, each immune response (innate and adaptive) can be classified as immunogenic (e.g., the immune response directed against pathogenic microorganisms) or tolerogenic (for example, the immunosuppression of autoreactive processes). Overall, the immune system aims to establish a dynamic and continuously regulated balance between these two opposing forces: reactivity to foreign molecules and self-antigen tolerance [1,2]. Interestingly, the same cell type may be involved in both activities, depending on factors like the molecular environment, their activation, and the patterns of gene expression [3]. Most of the known innate and adaptive immune cell populations are presented in Figure 1, showing their immunogenic and tolerogenic counterparts. Finding elements common to both sides of the balance, and that influence the general status of the immune system, will be interesting to better understand pathologies with a predominantly immune component, such as cancer and autoimmunity. In this review, we will discuss the possibility of considering the Helios transcription factor (encoded by the IKZF2 gene) as a potential biomarker and therapeutic target in systemic lupus erythematosus (SLE), a relatively common autoimmune disease.
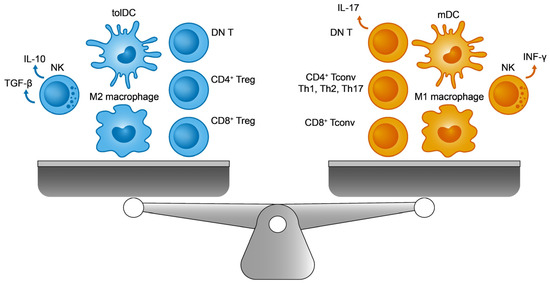
Figure 1.
The immune balance. On the (left), the immunosuppressive cells include M2 macrophages, regulatory T cells (Tregs), tolerogenic dendritic cells (tolDCs), double negative T cells (DN T), and TGF-β producing natural killer (NK) cells. On the (right), their immunogenic counterparts are M1 macrophages, conventional T lymphocytes (Tconv), immunogenic mature DCs (mDCs), IL-17 producing DN T cells, and INF-γ producing NK cells.
1.1. The Adaptive Immune Equilibrium
In addition to T cells, dendritic cells (DCs) play a crucial role in lymphoid organs (thymus, lymph nodes, and spleen), orchestrating the adaptive immune system. In this sense, here we will review the role and identity of the different tolerogenic and immunogenic immune subpopulations, focusing on T cell and DC repertoires.
1.1.1. Conventional CD4+ and CD8+ T Cells
After their generation in bone marrow from hematopoietic progenitors, in both humans and mice, immature T cells move to the thymus to complete their development. Once there, and during their maturation, thymocytes reorganize their TCR by genetic recombination to generate a specific repertoire of immune cells with distinct affinities for the MHC molecules associated to their peptides. Subsequently, after positive and negative selection, the remaining fraction of TCRαβ+CD4+CD8+ cells silence one of their co-receptors, CD8 or CD4 depending on the interaction with antigen presenting cells (APCs), through their MHCI or MHCII molecules, respectively [4,5]. Thus, naive TCRαβ+CD4+CD8− (restricted MHCII cells) and TCRαβ+CD4−CD8+ (restricted MHCI cells) T lymphocytes arise in the thymus, exhibiting tolerance for self-antigens towards peripheral lymphoid tissues to be activated by APCs (mainly, DCs), and they then trigger the cellular adaptive response. During this process, T cells undergo some notorious changes in the expression of several well-known immune markers, from naïve to effector, and to central or effector memory, which can easily be analyzed by flow cytometry (Table 1) [6,7].

Table 1.
Main human and mouse markers for different T cell subsets in terms of the activation of memory status.
Globally, these mature T lymphocytes are defined as conventional T cells (Tconv), and they are responsible for the positive and immunogenic responses to foreign antigens. Yet what are the homeostatic mechanisms that avoid the potentially autoreactive reactions of Tconv cells that escape the process of negative selection in the thymus? Other subpopulations of immune cells play a pivotal role in readdressing the immune equilibrium through immunosuppression, cells known as T regulatory cells (Tregs). Among these cells, most research has focused on CD4+ Tregs, and in particular, on a CD4+ Treg population derived from the thymus (CD4+ tTreg). These cells survive the process of negative selection and they exhibit high affinity for MHCII-self-peptide complexes, expressing higher levels of CD25 and FoxP3 [8]. Nevertheless, incipient CD8+ Tregs that express the receptors of natural killer (NK) cells are currently generating much interest [9].
1.1.2. Regulatory CD4+ T Cells
The discovery of the FoxP3 transcription factor as a marker and an essential regulator that maintains CD4+ tTreg’s immunosuppressive activity implied a paradigm shift in the field of immunology [10,11]. After years of intense research in this topic, a CD4+ tTreg population (CD25+ FoxP3+) has been clearly shown to maintain the peripheral immune equilibrium by controlling aberrant and exacerbated autoreactive responses against self-antigens, microorganisms, and environmental antigens [12,13]. In this regard, several allergies and autoimmune pathologies are associated with certain alterations to CD4+ tTregs, revealing the true importance of their correct function [14]. In addition, some roles of CD4+ tTregs in other body systems beyond the immune system have also been reported. For example, the close links between these lymphocytes and tissue regeneration [15], or other metabolic diseases like obesity [16], have been studied.
Although CD4+ tTreg lymphocytes represent the most abundant fraction of Tregs in the periphery, there are also other regulatory CD4+ cell types of different origins that reliably express FoxP3 and that participate in establishing tolerance. For example, peripheral CD4+ Tregs (CD4+ pTregs) are generated from CD4+ Tconvs outside the thymus in tolerogenic contexts, such as in the intestinal mucosa. Moreover, CD4+ iTregs can be induced in vitro from CD4+ Tconvs under specific culture conditions and these cells are currently being tested in clinical trials as potential autologous cell therapy for patients affected by several autoimmune diseases [17]. Despite their differences, these CD4+ tTregs, pTregs, and iTregs employ some common immunosuppressive mechanisms.
Given the intrinsic molecular and cellular complexity of the immune system, CD4+ Treg lymphocytes need to act on several elements and processes, of both the innate and adaptive responses, in order to ensure effective peripheral tolerance in a coordinated manner [18,19,20] (see Figure 2 for a summary of the main immunosuppressive strategies identified in CD4+ Tregs). Among these events, the overexpression of CD25 (the α subunit of the interleukin 2 receptor or IL-2R) facilitates the capture and sequestering of the available IL-2. The reduced availability of IL-2 specifically dampens the activation and proliferation of CD8+ Tconv cells that depends on this cytokine [21]. From a metabolic perspective, the surface ectoenzymes CD39 and CD73 transform extracellular ATP into adenosine molecules, the latter potentially inhibiting effector T and myeloid cells (DCs and macrophages) [22,23]. Furthermore, these Tregs produce tolerogenic cytokines like IL-10 and TGF-β that exert multiple effects on immune cells. By contrast, IL-10 and TGF-β inhibit T and B lymphocyte proliferation and stimulation [24]. Moreover, these cytokines would polarize DCs towards a more tolerogenic phenotype, which promotes the induction of pTregs [25]. Finally, Tregs and DCs interact directly through two different signaling systems that could induce a tolerogenic state in DCs: PD-1 (Treg)—PDL-1 (DCs) [26], and CTLA-4 (Treg)—CD80/86 (DCs) [27].
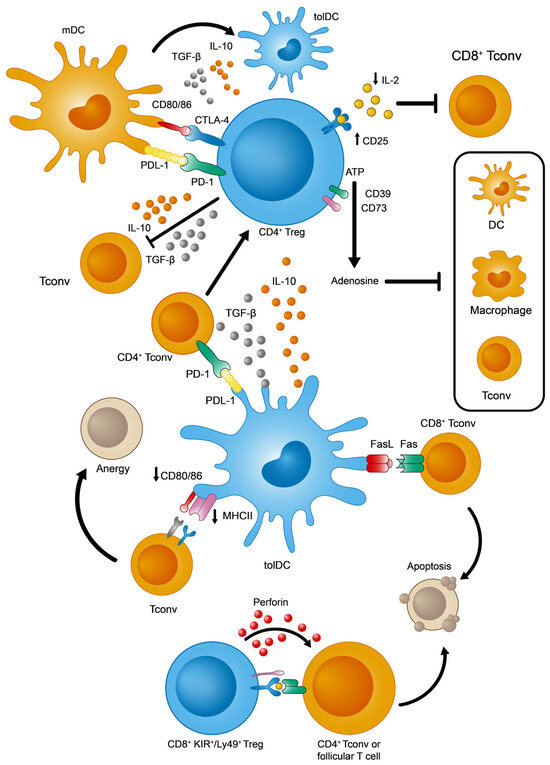
Figure 2.
Main immunosuppressive mechanisms adopted by tolDCs, CD4+, and KIR+/Ly49+ CD8+ Tregs. Adapted from [24,28,29].
1.1.3. Regulatory CD8+ Tregs
Globally, although most biomedical research into Treg cells has focused on the CD4+ Treg compartment, a recently described subset of CD8+ Treg lymphocytes that expresses inhibitory markers from NK cells is being investigated intensively in mice and humans. Previously, a wide range of different CD8+ T cells exhibiting immunomodulatory properties were classified as CD8+ Tregs, yet with heterogeneous phenotypes: CD8+ FoxP3+ [30], CD8+ CD28− [31], CD8+ CD103+ [32], and CD8+ CD122+ CD49dlow [33]. However, unlike CD4+ tTreg cells, these populations were found peripherally in very small proportions or in experimental contexts of antigenic exposure in mice. In other words, these subsets were never found naturally in significant numbers in healthy and young mice without prior manipulation [34,35].
This situation changed with the discovery of a new regulatory subpopulation of CD8+ T cells in naïve mice without any immune disruption, reliably defined by the expression of three markers: CD44+, CD122+, and Ly49+ (Ly49+ CD8+ Tregs from here on). This population has the ability to modulate the adaptive immune response by eliminating autoreactive CD4+ Tconv and follicular T lymphocytes [36,37,38]. Since then, other properties of this population have been clarified, such as (1) the absence of intranuclear FoxP3 expression; (2) high levels of surface CD127 (in contrast to CD4+ Treg cells) [38]; (3) homeostatic regulation, mainly through IL-15 and TGF-β [39,40,41]; (4) phenotypic acquisition induced peripherally after thymic maturation [42]; and (5) finally, the fact that these cells have a non-redundant role in controlling autoreactive antibody titers by acting in germinal centers from lymphoid organs [37,41], thereby regulating autoimmune phenomena [40,41,43]. In terms of their functionality, the best-defined effector mechanism used by CD8+ Ly49+ Tregs is the cytotoxic killing of autoreactive CD4+ T cells, regardless of IL-10 production [36] (Figure 2).
1.1.4. Tolerogenic Dendritic Cells (tolDCs)
As previously stated, DCs are the primary APCs that initiate the adaptive immune responses of T lymphocytes in response to foreign antigens in the periphery. However, in addition to this crucial role, they also contribute to immune tolerance in the organism in two other ways. Firstly, DCs mediate the negative selection and differentiation of tTregs in the thymus (central tolerance), while they also maintain peripheral tolerance by elimination, anergic induction, or conversion of autoreactive T cells to pTregs [44]. While immature DCs (imDCs) were simply believed to perform a tolerogenic function and mature DCs (mDCs) an immunogenic function, increasing experimental evidence suggests that this functional divide extends beyond the maturation state of DCs. In fact, maturation is necessary for their optimal tolerogenic function, even under basal conditions with no exogenous stimulation. Thus, we will refer to them functionally as imDCs, mDCs, and tolerogenic mature DCs (tolDCs) [45,46].
Apart from these states of maturation, distinct DC subpopulations fulfill specific roles in the immune response. These subpopulations are well-defined in both mouse models and humans, and include conventional (cDCs) and plasmacytoid dendritic cells (pDCs) [47] (Table 2). Type 1 (cDC1s) and type 2 (cDC2s) DCs are defined by their developmental transcriptional program, presenting MHCI antigens for CD8+ T cells or MHCII for CD4+ T cells, respectively. Furthermore, pDCs specialize in the production of type I interferons upon exposure to viral antigens [48], as well as helping to establish immune tolerance [49]. In line with this, it seems that cDC1 cells, with their inherent capacity to present antigens via MHCI molecules, also play a crucial role in ensuring peripheral tolerance to self-antigens [44,46,50,51,52,53].

Table 2.
Main human and mouse markers for plasmacytoid (pDC), conventional type 1 (cDC1), and conventional type 2 (cDC2) dendritic cells.
Focusing on their tolerogenic functions, tolDCs capture and process self-antigens, whether soluble or derived from apoptotic bodies, for their presentation to peripheral T cells [28]. Among the numerous mechanisms described, some are worthy of particular attention (Figure 2): (1) tolDCs promote the generation of CD4+ pTregs from CD4+ Tconvs in two ways, by secreting immunosuppressive cytokines (IL-10 and TGF-β) or by signaling through PDL-1 molecules [54,55]; (2) tolDCs eliminate autoreactive CD8+ T cells via the Fas/FasL apoptotic system [56]; and (3) tolDCs favor an anergic state in T cells due to the weak expression of MHCII and CD80/CD86 [57].
Also, as for iTreg lymphocytes, it is also possible to induce an immunosuppressive state in mouse and human DCs in vitro for their use in cell-based immunotherapies. This polarization towards tolerance is achieved using different immunomodulators, including glucocorticoids (e.g., dexamethasone), cytokines (e.g., IL-10 and TGF-β), rapamycin, or vitamin D3 [28].
1.2. Autoimmune Alterations in the Adaptive Immune Balance
In the case of cancer, the immune balance is inclined in favor of immunosuppression as a strategy to help the tumor evade active immune responses. By contrast, in the case of autoimmunity, the immune equilibrium is oriented towards immunogenicity against self-antigens. In this respect, there is considerable evidence of defects and alterations in number, frequency, and immunosuppressive function of CD4+ Tregs from patients with allergies, and other inflammatory or autoimmune diseases [13,14,58]. Although some inconsistencies have been noted depending on the patient cohort, the pathological state, and the different flow cytometry panels employed to define and phenotype Treg subsets, some common defects have been reported globally in CD4+ T lymphocytes in autoimmune contexts: (1) the conversion of CD4+ Tregs into effector CD4+ Tconvs; (2) the reduced ability of CD4+ Tregs to promote tolerogenic effects; and (3) the resistance to immunosuppression observed in some activated CD4+ Tconvs. By contrast, in type I diabetes, autoreactive CD8+ Tconv cells have been extensively described as an important inducer of cytotoxicity of pancreatic β cell death [59], and implicated in other autoimmune pathologies [60]. There is an increase in the number of CD8+ KIR+ Tregs in both blood and at inflammatory sites in autoimmune patients, and it was proposed that these lymphocytes eliminate pathogenic CD4+ T cells to avoid their exacerbated response [9].
Finally, the role of DCs in autoimmune progression has also been analyzed in depth [61,62], establishing that they promote an immunogenic environment due to inflammatory cytokine production. For example, IL-6, IL-12, and IL-23 secretion by immunogenic mDCs facilitates the polarization of self-reactive T cells. In the case of SLE, type-I interferons (IFN-I) mainly produced by pDCs deserve a special mention because they represent one of the most important molecular markers for this disease (see Section 4.4.2).
In summary, the same adaptive cell type can present either tolerogenic or immunogenic properties depending on their phenotype, the stimuli to which they are exposed, and their molecular context. The final cell outcome will depend on different signaling pathways that culminate in gene expression patterns maintained by specific transcription factors. In this regard, it is important to determine if there are any T linage-related transcription factors that contribute to the immunogenic or tolerogenic activity of CD8+ and CD4+ T cells.
2. Helios in the Immune System
One element in this delicate immune balance is the transcription factor Helios (Ikzf2 in mice and IKZF2 in humans), a member of the Ikaros family of transcription factors that appears to fulfill a critical role in maintaining a regulatory profile in the T-cell lineage. Helios deficiency, either in mice lacking Helios in all cells [38] or specifically in FoxP3+ cells [63] (mostly CD4+ Tregs), leads to an autoimmune phenotype at 5–6 months of age that is associated with distinct symptoms, reproducing some canonical immunological manifestations of lupus: hypergammaglobulinemia against nuclear antigens, splenomegaly, glomerulonephritis, an enhanced presence of activated Tconvs, and an expansion of follicular T lymphocytes (see below). Indeed, a germline mutation in IKZF2 was recently described in an SLE patient that was responsible for severe immune dysregulation [64]. This raises the question as to what the role of Helios is in each immune cell population.
2.1. Helios in CD4+ T Cells
Initially, Helios was identified in mouse models as a specific marker of CD4+ tTregs through a string of discoveries. (1) Until the first week of life, nearly all Tregs (CD4+ FoxP3+ T lymphocytes) that migrate from the thymus express Helios. (2) FoxP3+ Helios− CD4+ T cells only appear from the second week after weaning. (3) In adult animals, around 70% of the CD4+ Treg compartment expresses Helios. (4) Helios is not expressed in either iTregs or pTreg lymphocytes in an experimental tolerance model [65,66]. However, the validity of this latter assumption has become increasingly controversial due to the more recent detection of Helios in CD4+ pTregs [67], in activated and exhausted CD4+ Tconvs [68,69,70], and in iTregs [71]. Nonetheless, few studies have assessed the relative expression of Helios in specific subpopulations of immune cells beyond simply distinguishing between Helios+ and Helios− cells. In this sense, the merits of distinguishing up to three biologically relevant levels of Helios expression is currently being explored (Helioshigh, Heliosmid, and Helioslow) [72,73].
Beyond its proposed function as a unique molecular marker for tTregs, it is apparent that Helios+ and Helios− CD4+ Treg lymphocytes have notable phenotypic, genetic, and functional differences [74,75]. CD4+ Tregs expressing Helios exhibit a more activated phenotype, with a higher percentage of effector (CD44+ CD62L−) cells. Moreover, in vitro studies indicate that these Tregs have a stronger immunosuppressive capacity relative to CD4+ Tregs lacking Helios [76]. Furthermore, experiments on lymphopenic animals have shown that CD4+ Tregs expressing Helios in vivo express FoxP3 more stably. Indeed, the combined use of FoxP3 and Helios has been proposed to identify bona fide CD4+ Tregs in humans [77].
Interestingly, the deletion of Helios in mice, both in all cells [38] and specifically in FoxP3+ cells [63], provokes an autoimmune phenotype at 6 months of age, demonstrating the role of this transcription factor in stabilizing regulatory activity. However, the absence of Helios in the entire CD4+ T cell repertoire, including both Tconvs and Tregs, does not apparently evoke autoreactivity [38,65]. This curiosity may suggest that Helios is involved in both sides of the immune balance and it leads us to draw two hypotheses. (1) Helios expression in CD4+ Tconvs plays a significant role in the autoimmune reaction; and (2) other cell types with Helios-dependent regulatory activity impede autoreactive immune responses.
2.2. Helios in KIR+/Ly49+ CD8+
If there is one thing that CD4+ Tregs and CD8+ Ly49+ Tregs have in common, it is the expression of Helios as a maker of their regulatory identity. Nevertheless, research on CD8+ Ly49+ Tregs is still in its infancy compared to that on CD4+ Tregs. Evidence for the physiological relevance of Helios in CD8+ Ly49+ Tregs and their immunosuppressive function comes from the inability of CD8+ Ly49+ Helios− T lymphocytes to inhibit the follicular T cell response relative to CD8+ Ly49+ Helios+ cells. Moreover, unlike their Helios+ counterparts, CD8+ Ly49+ Helios− Treg lymphocytes exert a stronger effector phenotype (CD127low) and worse survival under inflammatory conditions [38]. Independently, weaker Helios expression has also been found in the CD8+ Ly49+ Tregs, correlating with autoimmune progression in a murine model that replicates the typical SLE symptomatology from the age of 5 months (including splenomegaly and germinal center reactivity). From this model, with deficient TGF-β signaling, expression of this cytokine may influence the Helios expressed by CD8+ T cells [41]. In addition, we found that both the reduction in the proportion of Ly49+ CD8+ T cells and the weaker Helios expression by these cells was correlated to disease progression in two mouse models of lupus (MRL/MPJ and MRL/LPR) [73].
A recent article in Science characterized the function and phenotype of the human equivalent of Ly49+ CD8+ Tregs in infectious and autoimmune contexts [9]. Although the murine family of Ly49 receptors lacks a genetic human homologue in terms of sequence, the members of the human Killer cell Immunoglobulin-like Receptor (KIR) family are functionally equivalent to these. Both populations, KIR+ CD8+ and their murine Ly49+ CD8+ T cell counterparts, can eliminate autoreactive CD4+ T cells in autoimmune and viral contexts, and they are characterized by their Helios+ phenotype.
As demonstrated, altered Helios expression in the T lymphocyte pool is linked to an autoimmune phenotype that replicates the typical symptoms observed in SLE, an archetypal condition of systemic autoimmunity. Thus, while the “mystery” surrounding CD8+ regulatory T cells is slowly lifting [34], many questions remain, such as what the origin of these KIR+/Ly49+ CD8+ T lymphocytes is, and what is their relationship to other cells in the immune system, like DCs.
2.3. Helios in Other Immune Cells: Double Negative (DN) T and NK Cells
2.3.1. Helios in DN T Cells
In addition to CD4+ and CD8+ T cells, there is a specific subpopulation of T lymphocytes with a TCRαβ+ profile that expresses neither CD4 nor the CD8 co-receptor. In terms of their possible origin, the model that currently seems to attract the most interest, according to recent studies, suggests that double negative (DN) T cells are derived from autoreactive CD8+ T cells that have lost their CD8 co-receptor [78,79]. In this context, DN T lymphocytes would be characterized by the expression of PD-1 at their surface and by the transient presence of Helios in their nucleus [78]. The pathological role of these PD-1+ DN T cells is witnessed by their proinflammatory effector phenotype (CD127low) and by the production of the IL-17 cytokine, which is tightly correlated with the autoimmune phenomenon [80,81]. However, the regulatory properties of the DN T repertoire cannot be ignored, and they have also been reported on the other side of the immunological equilibrium, in non-autoimmune contexts and in organ transplantation [82,83,84]. Again, this dual behavior of DN T cells would reflect the complexity of the immune system, the existence of specific subpopulations characterized by the expression of markers yet to be defined, and the influence of the molecular context (autoimmune or non-autoimmune).
The transcriptional profile of DN T cells from mice was recently analyzed [85], revealing different DN T subpopulations to be distinguished by the Helios gene (Ikzf2). Indeed, activated and non-activated DN T lymphocytes could be identified through this biomarker. Helios expression is enhanced in naïve DN T cells, whereas activated DN T cells express this transcription factor weakly.
2.3.2. Helios in NK Cells
In humans, after CD4+ Tregs and mucosal-associated invariant T (MAIT) lymphocytes, NK cells are the immune cell population with the strongest Helios expression (supplemental Figure S3 from [64]). Nevertheless, there are few data regarding Helios expression in the NK compartment. Helios has been proposed to regulate the activity of hyperreactive NK cells in mice resistant to viral infection [86]. Moreover, Helios downregulation was observed in some subsets of “memory-like” NK cells from cytomegalovirus-infected individuals [87], and finally, the proportion of CD16+ CD56dim NK cells was reduced in a lupus patient due to germline mutation of the IKZF2 gene [64]. Together, Helios appears to be an important transcription factor that controls the activity and homeostasis of NK cells, which are involved in autoimmunity [88] and lupus [89]. Further studies will be needed to examine the expression of Helios in autoimmune contexts and in different immune cells.
3. Systemic Lupus Erythematosus (SLE)
3.1. SLE Patients
SLE is a multifaceted autoimmune disease with both environmental and genetic influences. It may present in various ways, affecting different organs like the joints, nervous system, skin, kidneys, and heart (Figure 3) [90,91]. One prevalent feature in SLE patients is the presence of circulating antibodies against nuclear antigens, such as dsDNA, nucleosome proteins, and RNA-associated proteins. These antibodies can accumulate in various tissues, provoking damage and promoting an autoimmune response [92]. In general, like other autoimmune diseases, SLE exhibits a cyclic pattern of activity, stability, and occasionally, remission [90,93]. Thus, the diverse manifestations, varying in both form and degree of involvement, lead to heterogeneity in the proposed prevalence rates depending on the method or criteria used to assess patients. Fortunately, more intensive research into this disease is now translating into new, more sensitive, and specific diagnostic criteria for SLE that are being used in clinical practice (EULAR/ACR 2019 criteria vs. ACR 1997 criteria) [94]. SLE shows a clear gender bias, affecting nine females for every one male, making it one of the most extreme gender-biased diseases in medicine [95]. Therefore, finding new reliable markers to diagnose and predict this pathology is an urgent unmet medical need. This achievement would help unify the patient community worldwide and would ideally identify preclinical or incomplete cases [96]. Yet it remains unclear how basic research can address this issue.
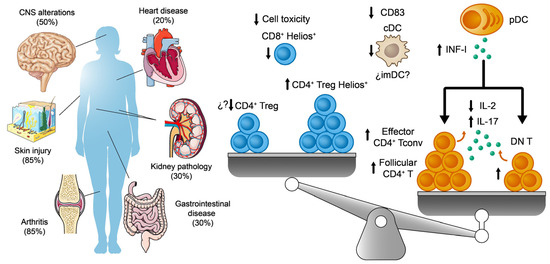
Figure 3.
On the left, the main systems affected in SLE are shown. The percentages indicate their prevalence among lupus patients. On the right, a scheme summarizes the major imbalances and alterations to Treg, Tconv, and DC populations associated with SLE.
3.2. Mouse Models of SLE
Many mouse models are available to be used in basic and preclinical research that recapitulate some of the main features of SLE [97,98]: spontaneous, inducible, transgenic, and humanized models. Compared to other spontaneous models, the MRL strain is characterized by (1) the accumulation of a broad repertoire of circulating antibodies against nuclear antigens (anti-dsDNA, anti-Ro, anti-La…), (2) a pronounced sex bias, and (3) by the presentation of additional SLE symptoms like glomerulonephritis, arthritis, nervous system inflammation, skin rashes, and vasculitis. There are two variants of the MRL strain: MRL/LPR mice that present a much more severe pathology with an early onset (at 3–4 months of age), provoked by homozygous mutation in the Fas protein encoding gene (CD95); and MRL/MPJ mice with no such genetic alteration and with a milder pathological phenotype that appears later in life (at 6–12 months of age), reflecting a genetic background prone to the autoimmune phenomenon [99]. Below we will discuss the main alterations in the aforementioned adaptive immune cells in relation to SLE and focusing on the role of Helios.
4. Adaptive Immune Disequilibrium in SLE
As we have seen, the immune balance between T cells (regulatory and conventional) and DCs is a complex homeostatic equilibrium that, when disturbed, could explain part of the autoimmune phenomena of the SLE pathology through either a shift towards a loss of tolerance to self-antigens or towards an exacerbated response [100]. Here we shall consider some findings related to the role of these cell types in SLE, with specific reference to the Helios expression in these populations (Figure 3).
4.1. CD4+ T Cells in SLE
4.1.1. CD4+ Tconvs and Tregs in SLE
SLE patients have an expanded population of effector memory CD4+ T cells relative to healthy individuals [101]. Thus, the production of IL-2 and IL-17 is altered in this cellular compartment, the two best studied cytokines in the immune balance. Specifically, there is a notorious reduction in IL-2 (essential for maintaining regulatory function) and enhanced IL-17 production (a critical cytokine in promoting autoimmunity), as seen in CD4+ Tconv cells from patients and in animal models [102,103]. In addition, there is an expansion of follicular and extrafollicular CD4+ T cell populations in SLE, promoting maturation of B cells and antibody release, and correlated with pathological severity [104,105].
There are conflicting data regarding CD4+ Tregs in the literature. While some studies demonstrated a smaller proportion of circulating regulatory cells in SLE patients [106], elsewhere, the opposite effect was reported: a significant increase in this immune subpopulation in the periphery as a homeostatic compensatory mechanism aimed at re-establishing the immunological equilibrium [107,108]. This controversy regarding the number and proportion of CD4+ Tregs in SLE patients was highlighted in a recent meta-analysis of 18 independent studies [109]. The global analysis revealed a slight decrease in the proportion of CD4+ Tregs, accompanied by very strong heterogeneity. It was proposed that the origin of these discrepancies might reside in (1) the criteria used to evaluate and diagnose active and inactive forms of the pathology; (2) the flow cytometry strategy used to define CD4+ Tregs; and (3) the treatment type administered to the patients. According to the same meta-analysis [109], there is also no consensus regarding possible defects in the regulatory activity of these cells. While two studies show that CD4+ Tregs from SLE patients have an altered immunosuppressive activity [110,111], another study showed no significant difference [112].
4.1.2. CD4+ Helios+ T Cells in SLE
Against this background, to further investigate the function and phenotype of CD4+ Tregs lymphocytes, and to avoid such heterogeneity and disparity of results, an analysis of the Helios transcription factor by flow cytometry has begun to be included in clinical research into autoimmune diseases [77,113]. It is worth noting the consistent increase in the proportion of CD4+ Helios+ Treg lymphocytes in SLE patients in three independent studies correlated with disease activity [108,114,115]. Because these CD4+ Helios+ Tregs present a more active immunosuppressive function than CD4+ Helios− Tregs, they could represent a component of the immune compensation system and respond to acute immune imbalances that need to be addressed.
4.2. CD8+ T Cells in SLE
4.2.1. CD8+ Tconvs and Tregs in SLE
Although less well-studied than their CD4+ counterparts, CD8+ T cells are receiving increasing attention as agents involved in the development of autoimmunity, and particularly SLE, due to both their inflammatory and protective functions [103,116,117]. One of the best studied characteristics of CD8+ T cells is their ability to induce targeted cell death via the secretion of granzyme and perforin, thereby halting infectious and tumor processes. CD8+ T cells from SLE patients exhibit compromised cytotoxicity, as witnessed by the reduced granzyme B and perforin [118]. This phenomenon may explain why viral infections are a leading cause of mortality in SLE patients [119]. In parallel, cytotoxicity is crucial for the optimal regulation of the humoral adaptive response by KIR+/Ly49+ CD8+ Tregs, eliminating autoreactive follicular T lymphocytes. The murine model of SLE in which perforin is lacking provides evidence for this, in which autoimmune symptoms are accelerated [120].
4.2.2. CD8+ Helios+ T Cells in SLE
As mentioned, the effects of Helios in CD8+ T cells have been almost exclusively studied in KIR+/Ly49+ CD8+ Tregs from autoimmune models that mimic SLE. Nevertheless, weaker Helios expression in the CD8+ T compartment and smaller numbers of these cells per unit volume have been described recently in the blood of lupus patients [41,121].
4.3. DN T in SLE
The association of DN T cells with SLE and with other autoimmune diseases has been well defined [100,118,122,123]. For example, these cells have been implicated in the production of autoreactive antibodies against dsDNA by B lymphocytes in humans [124]. In addition, an increase in the number of DN T cells has also been found in both the peripheral blood and kidneys of patients, and in mouse models of lupus, suggesting their role in tissue damage [125,126]. Also, the proportions of DN T cells were enhanced during autoimmune activity as opposed to remission in young lupus patients [127].
4.4. DCs in SLE
As previously discussed, pDCs and cDCs modulate immunogenic and immunosuppressive responses, fulfilling important roles in both innate and adaptive immunity. Therefore, it is not surprising that these cells have received attention for their potential contributions to our understanding of the pathophysiology of SLE [128,129,130]. Overall, the DC repertoire exhibits changes in number, frequency, phenotype, and function as lupus progresses. However, it is difficult to obtain uniform results regarding SLE in clinical practice due to its marked heterogeneity in terms of manifestations and activity. In addition, the different immunosuppressive drugs widely used to treat SLE affect the biology of DCs [131]. Furthermore, it is unclear whether SLE is a result of an intrinsic defect in DCs or if this pathology alters the behavior and phenotype of DCs as a secondary consequence of the molecular context produced mainly by circulating cytokines [132].
4.4.1. cDCs in SLE
Studies analyzing cDC frequencies have produced inconsistent data [128], although most seem to indicate that SLE patients have fewer cDCs in their peripheral blood, which correlates with a stronger infiltration of immature cDCs in the kidneys [133]. The few studies that have focused on the maturation and function of cDCs in SLE have also produced conflicting results. On the one hand, early data seemed to indicate that DCs from SLE patients have fewer co-stimulatory molecules and a weaker capacity to induce T lymphocyte proliferation relative to healthy individuals [134]. By contrast, during active stages of the disease, lower PD-L1 expression and higher CD80 and CD86 expression were detected in cDCs [135].
Due to their scarcity in circulation, information has been gained from DCs differentiated in vitro from monocytes, albeit again with conflicting results. While it was postulated that DCs derived from SLE patient monocytes have a less proinflammatory profile, a reduced capacity to stimulate T cells, and altered maturation [136], elsewhere, DCs were proposed to express CD80 and CD86 more strongly, and with enhanced proinflammatory cytokine secretion [137]. Nevertheless, there seems to be a consensus regarding the decrease in CD83 in the membrane of DCs, a marker of dendritic maturation and activation [138].
4.4.2. pDCs in SLE
One of the most widely accepted models for the initiation of SLE proposes that a malfunction in phagocytosis and in the elimination of circulating apoptotic bodies could lead to their accumulation in organs and tissues [139]. In this context, nucleic acid fragments in this cell debris could activate TLR7 and TLR9 receptors of pDCs, thereby boosting the production and release of type I IFNs into the bloodstream. This cytokine family would activate both the innate and adaptive immune systems, inducing what is known as the IFN signature [140]. The significance of these molecular mediators is evident since over 50% of adult SLE patients—and 90% of affected children—show a marked expression of type I IFN response genes [141]. Indeed, the selective removal of pDCs in SLE patients has beneficial effects on skin symptomatology [142]. Thus, the intricate cellular and molecular interplay among immune cells makes autoimmune diseases a heterogeneous group of pathologies that are difficult to manage. That is why their optimal treatment remains a challenge in contemporary medicine.
5. Helios as a Potential Biomarker for SLE: The Link between tolDCs and CD8+ Tregs
5.1. Helios as a Potential Biomarker in SLE
At the cellular and molecular level, many studies have focused on understanding the homeostatic equilibrium between T cells and DCs in SLE and other autoimmune diseases. Nevertheless, this only represents a small piece in the complete immune system puzzle and many questions remain due to the difficulties in fitting all the pieces together. Resolving these questions could open new opportunities for the treatment of these heterogeneous diseases, among which abundant subclinical cases exist, and new findings are driving constant changes in diagnosis criteria (e.g., in SLE) [143]. Hence, “the search for a specific SLE biomarker” [144] is still one of the most important challenges in relation to this disease. In terms of the ideal SLE biomarker, certain characteristics must be achieved. (1) It must have a clear implication in the pathophysiology of the disease; (2) it must have a high predictive value, specificity, and sensitivity; (3) it must be a biomarker with measurable variation that precedes periods of pathological activity, or in other words, it must be able to monitor the subclinical and incomplete evolution of SLE; (4) finally, this “ideal” biomarker must be reliably identified in biological samples using standardized assays that can be easily reproduced, interpreted, and performed by most laboratories at a reasonable cost [144]. Does Helios fulfill these requirements?
In terms of the first point, Helios could indeed play an essential role in the pathophysiology of lupus, both in humans and in mouse models. First, in mice, the global absence of Helios triggers an autoimmune phenomenon reminiscent of SLE [38,63]. Moreover, in classic murine SLE models like the MRL/LPR and MRL/MPJ mice, there are low levels of Helios in the Ly49+ CD8+ T cell populations [73]. Very recently, a lupus-like phenotype was diagnosed in a patient with a germline mutation in the IKZF2 gene [64]. Moreover, a genome-wide association study (GWAS) [145] of more than 7000 European SLE patients identified the IKZF2 gene as a susceptibility locus. Interestingly, this study identified other transcription factors, including other members of the Ikaros family (IKZF1 and IKZF3), among the candidate predisposition genes for lupus. This may suggest that the risk of SLE is the consequence of alterations to a gene expression network in multiple compartments of the immune system. Regarding the second point, a study in humans demonstrated the predictive value of the IKZF2 gene as a biomarker for the diagnosis of a prevalent SLE manifestation known as lupus nephritis [146]. As all analyses of Helios expression have been undertaken in patients with an evident pathology, its subclinical value is difficult to ascertain. Nevertheless, in asymptomatic MRL/MPJ and MRL/LPR animals (see Section 5.2), differential expression of Helios can be seen prior to disease manifestation and reflecting the future disease course: slow and mild (MRL/MPJ), or fast and severe (MRL/LPR) [73]. Finally, flow cytometry is a well-established experimental procedure in hospitals and immunology laboratories, and it is permanently undergoing advances [147]. Helios could be easily assessed simultaneously in different cell repertoires from a blood sample using this technique.
5.2. Helios Expression in T Cell Repertoires from Two Murine Models of SLE
Apart from the aforementioned evidence, a fifth property of Helios could be considered, as its expression follows different dynamics in each T subpopulation. This behavior could provide added value to this transversal T linage biomarker. For example, Helios was expressed more strongly in CD4+ Treg and Tconv populations in two murine models of lupus, yet more weakly by γδ T, DN T, and Ly49+ CD8+ Treg cells during the progression of the disease [73]. Perhaps the Helioshi profile of the CD4+ Treg fraction reflects a stronger activation and effector phenotype of these cells, a compensatory mechanism that is unable to combat the autoimmune imbalance. Conversely, this immune disequilibrium could be facilitated by a dampening of the immunosuppressive function of DN T and Ly49+ CD8+ Treg cells that express Helios weakly.
Remarkably, this biomarker proves to be a better criterion to classify both mouse models (MRL/MPJ vs. MRL/LPR) and disease states (healthy or predisease vs. diseased) relative to traditional criteria widely used in preclinical research, such as the relative proportion of effector memory CD4+ T lymphocytes (CD44+ CD62L− CD25− CD69+/−). For example, the statistical modeling of our data shows that Helios expression in the CD4+ Treg fraction is as powerful as anti-dsDNA antibody quantification in blood to distinguish the different groups of mice (Figure 4), an analysis based on a previous study [148]. Regarding the statistical modelling, univariate logistic regression was used to evaluate the predictive ability of the different biomarkers. In cases where complete and quasi-complete separation led to some estimated regression coefficients being infinite, Firth’s univariate logistic regression model was implemented. Forest plots were used to show the odds ratio (OR) with a 95% confidence interval, together with their p-values.
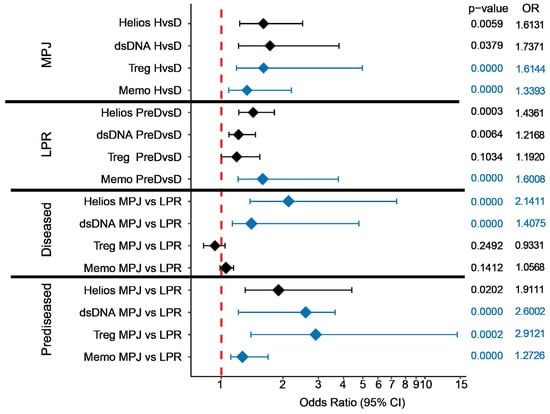
Figure 4.
A predictive biomarker analysis in MPJ and LPR mice of two lupus disease states (healthy (H), and prediseased (PreD) or diseased (D)), studying two variants of the mouse MRL background: MRL/MPJ (MPJ) and MRL/LPR (LPR). The data are presented as a Forest plot, with the odds ratios (ORs) and 95% confidence interval (95% CI) assessed by a univariate (black) or Firth’s univariate (blue) logistic regression. Unit changes for the continuous variables were assessed for the logistic regression. For circulating antibodies against double-stranded DNA molecules (dsDNA), their biological activity was divided by 10,000. Helios represents the proportion of Helios+ cells in the CD4+ Treg fraction; Treg, the proportion of Treg (FoxP3+CD25+) cells in the CD4+ compartment; and Memo, the proportion of CD44+ CD62L− CD25− cells in the CD4+ population. The data presented are derived from a previous study [73], and in addition, new flow cytometry analyses were taken into account. The analysis was performed and the graph obtained using the stats, logistf, and ggplot2 packages of R version 4.2.2.
5.3. Influence of tolDCs in Helios Expression on CD8+ T Regs
Trying to connect the aforementioned adaptive elements of this autoimmune puzzle, we found that the molecular and cellular environment could affect Helios expressions by CD8+ T populations. In vitro studies on bone marrow-derived DCs revealed that tolDCs were specifically able to boost the expression of Helios in OT-I CD8+ T cells in the presence of ovalbumin relative to imDCs and mDCs [73]. This result is in line with previous evidence supporting the link between tolDCs and CD8+ Tregs.
In an oncological scenario, an increase in the proportion of CD8+ FoxP3+ Treg cells has been associated with the presence of tolDCs generated by glucocorticoid exposure [149]. Furthermore, cDC1s were seen to be vital for maintaining peripheral tolerance against self-antigens through their activation of murine CD8+ FoxP3+ Treg cells [53]. Similarly, CD14+ monocytes promote the in vitro generation of immunosuppressive CD8+ FoxP3+ CD25+ Tregs from CD8+ CD25− cells in humans [150]. In terms of the potential mechanisms by which tolDCs could affect CD8+ T cell activity, the TGF-β produced by tolDCs may be important [151], as it is known to influence Helios expression in CD8+ Ly49+ Treg cells [41]. Moreover, the lack of DCs in mice leads to a significant decrease in the splenic CD8+ CD44+ T fraction, which is correlated with an autoimmune phenotype akin to lupus (e.g., splenomegaly, antibodies against circulating nuclear antigens; see [152]). Thus, it could be postulated that the CD8+ CD44+ T cell population largely corresponds to the CD8+ Ly49+ Treg cell fraction. If this were indeed the case, DCs would appear to play an integral role in regulating the differentiation and phenotype of a subset of CD8+ Ly49+ Treg lymphocytes, which helps maintain the immune balance and prevents the onset of systemic autoimmune phenomena. Together, our current understanding of the autoimmune imbalance in lupus supports the use of tolDCs and Tregs as therapeutic agents in cell-based immunotherapies. Nevertheless, this kind of approach still faces several limitations that may well be overcome in the next few years due to promising new advances through approaches like nanotechnology.
6. Cell-Based Immunotherapies for Autoimmune Diseases
Systemic and non-specific immunosuppressive treatments used to manage certain autoimmune disorders may have significant side effects, such as cancer and infections, as excessive immunosuppression may arise [153]. In recent years, efforts have been made to develop new immunotherapies targeting specific aspects of the autoimmune pathophysiology without disrupting the entire system. These therapies take advantage of the properties of the immune system itself as a biotechnological tool. In terms of immunotherapies, we will focus on cell-based approaches.
6.1. Cell-Based Immunotherapies
As the name suggests, cell immunotherapies are based on the autologous inoculation of immune cells—modified or not—to restore the immune imbalance depending on the disorder. Currently, ex vivo-generated CD4+ iTregs [154,155,156] and tolDCs [28,157] are being transferred into patients affected by diverse autoimmune diseases (multiple sclerosis, type I diabetes…) in clinical trials to test their therapeutic efficacy (Figure 5).

Figure 5.
Summary of two of the main strategies currently being tested in clinical trials to restore the altered immune balance in autoimmune diseases using cell transfer-based immunotherapy involving iTregs and tolDCs.
In the case of SLE, as there is currently no drug specifically designed to increase Helios levels in target cells, one potential treatment for this pathology could be the injection of therapeutical cells that either express enhanced levels of Helios (CD8+ Ly49+ iTreg cells) or promote its expression (tolDCs).
6.2. Limitations of Cell-Based Immunotherapies
As mentioned previously, DCs regulate adaptive immune responses in lymphoid tissues where they interact with different T cell populations. For successful immunomodulation, inoculated DCs need to overcome several barriers. (1) They must penetrate afferent lymphatic vessels; (2) they must then migrate towards a draining lymph node (LN); (3) and finally, they must be internalized into the LN structure [158]. These limitations would largely be responsible for the poor migratory efficiency of DCs in the target LN (less than 5%) [159]. Given that the magnitude of the T cell response is proportional to the number of injected DCs that reach the LNs [160], strategies to maximize the migration and mobilization of DCs to the LNs are being explored in both autoimmunity and cancer research. This concentration of therapeutic cells at specific targets could avoid the side effects derived from systemic and non-specific dissemination of DCs in the bloodstream. In this context, different approaches are being tested: (1) direct inoculation of DCs into the LNs [161]; (2) induction of enhanced CCR7 expression, a fundamental chemotaxis receptor on DCs [162]; and (3) the magnetic targeting of DCs coupled to iron oxide nanoparticles (IONPs) [163]. This latter strategy will be addressed below as a promising nanotechnology approach to biological problems. However, the therapeutic use of tolDCs for SLE [164] is currently at a preclinical stage, having been successfully tested in vivo in mouse models [165] and explored preliminarily in vitro with human cells [166,167]. However, there are still several obstacles that need to be tackled.
7. Nanobiomedicine
Step-by-step medicine is becoming more and more personalized, and nanotechnology is one of the areas revolutionizing biomedicine [168], a Key Enabling Technology recognized by the European Union [169]. The implementation of nanotechnology in biomedicine can be referred to as nanobiomedicine, based on the use of nanometric materials (with at least one dimension between 1 and 100 nm) [170] to diagnose, prevent, monitor, and/or treat disease [171]. At this nanometric scale, near a quantum level, common macroscopic materials could exhibit emerging physical and chemical properties that could be useful in medicine. The current repertoire of available nanoparticles (NPs) is tremendously diverse (e.g., polymeric, metallic, and ceramic NPs), each with unique properties and possibilities [172]. Some examples of metallic NPs include (1) the use of photothermic gold NPs to provoke tumor cell death by heating [173]; (2) the use of silver NPs as antibiotics [174]; (3) the use of magnetic IONPs as a contrast agent for magnetic resonance (MRI), as an inducer of tumor hyperthermia, and as an agent for cellular and molecular targeting (see Section 7.2).
7.1. Magnetic Iron Oxide Nanoparticles (MNPs)
As mentioned, magnetic iron oxide nanoparticles (MNPs) are among the best-known and widely studied nanomaterials. Because MNPs can be naturally processed and excreted via the main pathways of iron metabolism, they have been identified as one of the most suitable options for biomedicine in terms of biocompatibility [172,175,176]. Among the most common MNPs considered are MNPs based on magnetite (Fe3O4), maghemite (γ-Fe2O3), or mixed ferrites (Co, Mn, Ni, or Zn + Fe2O4). Overall, IONPs have two main elements: the iron oxide nucleus or core and the chemical coating. On the one hand, the “small” size of the nucleus (<50 nm) favors their superparamagnetic behavior, avoiding exacerbated aggregation of MNPs in the blood when subjected to an external magnetic field and once it is removed. On the other hand, the usually charged chemical coating plays a role in NP design: (1) improving core biocompatibility; (2) reducing immunogenicity; (3) preventing aggregation due to electrostatic repulsion; and (4) acting as an anchor to conjugate other molecules (drugs, antibodies, fluorescent dyes…) [177]. In addition, the coating composition affects the relationship between cells and MNPs. For instance, different coatings drive different types of intracellular trafficking and degradation depending on the cell type [178,179]. Likewise, because the coating has the potential to regulate the quantity of MNPs bound to the cell surface, the choice of coating could be critical to maximize the amount of iron per cell, and thus, their magnetic movement [180].
7.2. MNP Applications in Biomedicine
Among the numerous possibilities offered by MNPs in biomedicine, those that stand out include (Figure 6) (1) the localized delivery of molecules with biological activity (drugs, cytokines, antibodies, etc.) by means of magnetic fields to minimize the secondary effects related to their systemic dissemination [181,182]; (2) magnetic hyperthermia, which offers an option for oncological treatment through the heating of MNPs exposed to an alternating magnetic field [183,184,185]; (3) the use of MNPs as contrast agents for MRI [186,187]; (4) the magnetic targeting of immune cells using external magnetic fields as a solution for the previously indicated obstacles to cell-based immunotherapies with Tregs and tolDCs.
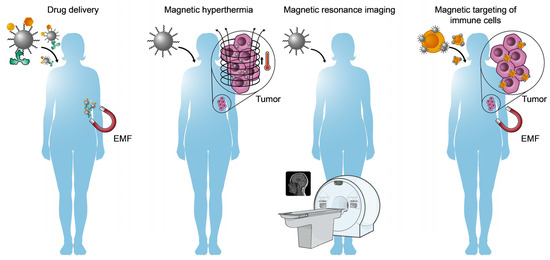
Figure 6.
Biomedical applications for magnetic iron oxide nanoparticles (MNPs). From the left, drug delivery via external magnetic fields (EMFs), cancer cell hyperthermia using MNPs and alternating magnetic fields, MNPs as contrast agents for magnetic resonance imaging, and magnetic targeting of immune cells with MNPs to optimize immunotherapy through EMFs.
Magnetic Targeting for Cell-Based Therapies
As indicated above, optimizing therapeutic cell migration to target tissues is key to realizing the full potential of cell-based therapies [188,189] (see Table 3 for examples of magnetic targeting of therapeutic MNPs to cells). Most research into the magnetic targeting of immune cells has focused on oncological therapies. Less attention has been paid to the other side of the immune balance, the potential use of MNPs for the magnetic targeting of immunosuppressive cells (tolDCs, iTregs…) towards lymphoid tissues as a strategy to improve cell-based immunotherapies in autoimmune contexts. What has been studied extensively is the interaction between DCs and different types of NPs [190,191,192], as well as the specific use of nanomaterials to induce the polarization of DCs towards tolDCs in vivo as a therapeutic approach for autoimmune pathologies [193,194]. What lessons can be learned from these studies to extend the idea of magnetic targeting to tolDCs?

Table 3.
Some successful examples of magnetic targeting of therapeutical cells.
7.3. MNPs and DCs
Among the vast repertoire of nanomaterials available, the possibility of modifying DCs with iron oxide MNPs has also been analyzed. From these studies, some general lessons were learned, regardless of the biomedical application for which they were intended: (1) at low concentrations (0–50 µg/mL Fe), MNPs do not exert significant cytotoxicity and they do not alter cell morphology, surface marker patterns, or the cytokine profile of previously matured DCs [202,203,204,205,206,207,208,209]. However, some reports did claim that MNPs can potentially reduce the viability of DCs at high concentrations (>400 µg/mL Fe), while not affecting their maturation [210]. In addition, DCs treated with MNPs can maintain a stable phenotype, but they may lose their optimal ability to stimulate CD4+ T lymphocytes [211]. In turn, MNPs can experience reduced migratory capacity at high concentrations and when they accumulate at micrometric instead of nanometric levels [206,212]. Regarding their surface electrical charge, positively charged MNPs favor antigen cross-presentation on DCs relative to those with an electrically negative coating [209,213]. This behavior is attributed to the chemical modification of their surface, which dictates their intracellular fate toward a particular compartment [178,179,213]. Also, negatively charged MNPs stimulate stronger production of the proinflammatory IL-1β than positively charged MNPs [209]. Finally, MNP treatment prior to in vitro DC maturation could alter this process [208]. Together, the literature cited suggests the use of positively charged superparamagnetic MNPs at low concentrations (<50 µg/mL) for the magnetic targeting of fully mature tolDCs towards LNs as a new cell-based immunotherapy approach.
8. Conclusions
Given the very heterogeneous manifestations of SLE in different cohorts of patients and the presence of abundant subclinical cases, the standard criteria for lupus diagnosis in clinical practice are constantly changing as new findings appear. Hence, the search for better biomarkers for SLE is one of the main challenges in the context of precision medicine for this disease. In this regard, Helios (encoded by IKZF2) is a transversal transcription factor that shapes the regulatory activity in the T cell compartment, and currently, it is being investigated intensively in autoimmune contexts. Indeed, its levels are independent and vary in different immune cell populations: DN T cells, CD4+ and KIR+/Ly49+ CD8+ T cells, and NKs. The predictive value of Helios in lupus and its role in maintaining the regulatory profile of T cell subpopulations make it a potential candidate to be considered in future studies into SLE classification.
On the other hand, in this review, we have discussed how Helios in CD8+ T cells could be influenced by their interaction with DCs, specifically tolDCs. Given that, in preclinical trials of cell-based immunotherapy, tolDCs do not fully restore the immune equilibrium to avoid exacerbated autoimmunity, efforts need to focus on strategies to overcome some of the main limitations associated with this approach. One of these obstacles is the very low proportion of therapeutic cells that reach the target tissue. As a potential solution, the possibility to drive the migration and retention of therapeutic cells by using MNPs and external magnetic fields (magnetic targeting) is currently being studied, with promising results.
Author Contributions
A.P.-M. (Andres París-Muñoz) and D.F.B. conceived and designed the review. A.P.-M. (Andres París-Muñoz) wrote most sections of the review and prepared most of the figures and tables. O.L.-T. was responsible for the statistical analysis. A.P.-M. (Antonio Pérez-Martínez) and D.F.B. coordinated, critically revised, modified, and completed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the grant PID2020-112685RB-100 funded by MCIN/AEI/10.13039/501100011033 (to D.F. Barber). A. París-Muñoz was a predoctoral fellow at the CNB-CSIC, supported by a fellowship from “la Caixa” Foundation (ID 100010434, fellowship code LCF/BQ/DE18/11670013), and is now a postdoctoral fellow at IdiPAZ and CNIO supported by “Progama Investigo” funded by NextGenerationEU.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank members of the laboratories led by Domingo F. Barber (CNB-CSIC, Madrid) and Antonio Pérez-Martínez (IdiPAZ-CNIO, Madrid) for their helpful comments and discussions. The authors are also grateful to Mark Sefton for English language editing of the manuscript.
Conflicts of Interest
The authors have no conflict of interest to declare.
References
- Eberl, G. Immunity by equilibrium. Nat. Rev. Immunol. 2016, 16, 524–532. [Google Scholar] [CrossRef]
- Eberl, G.; Pradeu, T. Towards a General Theory of Immunity? Trends Immunol. 2018, 39, 261–263. [Google Scholar] [CrossRef]
- Horwitz, D.A.; Fahmy, T.M.; Piccirillo, C.A.; La Cava, A. Rebalancing Immune Homeostasis to Treat Autoimmune Diseases. Trends Immunol. 2019, 40, 888–908. [Google Scholar] [CrossRef]
- Germain, R.N. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2002, 2, 309–322. [Google Scholar] [CrossRef]
- Taniuchi, I. CD4 Helper and CD8 Cytotoxic T Cell Differentiation. Annu. Rev. Immunol. 2018, 36, 579–601. [Google Scholar] [CrossRef]
- Rosenblum, M.D.; Way, S.S.; Abbas, A.K. Regulatory T cell memory. Nat. Rev. Immunol. 2016, 16, 90–101. [Google Scholar] [CrossRef]
- Golubovskaya, V.; Wu, L. Different Subsets of T Cells, Memory, Effector Functions, and CAR-T Immunotherapy. Cancers 2016, 8, 36. [Google Scholar] [CrossRef]
- Savage, P.A.; Klawon, D.E.J.; Miller, C.H. Regulatory T Cell Development. Annu. Rev. Immunol. 2020, 38, 421–453. [Google Scholar] [CrossRef]
- Li, J.; Zaslavsky, M.; Su, Y.; Guo, J.; Sikora, M.J.; van Unen, V.; Christophersen, A.; Chiou, S.H.; Chen, L.; Li, J.; et al. KIR(+)CD8(+) T cells suppress pathogenic T cells and are active in autoimmune diseases and COVID-19. Science 2022, 376, eabi9591. [Google Scholar] [CrossRef]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef]
- Josefowicz, S.Z.; Lu, L.F.; Rudensky, A.Y. Regulatory T cells: Mechanisms of differentiation and function. Annu. Rev. Immunol. 2012, 30, 531–564. [Google Scholar] [CrossRef]
- Sakaguchi, S.; Mikami, N.; Wing, J.B.; Tanaka, A.; Ichiyama, K.; Ohkura, N. Regulatory T Cells and Human Disease. Annu. Rev. Immunol. 2020, 38, 541–566. [Google Scholar] [CrossRef]
- Grant, C.R.; Liberal, R.; Mieli-Vergani, G.; Vergani, D.; Longhi, M.S. Regulatory T-cells in autoimmune diseases: Challenges, controversies and--yet--unanswered questions. Autoimmun. Rev. 2015, 14, 105–116. [Google Scholar] [CrossRef]
- Burzyn, D.; Benoist, C.; Mathis, D. Regulatory T cells in nonlymphoid tissues. Nat. Immunol. 2013, 14, 1007–1013. [Google Scholar] [CrossRef]
- Feuerer, M.; Herrero, L.; Cipolletta, D.; Naaz, A.; Wong, J.; Nayer, A.; Lee, J.; Goldfine, A.B.; Benoist, C.; Shoelson, S.; et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 2009, 15, 930–939. [Google Scholar] [CrossRef]
- Shevach, E.M. Foxp3(+) T Regulatory Cells: Still Many Unanswered Questions-A Perspective After 20 Years of Study. Front. Immunol. 2018, 9, 1048. [Google Scholar] [CrossRef]
- Vignali, D.A.; Collison, L.W.; Workman, C.J. How regulatory T cells work. Nat. Rev. Immunol. 2008, 8, 523–532. [Google Scholar] [CrossRef]
- Plitas, G.; Rudensky, A.Y. Regulatory T Cells: Differentiation and Function. Cancer Immunol. Res. 2016, 4, 721–725. [Google Scholar] [CrossRef]
- Shevyrev, D.; Tereshchenko, V. Treg Heterogeneity, Function, and Homeostasis. Front. Immunol. 2019, 10, 3100. [Google Scholar] [CrossRef]
- Chinen, T.; Kannan, A.K.; Levine, A.G.; Fan, X.; Klein, U.; Zheng, Y.; Gasteiger, G.; Feng, Y.; Fontenot, J.D.; Rudensky, A.Y. An essential role for the IL-2 receptor in Treg cell function. Nat. Immunol. 2016, 17, 1322–1333. [Google Scholar] [CrossRef]
- Ernst, P.B.; Garrison, J.C.; Thompson, L.F. Much ado about adenosine: Adenosine synthesis and function in regulatory T cell biology. J. Immunol. 2010, 185, 1993–1998. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L.; Jackson, E.K. Adenosine and prostaglandin e2 production by human inducible regulatory T cells in health and disease. Front. Immunol. 2013, 4, 212. [Google Scholar] [CrossRef]
- Schmidt, A.; Oberle, N.; Krammer, P.H. Molecular mechanisms of treg-mediated T cell suppression. Front. Immunol. 2012, 3, 51. [Google Scholar] [CrossRef]
- Wallet, M.A.; Sen, P.; Tisch, R. Immunoregulation of dendritic cells. Clin. Med. Res. 2005, 3, 166–175. [Google Scholar] [CrossRef]
- Gianchecchi, E.; Fierabracci, A. Inhibitory Receptors and Pathways of Lymphocytes: The Role of PD-1 in Treg Development and Their Involvement in Autoimmunity Onset and Cancer Progression. Front. Immunol. 2018, 9, 2374. [Google Scholar] [CrossRef]
- Onishi, Y.; Fehervari, Z.; Yamaguchi, T.; Sakaguchi, S. Foxp3+ natural regulatory T cells preferentially form aggregates on dendritic cells in vitro and actively inhibit their maturation. Proc. Natl. Acad. Sci. USA 2008, 105, 10113–10118. [Google Scholar] [CrossRef]
- Domogalla, M.P.; Rostan, P.V.; Raker, V.K.; Steinbrink, K. Tolerance through Education: How Tolerogenic Dendritic Cells Shape Immunity. Front. Immunol. 2017, 8, 1764. [Google Scholar] [CrossRef]
- Švajger, U.; Obermajer, N.; Jeras, M. Novel Findings in Drug-Induced Dendritic Cell Tolerogenicity. Int. Rev. Immunol. 2010, 29, 574–607. [Google Scholar] [CrossRef]
- Mayer, C.T.; Floess, S.; Baru, A.M.; Lahl, K.; Huehn, J.; Sparwasser, T. CD8+ Foxp3+ T cells share developmental and phenotypic features with classical CD4+ Foxp3+ regulatory T cells but lack potent suppressive activity. Eur. J. Immunol. 2011, 41, 716–725. [Google Scholar] [CrossRef]
- Vuddamalay, Y.; van Meerwijk, J.P. CD28(−) and CD28(low)CD8(+) Regulatory T Cells: Of Mice and Men. Front. Immunol. 2017, 8, 31. [Google Scholar] [CrossRef]
- Liu, Y.; Deng, W.; Meng, Q.; Qiu, X.; Sun, D.; Dai, C. CD8+iTregs attenuate glomerular endothelial cell injury in lupus-prone mice through blocking the activation of p38 MAPK and NF-kappaB. Mol. Immunol. 2018, 103, 133–143. [Google Scholar] [CrossRef]
- Akane, K.; Kojima, S.; Mak, T.W.; Shiku, H.; Suzuki, H. CD8+CD122+CD49dlow regulatory T cells maintain T-cell homeostasis by killing activated T cells via Fas/FasL-mediated cytotoxicity. Proc. Natl. Acad. Sci. USA 2016, 113, 2460–2465. [Google Scholar] [CrossRef]
- Mishra, S.; Srinivasan, S.; Ma, C.; Zhang, N. CD8(+) Regulatory T Cell—A Mystery to Be Revealed. Front. Immunol. 2021, 12, 708874. [Google Scholar] [CrossRef]
- Niederlova, V.; Tsyklauri, O.; Chadimova, T.; Stepanek, O. CD8(+) Tregs revisited: A heterogeneous population with different phenotypes and properties. Eur. J. Immunol. 2021, 51, 512–530. [Google Scholar] [CrossRef]
- Kim, H.J.; Verbinnen, B.; Tang, X.; Lu, L.; Cantor, H. Inhibition of follicular T-helper cells by CD8(+) regulatory T cells is essential for self tolerance. Nature 2010, 467, 328–332. [Google Scholar] [CrossRef]
- Kim, H.J.; Wang, X.; Radfar, S.; Sproule, T.J.; Roopenian, D.C.; Cantor, H. CD8+ T regulatory cells express the Ly49 Class I MHC receptor and are defective in autoimmune prone B6-Yaa mice. Proc. Natl. Acad. Sci. USA 2011, 108, 2010–2015. [Google Scholar] [CrossRef]
- Kim, H.J.; Barnitz, R.A.; Kreslavsky, T.; Brown, F.D.; Moffett, H.; Lemieux, M.E.; Kaygusuz, Y.; Meissner, T.; Holderried, T.A.; Chan, S.; et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science 2015, 350, 334–339. [Google Scholar] [CrossRef]
- Satooka, H.; Nagakubo, D.; Sato, T.; Hirata, T. The ERM Protein Moesin Regulates CD8(+) Regulatory T Cell Homeostasis and Self-Tolerance. J. Immunol. 2017, 199, 3418–3426. [Google Scholar] [CrossRef]
- Stocks, B.T.; Wilson, C.S.; Marshall, A.F.; Hoopes, E.M.; Moore, D.J. Regulation of Diabetogenic Immunity by IL-15-Activated Regulatory CD8 T Cells in Type 1 Diabetes. J. Immunol. 2019, 203, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Liao, W.; Liu, Y.; Yang, M.; Ma, C.; Wu, H.; Zhao, M.; Zhang, X.; Qiu, Y.; Lu, Q.; et al. TGF-beta and Eomes control the homeostasis of CD8+ regulatory T cells. J. Exp. Med. 2021, 218, e20200030. [Google Scholar] [CrossRef]
- Shytikov, D.; Rohila, D.; Li, D.; Wang, P.; Jiang, M.; Zhang, M.; Xu, Q.; Lu, L. Functional Characterization of Ly49(+)CD8 T-Cells in Both Normal Condition and During Anti-Viral Response. Front. Immunol. 2020, 11, 602783. [Google Scholar] [CrossRef]
- Saligrama, N.; Zhao, F.; Sikora, M.J.; Serratelli, W.S.; Fernandes, R.A.; Louis, D.M.; Yao, W.; Ji, X.; Idoyaga, J.; Mahajan, V.B.; et al. Opposing T cell responses in experimental autoimmune encephalomyelitis. Nature 2019, 572, 481–487. [Google Scholar] [CrossRef]
- Iberg, C.A.; Jones, A.; Hawiger, D. Dendritic Cells As Inducers of Peripheral Tolerance. Trends Immunol. 2017, 38, 793–804. [Google Scholar] [CrossRef]
- Lutz, M.B.; Schuler, G. Immature, semi-mature and fully mature dendritic cells: Which signals induce tolerance or immunity? Trends Immunol. 2002, 23, 445–449. [Google Scholar] [CrossRef]
- Iberg, C.A.; Hawiger, D. Natural and Induced Tolerogenic Dendritic Cells. J. Immunol. 2020, 204, 733–744. [Google Scholar] [CrossRef]
- Verheye, E.; Bravo Melgar, J.; Deschoemaeker, S.; Raes, G.; Maes, A.; De Bruyne, E.; Menu, E.; Vanderkerken, K.; Laoui, D.; De Veirman, K. Dendritic Cell-Based Immunotherapy in Multiple Myeloma: Challenges, Opportunities, and Future Directions. Int. J. Mol. Sci. 2022, 23, 904. [Google Scholar] [CrossRef]
- Li, S.; Wu, J.; Zhu, S.; Liu, Y.J.; Chen, J. Disease-Associated Plasmacytoid Dendritic Cells. Front. Immunol. 2017, 8, 1268. [Google Scholar] [CrossRef]
- Uto, T.; Takagi, H.; Fukaya, T.; Nasu, J.; Fukui, T.; Miyanaga, N.; Arimura, K.; Nakamura, T.; Choijookhuu, N.; Hishikawa, Y.; et al. Critical role of plasmacytoid dendritic cells in induction of oral tolerance. J. Allergy Clin. Immunol. 2018, 141, 2156–2167.e9. [Google Scholar] [CrossRef] [PubMed]
- Bonifaz, L.; Bonnyay, D.; Mahnke, K.; Rivera, M.; Nussenzweig, M.C.; Steinman, R.M. Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J. Exp. Med. 2002, 196, 1627–1638. [Google Scholar] [CrossRef] [PubMed]
- Belz, G.T.; Behrens, G.M.; Smith, C.M.; Miller, J.F.; Jones, C.; Lejon, K.; Fathman, C.G.; Mueller, S.N.; Shortman, K.; Carbone, F.R.; et al. The CD8alpha(+) dendritic cell is responsible for inducing peripheral self-tolerance to tissue-associated antigens. J. Exp. Med. 2002, 196, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Lutz, M.B.; Kurts, C. Induction of peripheral CD4+ T-cell tolerance and CD8+ T-cell cross-tolerance by dendritic cells. Eur. J. Immunol. 2009, 39, 2325–2330. [Google Scholar] [CrossRef] [PubMed]
- Joeris, T.; Gomez-Casado, C.; Holmkvist, P.; Tavernier, S.J.; Silva-Sanchez, A.; Klotz, L.; Randall, T.D.; Mowat, A.M.; Kotarsky, K.; Malissen, B.; et al. Intestinal cDC1 drive cross-tolerance to epithelial-derived antigen via induction of FoxP3(+)CD8(+) T(regs). Sci. Immunol. 2021, 6, eabd3774. [Google Scholar] [CrossRef] [PubMed]
- Francisco, L.M.; Salinas, V.H.; Brown, K.E.; Vanguri, V.K.; Freeman, G.J.; Kuchroo, V.K.; Sharpe, A.H. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J. Exp. Med. 2009, 206, 3015–3029. [Google Scholar] [CrossRef] [PubMed]
- Bourque, J.; Hawiger, D. Immunomodulatory Bonds of the Partnership between Dendritic Cells and T Cells. Crit. Rev. Immunol. 2018, 38, 379–401. [Google Scholar] [CrossRef] [PubMed]
- Kurts, C.; Heath, W.R.; Kosaka, H.; Miller, J.F.; Carbone, F.R. The peripheral deletion of autoreactive CD8+ T cells induced by cross-presentation of self-antigens involves signaling through CD95 (Fas, Apo-1). J. Exp. Med. 1998, 188, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Steinbrink, K.; Graulich, E.; Kubsch, S.; Knop, J.; Enk, A.H. CD4(+) and CD8(+) anergic T cells induced by interleukin-10-treated human dendritic cells display antigen-specific suppressor activity. Blood 2002, 99, 2468–2476. [Google Scholar] [CrossRef] [PubMed]
- Martin-Orozco, E.; Norte-Munoz, M.; Martinez-Garcia, J. Regulatory T Cells in Allergy and Asthma. Front. Pediatr. 2017, 5, 117. [Google Scholar] [CrossRef]
- Burrack, A.L.; Martinov, T.; Fife, B.T. T Cell-Mediated Beta Cell Destruction: Autoimmunity and Alloimmunity in the Context of Type 1 Diabetes. Front Endocrinol. 2017, 8, 343. [Google Scholar] [CrossRef]
- Deng, Q.; Luo, Y.; Chang, C.; Wu, H.; Ding, Y.; Xiao, R. The Emerging Epigenetic Role of CD8+T Cells in Autoimmune Diseases: A Systematic Review. Front. Immunol. 2019, 10, 856. [Google Scholar] [CrossRef]
- Ganguly, D.; Haak, S.; Sisirak, V.; Reizis, B. The role of dendritic cells in autoimmunity. Nat. Rev. Immunol. 2013, 13, 566–577. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Yang, F.; Zheng, Y.; Ye, T.; Yang, L. Immunomodulatory Role and Therapeutic Potential of Non-Coding RNAs Mediated by Dendritic Cells in Autoimmune and Immune Tolerance-Related Diseases. Front. Immunol. 2021, 12, 678918. [Google Scholar] [CrossRef]
- Sebastian, M.; Lopez-Ocasio, M.; Metidji, A.; Rieder, S.A.; Shevach, E.M.; Thornton, A.M. Helios Controls a Limited Subset of Regulatory T Cell Functions. J. Immunol. 2016, 196, 144–155. [Google Scholar] [CrossRef]
- Shahin, T.; Mayr, D.; Shoeb, M.R.; Kuehn, H.S.; Hoeger, B.; Giuliani, S.; Gawriyski, L.M.; Petronczki, O.Y.; Hadjadj, J.; Bal, S.K.; et al. Identification of germline monoallelic mutations in IKZF2 in patients with immune dysregulation. Blood Adv. 2022, 6, 2444–2451. [Google Scholar] [CrossRef]
- Thornton, A.M.; Korty, P.E.; Tran, D.Q.; Wohlfert, E.A.; Murray, P.E.; Belkaid, Y.; Shevach, E.M. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 2010, 184, 3433–3441. [Google Scholar] [CrossRef]
- Thornton, A.M.; Shevach, E.M. Helios: Still behind the clouds. Immunology 2019, 158, 161–170. [Google Scholar] [CrossRef]
- Gottschalk, R.A.; Corse, E.; Allison, J.P. Expression of Helios in peripherally induced Foxp3+ regulatory T cells. J. Immunol. 2012, 188, 976–980. [Google Scholar] [CrossRef]
- Akimova, T.; Beier, U.H.; Wang, L.; Levine, M.H.; Hancock, W.W. Helios expression is a marker of T cell activation and proliferation. PLoS ONE 2011, 6, e24226. [Google Scholar] [CrossRef]
- Crawford, A.; Angelosanto, J.M.; Kao, C.; Doering, T.A.; Odorizzi, P.M.; Barnett, B.E.; Wherry, E.J. Molecular and transcriptional basis of CD4(+) T cell dysfunction during chronic infection. Immunity 2014, 40, 289–302. [Google Scholar] [CrossRef]
- Ross, E.M.; Bourges, D.; Hogan, T.V.; Gleeson, P.A.; van Driel, I.R. Helios defines T cells being driven to tolerance in the periphery and thymus. Eur. J. Immunol. 2014, 44, 2048–2058. [Google Scholar] [CrossRef]
- Verhagen, J.; Wraith, D.C. Comment on “Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells”. J. Immunol. 2010, 185, 7129. [Google Scholar] [CrossRef]
- Lam, A.J.; Uday, P.; Gillies, J.K.; Levings, M.K. Helios is a marker, not a driver, of human Treg stability. Eur. J. Immunol. 2022, 52, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Paris-Munoz, A.; Aizpurua, G.; Barber, D.F. Helios Expression Is Downregulated on CD8(+) Treg in Two Mouse Models of Lupus During Disease Progression. Front. Immunol. 2022, 13, 922958. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, C.A.; Yeh, W.I.; Seay, H.R.; Saikumar Lakshmi, P.; Chopra, G.; Zhang, L.; Perry, D.J.; McClymont, S.A.; Yadav, M.; Lopez, M.C.; et al. Divergent Phenotypes of Human Regulatory T Cells Expressing the Receptors TIGIT and CD226. J. Immunol. 2015, 195, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Thornton, A.M.; Lu, J.; Korty, P.E.; Kim, Y.C.; Martens, C.; Sun, P.D.; Shevach, E.M. Helios(+) and Helios(-) Treg subpopulations are phenotypically and functionally distinct and express dissimilar TCR repertoires. Eur. J. Immunol. 2019, 49, 398–412. [Google Scholar] [CrossRef] [PubMed]
- Sugita, K.; Hanakawa, S.; Honda, T.; Kondoh, G.; Miyachi, Y.; Kabashima, K.; Nomura, T. Generation of Helios reporter mice and an evaluation of the suppressive capacity of Helios(+) regulatory T cells in vitro. Exp. Dermatol. 2015, 24, 554–556. [Google Scholar] [CrossRef] [PubMed]
- Morina, L.; Jones, M.E.; Oguz, C.; Kaplan, M.J.; Gangaplara, A.; Fitzhugh, C.D.; Kanakry, C.G.; Shevach, E.M.; Buszko, M. Co-expression of Foxp3 and Helios facilitates the identification of human T regulatory cells in health and disease. Front. Immunol. 2023, 14, 1114780. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rodriguez, N.; Apostolidis, S.A.; Penaloza-MacMaster, P.; Martin Villa, J.M.; Barouch, D.H.; Tsokos, G.C.; Crispin, J.C. Programmed cell death 1 and Helios distinguish TCR-alphabeta+ double-negative (CD4−CD8−) T cells that derive from self-reactive CD8 T cells. J. Immunol. 2015, 194, 4207–4214. [Google Scholar] [CrossRef]
- Rodriguez-Rodriguez, N.; Flores-Mendoza, G.; Apostolidis, S.A.; Rosetti, F.; Tsokos, G.C.; Crispin, J.C. TCR-alpha/beta CD4(−) CD8(-) double negative T cells arise from CD8(+) T cells. J. Leukoc. Biol. 2020, 108, 851–857. [Google Scholar] [CrossRef]
- Rodriguez-Rodriguez, N.; Apostolidis, S.A.; Fitzgerald, L.; Meehan, B.S.; Corbett, A.J.; Martin-Villa, J.M.; McCluskey, J.; Tsokos, G.C.; Crispin, J.C. Pro-inflammatory self-reactive T cells are found within murine TCR-alphabeta(+) CD4(−) CD8(−) PD-1(+) cells. Eur. J. Immunol. 2016, 46, 1383–1391. [Google Scholar] [CrossRef]
- Li, H.; Adamopoulos, I.E.; Moulton, V.R.; Stillman, I.E.; Herbert, Z.; Moon, J.J.; Sharabi, A.; Krishfield, S.; Tsokos, M.G.; Tsokos, G.C. Systemic lupus erythematosus favors the generation of IL-17 producing double negative T cells. Nat. Commun. 2020, 11, 2859. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Voelkl, S.; Heymann, J.; Przybylski, G.K.; Mondal, K.; Laumer, M.; Kunz-Schughart, L.; Schmidt, C.A.; Andreesen, R.; Mackensen, A. Isolation and characterization of human antigen-specific TCR alpha beta+ CD4(−)CD8- double-negative regulatory T cells. Blood 2005, 105, 2828–2835. [Google Scholar] [CrossRef] [PubMed]
- Voelkl, S.; Gary, R.; Mackensen, A. Characterization of the immunoregulatory function of human TCR-alphabeta+ CD4- CD8- double-negative T cells. Eur. J. Immunol. 2011, 41, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Juvet, S.C.; Zhang, L. Double negative regulatory T cells in transplantation and autoimmunity: Recent progress and future directions. J. Mol. Cell Biol. 2012, 4, 48–58. [Google Scholar] [CrossRef]
- Yang, L.; Zhu, Y.; Tian, D.; Wang, S.; Guo, J.; Sun, G.; Jin, H.; Zhang, C.; Shi, W.; Gershwin, M.E.; et al. Transcriptome landscape of double negative T cells by single-cell RNA sequencing. J. Autoimmun. 2021, 121, 102653. [Google Scholar] [CrossRef] [PubMed]
- Narni-Mancinelli, E.; Jaeger, B.N.; Bernat, C.; Fenis, A.; Kung, S.; De Gassart, A.; Mahmood, S.; Gut, M.; Heath, S.C.; Estelle, J.; et al. Tuning of natural killer cell reactivity by NKp46 and Helios calibrates T cell responses. Science 2012, 335, 344–348. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, T.; Hwang, I.; Kim, A.; Nitschke, L.; Kim, M.; Scott, J.M.; Kamimura, Y.; Lanier, L.L.; Kim, S. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 2015, 42, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Kucuksezer, U.C.; Aktas Cetin, E.; Esen, F.; Tahrali, I.; Akdeniz, N.; Gelmez, M.Y.; Deniz, G. The Role of Natural Killer Cells in Autoimmune Diseases. Front. Immunol. 2021, 12, 622306. [Google Scholar] [CrossRef]
- Spada, R.; Rojas, J.M.; Perez-Yague, S.; Mulens, V.; Cannata-Ortiz, P.; Bragado, R.; Barber, D.F. NKG2D ligand overexpression in lupus nephritis correlates with increased NK cell activity and differentiation in kidneys but not in the periphery. J. Leukoc. Biol. 2015, 97, 583–598. [Google Scholar] [CrossRef]
- Kaul, A.; Gordon, C.; Crow, M.K.; Touma, Z.; Urowitz, M.B.; van Vollenhoven, R.; Ruiz-Irastorza, G.; Hughes, G. Systemic lupus erythematosus. Nat. Rev. Dis. Primers 2016, 2, 16039. [Google Scholar] [CrossRef]
- Accapezzato, D.; Caccavale, R.; Paroli, M.P.; Gioia, C.; Nguyen, B.L.; Spadea, L.; Paroli, M. Advances in the Pathogenesis and Treatment of Systemic Lupus Erythematosus. Int. J. Mol. Sci. 2023, 24, 6578. [Google Scholar] [CrossRef] [PubMed]
- Pisetsky, D.S.; Lipsky, P.E. New insights into the role of antinuclear antibodies in systemic lupus erythematosus. Nat. Rev. Rheumatol. 2020, 16, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Tsokos, G.C. Autoimmunity and organ damage in systemic lupus erythematosus. Nat. Immunol. 2020, 21, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Aringer, M.; Costenbader, K.; Daikh, D.; Brinks, R.; Mosca, M.; Ramsey-Goldman, R.; Smolen, J.S.; Wofsy, D.; Boumpas, D.T.; Kamen, D.L.; et al. 2019 European League Against Rheumatism/American College of Rheumatology Classification Criteria for Systemic Lupus Erythematosus. Arthritis Amp. Rheumatol. 2019, 71, 1400–1412. [Google Scholar] [CrossRef] [PubMed]
- Laffont, S.; Guery, J.C. Deconstructing the sex bias in allergy and autoimmunity: From sex hormones and beyond. Adv. Immunol. 2019, 142, 35–64. [Google Scholar] [PubMed]
- Ugarte-Gil, M.F.; Gonzalez, L.A.; Alarcon, G.S. Lupus: The new epidemic. Lupus 2019, 28, 1031–1050. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Titov, A.A.; Morel, L. An update on lupus animal models. Curr. Opin. Rheumatol. 2017, 29, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.L.; Gilkeson, G. Mouse models of lupus: What they tell us and what they don’t. Lupus Sci. Med. 2018, 5, e000199. [Google Scholar] [CrossRef]
- Nguyen, C.; Limaye, N.; Wakeland, E.K. Susceptibility genes in the pathogenesis of murine lupus. Arthritis Res. 2002, 4 (Suppl. S3), S255–S263. [Google Scholar] [CrossRef]
- Chen, P.M.; Tsokos, G.C. T Cell Abnormalities in the Pathogenesis of Systemic Lupus Erythematosus: An Update. Curr. Rheumatol. Rep. 2021, 23, 12. [Google Scholar] [CrossRef]
- Hedrich, C.M.; Crispin, J.C.; Rauen, T.; Ioannidis, C.; Apostolidis, S.A.; Lo, M.S.; Kyttaris, V.C.; Tsokos, G.C. cAMP response element modulator α controls IL2 and IL17A expression during CD4 lineage commitment and subset distribution in lupus. Proc. Natl. Acad. Sci. USA 2012, 109, 16606–16611. [Google Scholar] [CrossRef] [PubMed]
- Moulton, V.R.; Tsokos, G.C. T cell signaling abnormalities contribute to aberrant immune cell function and autoimmunity. J. Clin. Investig. 2015, 125, 2220–2227. [Google Scholar] [CrossRef] [PubMed]
- Moulton, V.R.; Suarez-Fueyo, A.; Meidan, E.; Li, H.; Mizui, M.; Tsokos, G.C. Pathogenesis of Human Systemic Lupus Erythematosus: A Cellular Perspective. Trends Mol. Med. 2017, 23, 615–635. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, P.; Ma, L.; Shan, Y.; Jiang, Z.; Wang, J.; Jiang, Y. Increased Interleukin 21 and Follicular Helper T-like Cells and Reduced Interleukin 10+ B cells in Patients with New-onset Systemic Lupus Erythematosus. J. Rheumatol. 2014, 41, 1781–1792. [Google Scholar] [CrossRef]
- Choi, J.-Y.; Ho, J.H.-E.; Pasoto, S.G.; Bunin, V.; Kim, S.T.; Carrasco, S.; Borba, E.F.; Gonçalves, C.R.; Costa, P.R.; Kallas, E.G.; et al. Circulating Follicular Helper-Like T Cells in Systemic Lupus Erythematosus: Association With Disease Activity. Arthritis Rheumatol. 2015, 67, 988–999. [Google Scholar] [CrossRef]
- Singh, R.P.; Bischoff, D.S. Sex Hormones and Gender Influence the Expression of Markers of Regulatory T Cells in SLE Patients. Front. Immunol. 2021, 12, 619268. [Google Scholar] [CrossRef]
- Lin, S.C.; Chen, K.H.; Lin, C.H.; Kuo, C.C.; Ling, Q.D.; Chan, C.H. The quantitative analysis of peripheral blood FOXP3-expressing T cells in systemic lupus erythematosus and rheumatoid arthritis patients. Eur. J. Clin. Investig. 2007, 37, 987–996. [Google Scholar] [CrossRef]
- Ferreira, R.C.; Castro Dopico, X.; Oliveira, J.J.; Rainbow, D.B.; Yang, J.H.; Trzupek, D.; Todd, S.A.; McNeill, M.; Steri, M.; Orru, V.; et al. Chronic Immune Activation in Systemic Lupus Erythematosus and the Autoimmune PTPN22 Trp(620) Risk Allele Drive the Expansion of FOXP3(+) Regulatory T Cells and PD-1 Expression. Front. Immunol. 2019, 10, 2606. [Google Scholar] [CrossRef]
- Li, W.; Deng, C.; Yang, H.; Wang, G. The Regulatory T Cell in Active Systemic Lupus Erythematosus Patients: A Systemic Review and Meta-Analysis. Front. Immunol. 2019, 10, 159. [Google Scholar] [CrossRef]
- Venigalla, R.K.; Tretter, T.; Krienke, S.; Max, R.; Eckstein, V.; Blank, N.; Fiehn, C.; Ho, A.D.; Lorenz, H.M. Reduced CD4+,CD25- T cell sensitivity to the suppressive function of CD4+,CD25high,CD127-/low regulatory T cells in patients with active systemic lupus erythematosus. Arthritis Rheum. 2008, 58, 2120–2130. [Google Scholar] [CrossRef]
- Bonelli, M.; Savitskaya, A.; von Dalwigk, K.; Steiner, C.W.; Aletaha, D.; Smolen, J.S.; Scheinecker, C. Quantitative and qualitative deficiencies of regulatory T cells in patients with systemic lupus erythematosus (SLE). Int. Immunol. 2008, 20, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Ye, S.; Chen, G.; Kuang, M.; Shen, N.; Chen, S. Dysfunctional CD4+,CD25+ regulatory T cells in untreated active systemic lupus erythematosus secondary to interferon-alpha-producing antigen-presenting cells. Arthritis Rheum. 2008, 58, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.Q.; Ji, N.F.; Gu, C.J.; Wang, Y.L.; Huang, M.; Zhang, M.S. Coexpression of Helios in Foxp3(+) Regulatory T Cells and Its Role in Human Disease. Dis. Markers 2021, 2021, 5574472. [Google Scholar] [CrossRef] [PubMed]
- Alexander, T.; Sattler, A.; Templin, L.; Kohler, S.; Gross, C.; Meisel, A.; Sawitzki, B.; Burmester, G.R.; Arnold, R.; Radbruch, A.; et al. Foxp3+ Helios+ regulatory T cells are expanded in active systemic lupus erythematosus. Ann. Rheum. Dis. 2013, 72, 1549–1558. [Google Scholar] [CrossRef] [PubMed]
- Golding, A.; Hasni, S.; Illei, G.; Shevach, E.M. The percentage of FoxP3+Helios+ Treg cells correlates positively with disease activity in systemic lupus erythematosus. Arthritis Rheum. 2013, 65, 2898–2906. [Google Scholar] [CrossRef] [PubMed]
- Gravano, D.M.; Hoyer, K.K. Promotion and prevention of autoimmune disease by CD8+ T cells. J. Autoimmun. 2013, 45, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Fueyo, A.; Bradley, S.J.; Tsokos, G.C. T cells in Systemic Lupus Erythematosus. Curr. Opin. Immunol. 2016, 43, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.M.; Tsokos, G.C. The role of CD8+ T-cell systemic lupus erythematosus pathogenesis: An update. Curr. Opin. Rheumatol. 2021, 33, 586–591. [Google Scholar] [CrossRef]
- Fors Nieves, C.E.; Izmirly, P.M. Mortality in Systemic Lupus Erythematosus: An Updated Review. Curr. Rheumatol. Rep. 2016, 18, 21. [Google Scholar] [CrossRef]
- Peng, S.L.; Moslehi, J.; Robert, M.E.; Craft, J. Perforin protects against autoimmunity in lupus-prone mice. J. Immunol. 1998, 160, 652–660. [Google Scholar] [CrossRef]
- Holderried, T.A.; Kruse, M.; Serries, M.; Brossart, P.; Wolf, D.; Wolf, A.M. Helios-expressing CD8(+) T cells are decreased in patients with systemic lupus erythematosus. Lupus 2021, 30, 1022–1024. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tsokos, G.C. Double-negative T cells in autoimmune diseases. Curr. Opin. Rheumatol. 2021, 33, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zheng, Y.; Sheng, J.; Han, Y.; Yang, Y.; Pan, H.; Yao, J. CD3(+)CD4(−)CD8(−) (Double-Negative) T Cells in Inflammation, Immune Disorders and Cancer. Front. Immunol. 2022, 13, 816005. [Google Scholar] [CrossRef] [PubMed]
- Shivakumar, S.; Tsokos, G.C.; Datta, S.K. T cell receptor alpha/beta expressing double-negative (CD4−/CD8−) and CD4+ T helper cells in humans augment the production of pathogenic anti-DNA autoantibodies associated with lupus nephritis. J. Immunol. 1989, 143, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Crispin, J.C.; Oukka, M.; Bayliss, G.; Cohen, R.A.; Van Beek, C.A.; Stillman, I.E.; Kyttaris, V.C.; Juang, Y.T.; Tsokos, G.C. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J. Immunol. 2008, 181, 8761–8766. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.J.; Jacob, A.; Chang, A.; Quigg, R.J.; Jarvis, J.N. Double negative T cells, a potential biomarker for systemic lupus erythematosus. Precis. Clin. Med. 2020, 3, 34–43. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, Z.A.; El-Owaidy, R.H.; Mohamed, N.L.; Shehata, B.A. Alpha beta double negative T cells in children with systemic lupus erythematosus: The relation to disease activity and characteristics. Mod. Rheumatol. 2018, 28, 654–660. [Google Scholar] [CrossRef]
- Klarquist, J.; Zhou, Z.; Shen, N.; Janssen, E.M. Dendritic Cells in Systemic Lupus Erythematosus: From Pathogenic Players to Therapeutic Tools. Mediat. Inflamm. 2016, 2016, 5045248. [Google Scholar] [CrossRef]
- Kaewraemruaen, C.; Ritprajak, P.; Hirankarn, N. Dendritic cells as key players in systemic lupus erythematosus. Asian Pac. J. Allergy Immunol. 2020, 38, 225–232. [Google Scholar]
- Liu, J.; Zhang, X.; Cao, X. Dendritic cells in systemic lupus erythematosus: From pathogenesis to therapeutic applications. J. Autoimmun. 2022, 132, 102856. [Google Scholar] [CrossRef]
- Shodell, M.; Shah, K.; Siegal, F.P. Circulating human plasmacytoid dendritic cells are highly sensitive to corticosteroid administration. Lupus 2003, 12, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Blanco, P.; Palucka, A.K.; Gill, M.; Pascual, V.; Banchereau, J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science 2001, 294, 1540–1543. [Google Scholar] [CrossRef] [PubMed]
- Fiore, N.; Castellano, G.; Blasi, A.; Capobianco, C.; Loverre, A.; Montinaro, V.; Netti, S.; Torres, D.; Manno, C.; Grandaliano, G.; et al. Immature myeloid and plasmacytoid dendritic cells infiltrate renal tubulointerstitium in patients with lupus nephritis. Mol. Immunol. 2008, 45, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Scheinecker, C.; Zwolfer, B.; Koller, M.; Manner, G.; Smolen, J.S. Alterations of dendritic cells in systemic lupus erythematosus: Phenotypic and functional deficiencies. Arthritis Rheum. 2001, 44, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Mozaffarian, N.; Wiedeman, A.E.; Stevens, A.M. Active systemic lupus erythematosus is associated with failure of antigen-presenting cells to express programmed death ligand-1. Rheumatology 2008, 47, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Koller, M.; Zwolfer, B.; Steiner, G.; Smolen, J.S.; Scheinecker, C. Phenotypic and functional deficiencies of monocyte-derived dendritic cells in systemic lupus erythematosus (SLE) patients. Int. Immunol. 2004, 16, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Mehta, H.; McCune, W.J.; Kaplan, M.J. Aberrant phenotype and function of myeloid dendritic cells in systemic lupus erythematosus. J. Immunol. 2006, 177, 5878–5889. [Google Scholar] [CrossRef]
- Li, Z.; Ju, X.; Silveira, P.A.; Abadir, E.; Hsu, W.H.; Hart, D.N.J.; Clark, G.J. CD83: Activation Marker for Antigen Presenting Cells and Its Therapeutic Potential. Front. Immunol. 2019, 10, 1312. [Google Scholar] [CrossRef]
- Mahajan, A.; Herrmann, M.; Munoz, L.E. Clearance Deficiency and Cell Death Pathways: A Model for the Pathogenesis of SLE. Front. Immunol. 2016, 7, 35. [Google Scholar] [CrossRef]
- Sim, T.M.; Ong, S.J.; Mak, A.; Tay, S.H. Type I Interferons in Systemic Lupus Erythematosus: A Journey from Bench to Bedside. Int. J. Mol. Sci. 2022, 23, 2505. [Google Scholar] [CrossRef]
- Rönnblom, L.; Leonard, D. Interferon pathway in SLE: One key to unlocking the mystery of the disease. Lupus Sci. Med. 2019, 6, e000270. [Google Scholar] [CrossRef]
- Karnell, J.L.; Wu, Y.; Mittereder, N.; Smith, M.A.; Gunsior, M.; Yan, L.; Casey, K.A.; Henault, J.; Riggs, J.M.; Nicholson, S.M.; et al. Depleting plasmacytoid dendritic cells reduces local type I interferon responses and disease activity in patients with cutaneous lupus. Sci. Transl. Med. 2021, 13, eabf8442. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, F.; Natalucci, F.; Picciariello, L.; Ciancarella, C.; Dolcini, G.; Gattamelata, A.; Alessandri, C.; Conti, F. Application of Machine Learning Models in Systemic Lupus Erythematosus. Int. J. Mol. Sci. 2023, 24, 4514. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.A.; Ugarte-Gil, M.F.; Alarcon, G.S. Systemic lupus erythematosus: The search for the ideal biomarker. Lupus 2021, 30, 181–203. [Google Scholar] [CrossRef] [PubMed]
- Bentham, J.; Morris, D.L.; Graham, D.S.C.; Pinder, C.L.; Tombleson, P.; Behrens, T.W.; Martín, J.; Fairfax, B.P.; Knight, J.C.; Chen, L.; et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015, 47, 1457–1464. [Google Scholar] [CrossRef]
- Zhou, M.; Kang, Y.; Li, J.; Li, R.; Lu, L. Omics-based integrated analysis identified IKZF2 as a biomarker associated with lupus nephritis. Sci. Rep. 2022, 12, 9612. [Google Scholar] [CrossRef]
- Robinson, J.P.; Ostafe, R.; Iyengar, S.N.; Rajwa, B.; Fischer, R. Flow Cytometry: The Next Revolution. Cells 2023, 12, 1875. [Google Scholar] [CrossRef]
- Shipa, M.; Santos, L.R.; Nguyen, D.X.; Embleton-Thirsk, A.; Parvaz, M.; Heptinstall, L.L.; Pepper, R.J.; Isenberg, D.A.; Gordon, C.; Ehrenstein, M.R. Identification of biomarkers to stratify response to B-cell-targeted therapies in systemic lupus erythematosus: An exploratory analysis of a randomised controlled trial. Lancet Rheumatol. 2023, 5, e24–e35. [Google Scholar] [CrossRef]
- Chen, L.; Hasni, M.S.; Jondal, M.; Yakimchuk, K. Modification of anti-tumor immunity by tolerogenic dendritic cells. Autoimmunity 2017, 50, 370–376. [Google Scholar] [CrossRef]
- Mahic, M.; Henjum, K.; Yaqub, S.; Bjornbeth, B.A.; Torgersen, K.M.; Tasken, K.; Aandahl, E.M. Generation of highly suppressive adaptive CD8(+)CD25(+)FOXP3(+) regulatory T cells by continuous antigen stimulation. Eur. J. Immunol. 2008, 38, 640–646. [Google Scholar] [CrossRef]
- Seeger, P.; Musso, T.; Sozzani, S. The TGF-beta superfamily in dendritic cell biology. Cytokine Growth Factor. Rev. 2015, 26, 647–657. [Google Scholar] [CrossRef]
- Ohnmacht, C.; Pullner, A.; King, S.B.; Drexler, I.; Meier, S.; Brocker, T.; Voehringer, D. Constitutive ablation of dendritic cells breaks self-tolerance of CD4 T cells and results in spontaneous fatal autoimmunity. J. Exp. Med. 2009, 206, 549–559. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, X.; Cao, F.; Bellanti, J.A.; Zhou, J.; Zheng, S.G. Prospects of the Use of Cell Therapy to Induce Immune Tolerance. Front. Immunol. 2020, 11, 792. [Google Scholar] [CrossRef]
- Wang, X.; Lu, L.; Jiang, S. Regulatory T cells: Customizing for the clinic. Sci. Transl. Med. 2011, 3, 83ps19. [Google Scholar] [CrossRef]
- Dall’Era, M.; Pauli, M.L.; Remedios, K.; Taravati, K.; Sandova, P.M.; Putnam, A.L.; Lares, A.; Haemel, A.; Tang, Q.; Hellerstein, M.; et al. Adoptive Treg Cell Therapy in a Patient with Systemic Lupus Erythematosus. Arthritis Rheumatol. 2019, 71, 431–440. [Google Scholar] [CrossRef]
- Ghobadinezhad, F.; Ebrahimi, N.; Mozaffari, F.; Moradi, N.; Beiranvand, S.; Pournazari, M.; Rezaei-Tazangi, F.; Khorram, R.; Afshinpour, M.; Robino, R.A.; et al. The emerging role of regulatory cell-based therapy in autoimmune disease. Front. Immunol. 2022, 13, 1075813. [Google Scholar] [CrossRef]
- Passeri, L.; Marta, F.; Bassi, V.; Gregori, S. Tolerogenic Dendritic Cell-Based Approaches in Autoimmunity. Int. J. Mol. Sci. 2021, 22, 8415. [Google Scholar] [CrossRef] [PubMed]
- Seyfizadeh, N.; Muthuswamy, R.; Mitchell, D.A.; Nierkens, S.; Seyfizadeh, N. Migration of dendritic cells to the lymph nodes and its enhancement to drive anti-tumor responses. Crit. Rev. Oncol./Hematol. 2016, 107, 100–110. [Google Scholar] [CrossRef]
- Adema, G.J.; de Vries, I.J.; Punt, C.J.; Figdor, C.G. Migration of dendritic cell based cancer vaccines: In vivo veritas? Curr. Opin. Immunol. 2005, 17, 170–174. [Google Scholar] [CrossRef]
- MartIn-Fontecha, A.; Sebastiani, S.; Höpken, U.E.; Uguccioni, M.; Lipp, M.; Lanzavecchia, A.; Sallusto, F. Regulation of dendritic cell migration to the draining lymph node: Impact on T lymphocyte traffic and priming. J. Exp. Med. 2003, 198, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Mansilla, M.J.; Hilkens, C.M.U.; Martinez-Caceres, E.M. Challenges in tolerogenic dendritic cell therapy for autoimmune diseases: The route of administration. Immunother. Adv. 2023, 3, ltad012. [Google Scholar] [CrossRef]
- Okada, N.; Mori, N.; Koretomo, R.; Okada, Y.; Nakayama, T.; Yoshie, O.; Mizuguchi, H.; Hayakawa, T.; Nakagawa, S.; Mayumi, T.; et al. Augmentation of the migratory ability of DC-based vaccine into regional lymph nodes by efficient CCR7 gene transduction. Gene Ther. 2005, 12, 129–139. [Google Scholar] [CrossRef]
- Jin, H.; Qian, Y.; Dai, Y.; Qiao, S.; Huang, C.; Lu, L.; Luo, Q.; Chen, J.; Zhang, Z. Magnetic Enrichment of Dendritic Cell Vaccine in Lymph Node with Fluorescent-Magnetic Nanoparticles Enhanced Cancer Immunotherapy. Theranostics 2016, 6, 2000–2014. [Google Scholar] [CrossRef]
- Mohammadi, B.; Saghafi, M.; Abdulsattar Faraj, T.; Kamal Kheder, R.; Sajid Abdulabbas, H.; Esmaeili, S.A. The role of tolerogenic dendritic cells in systematic lupus erythematosus progression and remission. Int. Immunopharmacol. 2023, 115, 109601. [Google Scholar] [CrossRef]
- Funes, S.C.; Rios, M.; Gomez-Santander, F.; Fernandez-Fierro, A.; Altamirano-Lagos, M.J.; Rivera-Perez, D.; Pulgar-Sepulveda, R.; Jara, E.L.; Rebolledo-Zelada, D.; Villarroel, A.; et al. Tolerogenic dendritic cell transfer ameliorates systemic lupus erythematosus in mice. Immunology 2019, 158, 322–339. [Google Scholar] [CrossRef]
- Wu, H.J.; Lo, Y.; Luk, D.; Lau, C.S.; Lu, L.; Mok, M.Y. Alternatively activated dendritic cells derived from systemic lupus erythematosus patients have tolerogenic phenotype and function. Clin. Immunol. 2015, 156, 43–57. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Obreque, J.; Vega, F.; Torres, A.; Cuitino, L.; Mackern-Oberti, J.P.; Viviani, P.; Kalergis, A.; Llanos, C. Autologous tolerogenic dendritic cells derived from monocytes of systemic lupus erythematosus patients and healthy donors show a stable and immunosuppressive phenotype. Immunology 2017, 152, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Tinkle, S.; McNeil, S.E.; Muhlebach, S.; Bawa, R.; Borchard, G.; Barenholz, Y.C.; Tamarkin, L.; Desai, N. Nanomedicines: Addressing the scientific and regulatory gap. Ann. N. Y Acad. Sci. 2014, 1313, 35–56. [Google Scholar] [CrossRef] [PubMed]
- Pita, R.; Ehmann, F.; Papaluca, M. Nanomedicines in the EU-Regulatory Overview. AAPS J. 2016, 18, 1576–1582. [Google Scholar] [CrossRef]
- Sousa De Almeida, M.; Susnik, E.; Drasler, B.; Taladriz-Blanco, P.; Petri-Fink, A.; Rothen-Rutishauser, B. Understanding nanoparticle endocytosis to improve targeting strategies in nanomedicine. Chem. Soc. Rev. 2021, 50, 5397–5434. [Google Scholar] [CrossRef]
- Soares, S.; Sousa, J.; Pais, A.; Vitorino, C. Nanomedicine: Principles, Properties, and Regulatory Issues. Front. Chem. 2018, 6, 360. [Google Scholar] [CrossRef]
- Altammar, K.A. A review on nanoparticles: Characteristics, synthesis, applications, and challenges. Front. Microbiol. 2023, 14, 1155622. [Google Scholar] [CrossRef]
- Sztandera, K.; Gorzkiewicz, M.; Klajnert-Maculewicz, B. Gold Nanoparticles in Cancer Treatment. Mol. Pharm. 2019, 16, 1–23. [Google Scholar] [CrossRef]
- Bruna, T.; Maldonado-Bravo, F.; Jara, P.; Caro, N. Silver Nanoparticles and Their Antibacterial Applications. Int. J. Mol. Sci. 2021, 22, 7202. [Google Scholar] [CrossRef]
- Kolosnjaj-Tabi, J.; Lartigue, L.; Javed, Y.; Luciani, N.; Pellegrino, T.; Wilhelm, C.; Alloyeau, D.; Gazeau, F. Biotransformations of magnetic nanoparticles in the body. Nano Today 2016, 11, 280–284. [Google Scholar] [CrossRef]
- Rojas, J.M.; Gavilan, H.; Del Dedo, V.; Lorente-Sorolla, E.; Sanz-Ortega, L.; da Silva, G.B.; Costo, R.; Perez-Yague, S.; Talelli, M.; Marciello, M.; et al. Time-course assessment of the aggregation and metabolization of magnetic nanoparticles. Acta Biomater. 2017, 58, 181–195. [Google Scholar] [CrossRef]
- Schubert, J.; Chanana, M. Coating Matters: Review on Colloidal Stability of Nanoparticles with Biocompatible Coatings in Biological Media, Living Cells and Organisms. Curr. Med. Chem. 2018, 25, 4553–4586. [Google Scholar] [CrossRef]
- Portilla, Y.; Mellid, S.; Paradela, A.; Ramos-Fernández, A.; Daviu, N.; Sanz-Ortega, L.; Pérez-Yagüe, S.; Morales, M.P.; Barber, D.F. Iron Oxide Nanoparticle Coatings Dictate Cell Outcomes Despite the Influence of Protein Coronas. ACS Appl. Mater. Interfaces 2021, 13, 7924–7944. [Google Scholar] [CrossRef]
- Portilla, Y.; Mulens-Arias, V.; Paradela, A.; Ramos-Fernández, A.; Pérez-Yagüe, S.; Morales, M.P.; Barber, D.F. The surface coating of iron oxide nanoparticles drives their intracellular trafficking and degradation in endolysosomes differently depending on the cell type. Biomaterials 2022, 281, 121365. [Google Scholar] [CrossRef]
- Sanz-Ortega, L.; Rojas, J.M.; Marcos, A.; Portilla, Y.; Stein, J.V.; Barber, D.F. T cells loaded with magnetic nanoparticles are retained in peripheral lymph nodes by the application of a magnetic field. J. Nanobiotechnology 2019, 17, 14. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Barua, S.; Sharma, G.; Dey, S.K.; Rege, K. Inorganic nanoparticles for cancer imaging and therapy. J. Control Release 2011, 155, 344–357. [Google Scholar] [CrossRef]
- Bao, G.; Mitragotri, S.; Tong, S. Multifunctional nanoparticles for drug delivery and molecular imaging. Annu. Rev. Biomed. Eng. 2013, 15, 253–282. [Google Scholar] [CrossRef]
- Gupta, R.; Sharma, D. Evolution of Magnetic Hyperthermia for Glioblastoma Multiforme Therapy. ACS Chem. Neurosci. 2019, 10, 1157–1172. [Google Scholar] [CrossRef]
- Mejías, R.; Hernández Flores, P.; Talelli, M.; Tajada-Herráiz, J.L.; Brollo, M.E.F.; Portilla, Y.; Morales, M.P.; Barber, D.F. Cell-Promoted Nanoparticle Aggregation Decreases Nanoparticle-Induced Hyperthermia under an Alternating Magnetic Field Independently of Nanoparticle Coating, Core Size, and Subcellular Localization. ACS Appl. Mater. Interfaces 2019, 11, 340–355. [Google Scholar] [CrossRef]
- Egea-Benavente, D.; Ovejero, J.G.; Morales, M.D.P.; Barber, D.F. Understanding MNPs Behaviour in Response to AMF in Biological Milieus and the Effects at the Cellular Level: Implications for a Rational Design That Drives Magnetic Hyperthermia Therapy toward Clinical Implementation. Cancers 2021, 13, 4583. [Google Scholar] [CrossRef]
- Qiao, R.; Yang, C.; Gao, M. Superparamagnetic iron oxide nanoparticles: From preparations to in vivo MRI applications. J. Mater. Chem. 2009, 19, 6274–6293. [Google Scholar] [CrossRef]
- Avasthi, A.; Caro, C.; Pozo-Torres, E.; Leal, M.P.; García-Martín, M.L. Magnetic Nanoparticles as MRI Contrast Agents. Top Curr. Chem. 2020, 378, 40. [Google Scholar] [CrossRef]
- Sanz-Ortega, L.; Rojas, J.M.; Barber, D.F. Improving Tumor Retention of Effector Cells in Adoptive Cell Transfer Therapies by Magnetic Targeting. Pharmaceutics 2020, 12, 812. [Google Scholar] [CrossRef]
- Chen, Y.; Hou, S. Application of magnetic nanoparticles in cell therapy. Stem Cell Res. Ther. 2022, 13, 135. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Zhang, Y.; Xin, Y.; Jiang, C.; Yan, B.; Zhai, S. Interactions Between Nanoparticles and Dendritic Cells: From the Perspective of Cancer Immunotherapy. Front. Oncol. 2018, 8, 404. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Dobrovolskaia, M.A. Immunological effects of iron oxide nanoparticles and iron-based complex drug formulations: Therapeutic benefits, toxicity, mechanistic insights, and translational considerations. Nanomedicine 2018, 14, 977–990. [Google Scholar] [CrossRef]
- Chung, S.; Revia, R.A.; Zhang, M. Iron oxide nanoparticles for immune cell labeling and cancer immunotherapy. Nanoscale Horiz. 2021, 6, 696–717. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.G.; Sun, T. Nanoparticle-Based Drug Delivery Systems for Induction of Tolerance and Treatment of Autoimmune Diseases. Front. Bioeng. Biotechnol. 2022, 10, 889291. [Google Scholar] [CrossRef]
- Cifuentes-Rius, A.; Desai, A.; Yuen, D.; Johnston, A.P.R.; Voelcker, N.H. Inducing immune tolerance with dendritic cell-targeting nanomedicines. Nat. Nanotechnol. 2021, 16, 37–46. [Google Scholar] [CrossRef]
- Polyak, B.; Fishbein, I.; Chorny, M.; Alferiev, I.; Williams, D.; Yellen, B.; Friedman, G.; Levy, R.J. High field gradient targeting of magnetic nanoparticle-loaded endothelial cells to the surfaces of steel stents. Proc. Natl. Acad. Sci. USA 2008, 105, 698–703. [Google Scholar] [CrossRef]
- Tukmachev, D.; Lunov, O.; Zablotskii, V.; Dejneka, A.; Babic, M.; Sykova, E.; Kubinova, S. An effective strategy of magnetic stem cell delivery for spinal cord injury therapy. Nanoscale 2015, 7, 3954–3958. [Google Scholar] [CrossRef]
- Luo, Z.; Luo, L.; Lu, Y.; Zhu, C.; Qin, B.; Jiang, M.; Li, X.; Shi, Y.; Zhang, J.; Liu, Y.; et al. Dual-binding nanoparticles improve the killing effect of T cells on solid tumor. J. Nanobiotechnology 2022, 20, 261. [Google Scholar] [CrossRef]
- Jang, E.-S.; Shin, J.-H.; Ren, G.; Park, M.-J.; Cheng, K.; Chen, X.; Wu, J.C.; Sunwoo, J.B.; Cheng, Z. The manipulation of natural killer cells to target tumor sites using magnetic nanoparticles. Biomaterials 2012, 33, 5584–5592. [Google Scholar] [CrossRef] [PubMed]
- Boosz, P.; Pfister, F.; Stein, R.; Friedrich, B.; Fester, L.; Band, J.; Mühlberger, M.; Schreiber, E.; Lyer, S.; Dudziak, D.; et al. Citrate-Coated Superparamagnetic Iron Oxide Nanoparticles Enable a Stable Non-Spilling Loading of T Cells and Their Magnetic Accumulation. Cancers 2021, 13, 4143. [Google Scholar] [CrossRef]
- Pfister, F.; Dorrie, J.; Schaft, N.; Buchele, V.; Unterweger, H.; Carnell, L.R.; Schreier, P.; Stein, R.; Kubankova, M.; Guck, J.; et al. Human T cells loaded with superparamagnetic iron oxide nanoparticles retain antigen-specific TCR functionality. Front. Immunol. 2023, 14, 1223695. [Google Scholar] [CrossRef]
- Muhlberger, M.; Janko, C.; Unterweger, H.; Friedrich, R.P.; Friedrich, B.; Band, J.; Cebulla, N.; Alexiou, C.; Dudziak, D.; Lee, G.; et al. Functionalization of T Lymphocytes with Citrate-Coated Superparamagnetic Iron Oxide Nanoparticles for Magnetically Controlled Immune Therapy. Int. J. Nanomed. 2019, 14, 8421–8432. [Google Scholar] [CrossRef]
- Briley-Saebo, K.C.; Leboeuf, M.; Dickson, S.; Mani, V.; Fayad, Z.A.; Palucka, A.K.; Banchereau, J.; Merad, M. Longitudinal tracking of human dendritic cells in murine models using magnetic resonance imaging. Magn. Reson. Med. 2010, 64, 1510–1519. [Google Scholar] [CrossRef]
- Mou, Y.; Chen, B.; Zhang, Y.; Hou, Y.; Xie, H.; Xia, G.; Tang, M.; Huang, X.; Ni, Y.; Hu, Q. Influence of synthetic superparamagnetic iron oxide on dendritic cells. Int. J. Nanomed. 2011, 6, 1779–1786. [Google Scholar][Green Version]
- Mou, Y.; Hou, Y.; Chen, B.; Hua, Z.; Zhang, Y.; Xie, H.; Xia, G.; Wang, Z.; Huang, X.; Han, W.; et al. In vivo migration of dendritic cells labeled with synthetic superparamagnetic iron oxide. Int. J. Nanomed. 2011, 6, 2633–2640. [Google Scholar][Green Version]
- de Chickera, S.; Willert, C.; Mallet, C.; Foley, R.; Foster, P.; Dekaban, G.A. Cellular MRI as a suitable, sensitive non-invasive modality for correlating in vivo migratory efficiencies of different dendritic cell populations with subsequent immunological outcomes. Int. Immunol. 2011, 24, 29–41. [Google Scholar] [CrossRef][Green Version]
- de Chickera, S.N.; Snir, J.; Willert, C.; Rohani, R.; Foley, R.; Foster, P.J.; Dekaban, G.A. Labelling dendritic cells with SPIO has implications for their subsequent in vivo migration as assessed with cellular MRI. Contrast Media Mol. Imaging 2011, 6, 314–327. [Google Scholar] [CrossRef]
- Su, H.; Mou, Y.; An, Y.; Han, W.; Huang, X.; Xia, G.; Ni, Y.; Zhang, Y.; Ma, J.; Hu, Q. The migration of synthetic magnetic nanoparticle labeled dendritic cells into lymph nodes with optical imaging. Int. J. Nanomed. 2013, 8, 3737–3744. [Google Scholar][Green Version]
- Xu, Y.; Wu, C.; Zhu, W.; Xia, C.; Wang, D.; Zhang, H.; Wu, J.; Lin, G.; Wu, B.; Gong, Q.; et al. Superparamagnetic MRI probes for in vivo tracking of dendritic cell migration with a clinical 3 T scanner. Biomaterials 2015, 58, 63–71. [Google Scholar] [CrossRef]
- Liu, H.; Dong, H.; Zhou, N.; Dong, S.; Chen, L.; Zhu, Y.; Hu, H.M.; Mou, Y. SPIO Enhance the Cross-Presentation and Migration of DCs and Anionic SPIO Influence the Nanoadjuvant Effects Related to Interleukin-1beta. Nanoscale Res. Lett. 2018, 13, 409. [Google Scholar] [CrossRef] [PubMed]
- Dekaban, G.A.; Snir, J.; Shrum, B.; de Chickera, S.; Willert, C.; Merrill, M.; Said, E.A.; Sekaly, R.P.; Foster, P.J.; O’Connell, P.J. Semiquantitation of mouse dendritic cell migration in vivo using cellular MRI. J. Immunother. 2009, 32, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Blank, F.; Gerber, P.; Rothen-Rutishauser, B.; Sakulkhu, U.; Salaklang, J.; De Peyer, K.; Gehr, P.; Nicod, L.P.; Hofmann, H.; Geiser, T.; et al. Biomedical nanoparticles modulate specific CD4+ T cell stimulation by inhibition of antigen processing in dendritic cells. Nanotoxicology 2011, 5, 606–621. [Google Scholar] [CrossRef]
- Verdijk, P.; Scheenen, T.W.; Lesterhuis, W.J.; Gambarota, G.; Veltien, A.A.; Walczak, P.; Scharenborg, N.M.; Bulte, J.W.; Punt, C.J.; Heerschap, A.; et al. Sensitivity of magnetic resonance imaging of dendritic cells for in vivo tracking of cellular cancer vaccines. Int. J. Cancer 2007, 120, 978–984. [Google Scholar] [CrossRef]
- Mou, Y.; Xing, Y.; Ren, H.; Cui, Z.; Zhang, Y.; Yu, G.; Urba, W.J.; Hu, Q.; Hu, H. The Effect of Superparamagnetic Iron Oxide Nanoparticle Surface Charge on Antigen Cross-Presentation. Nanoscale Res. Lett. 2017, 12, 52. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).