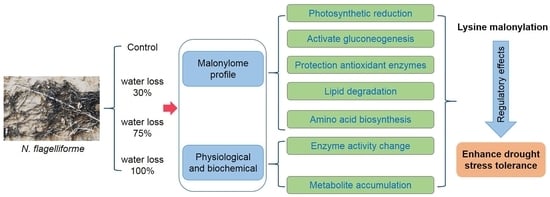
The Enzyme Lysine Malonylation of Calvin Cycle and Gluconeogenesis Regulated Glycometabolism in Nostoc flagelliforme to Adapt to Drought Stress
Abstract
1. Introduction
2. Results
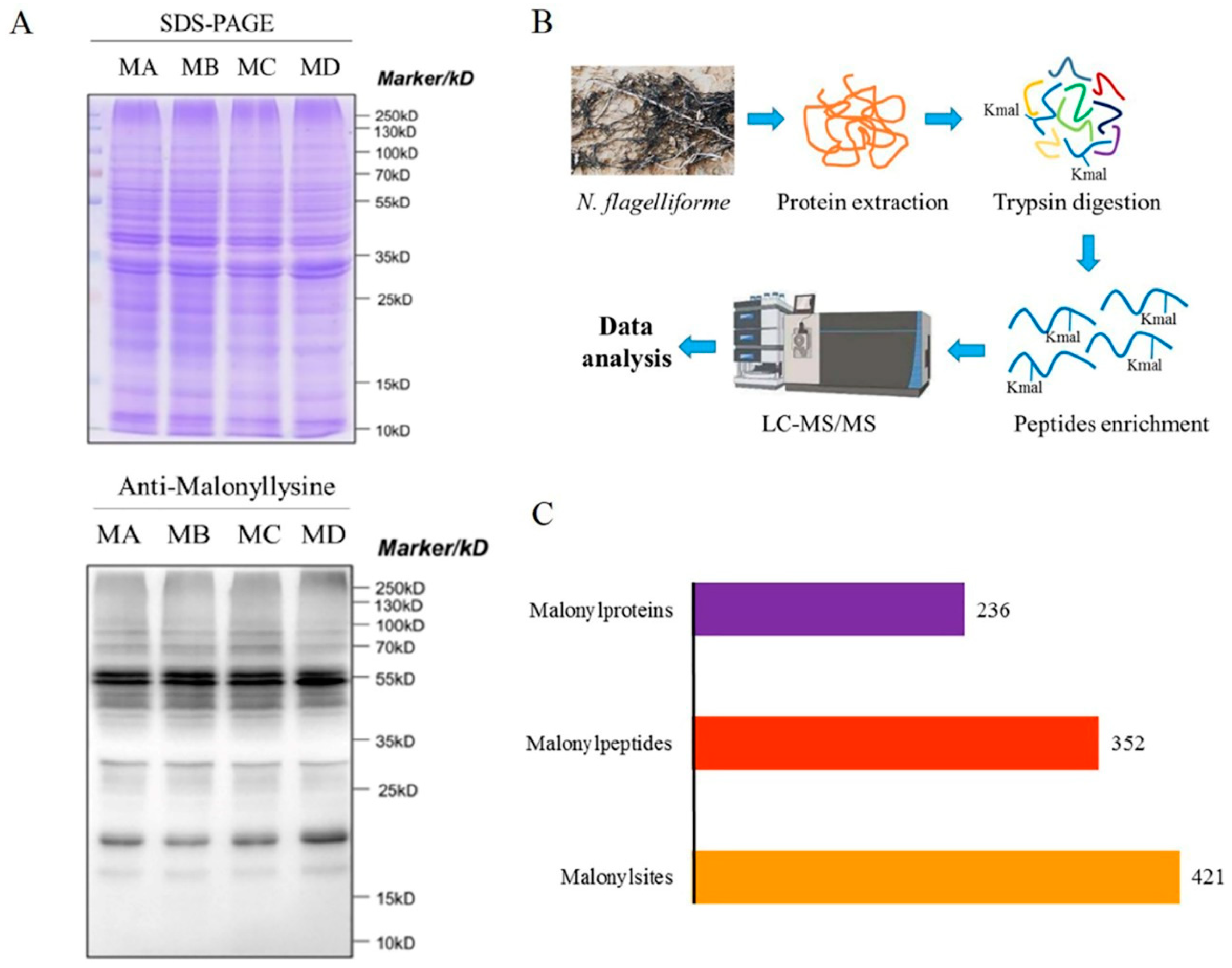
2.1. Determination of Kmal in N. flagelliforme
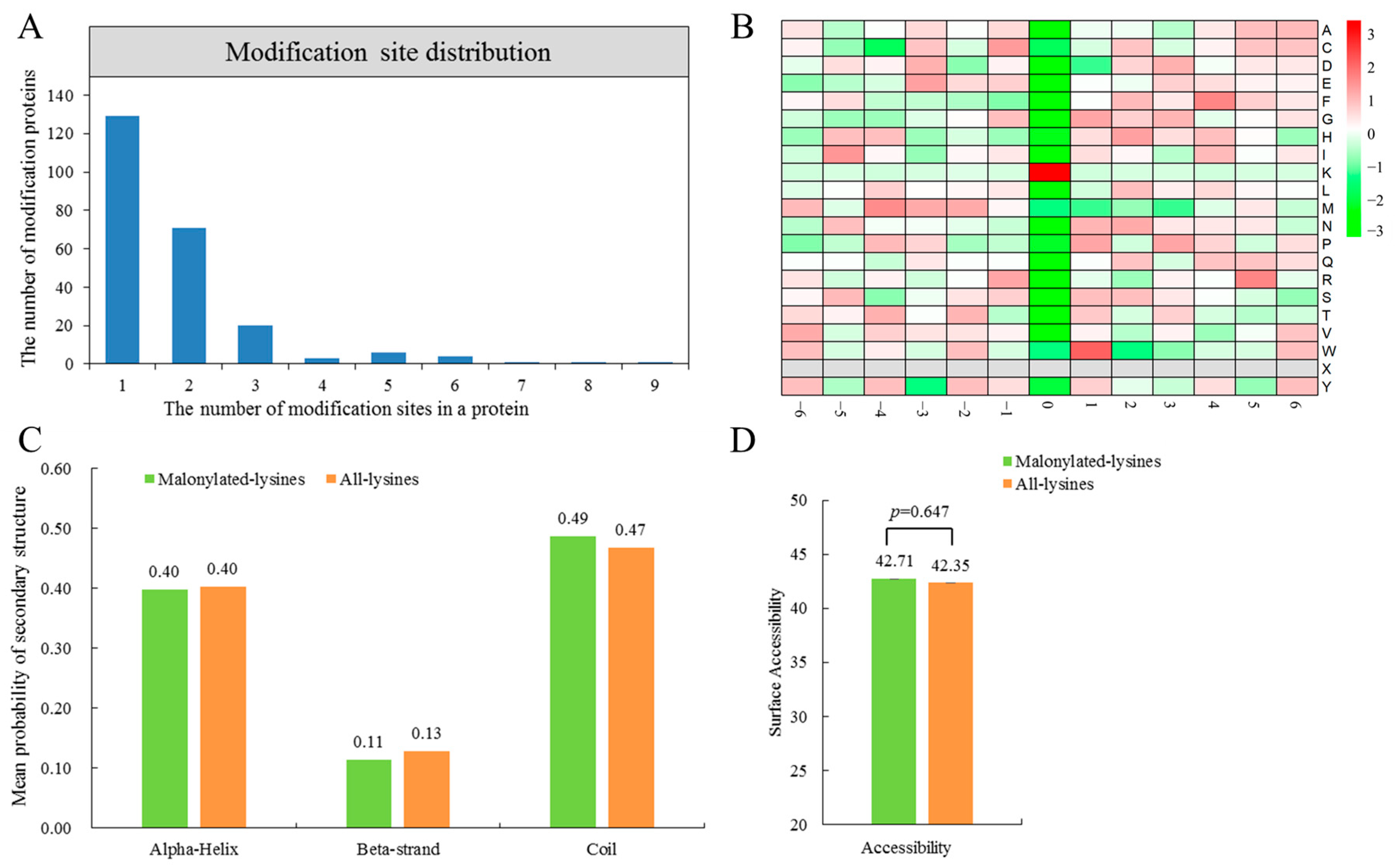
2.2. Analysis of Malonylated Lysine Residues in N. flagelliforme
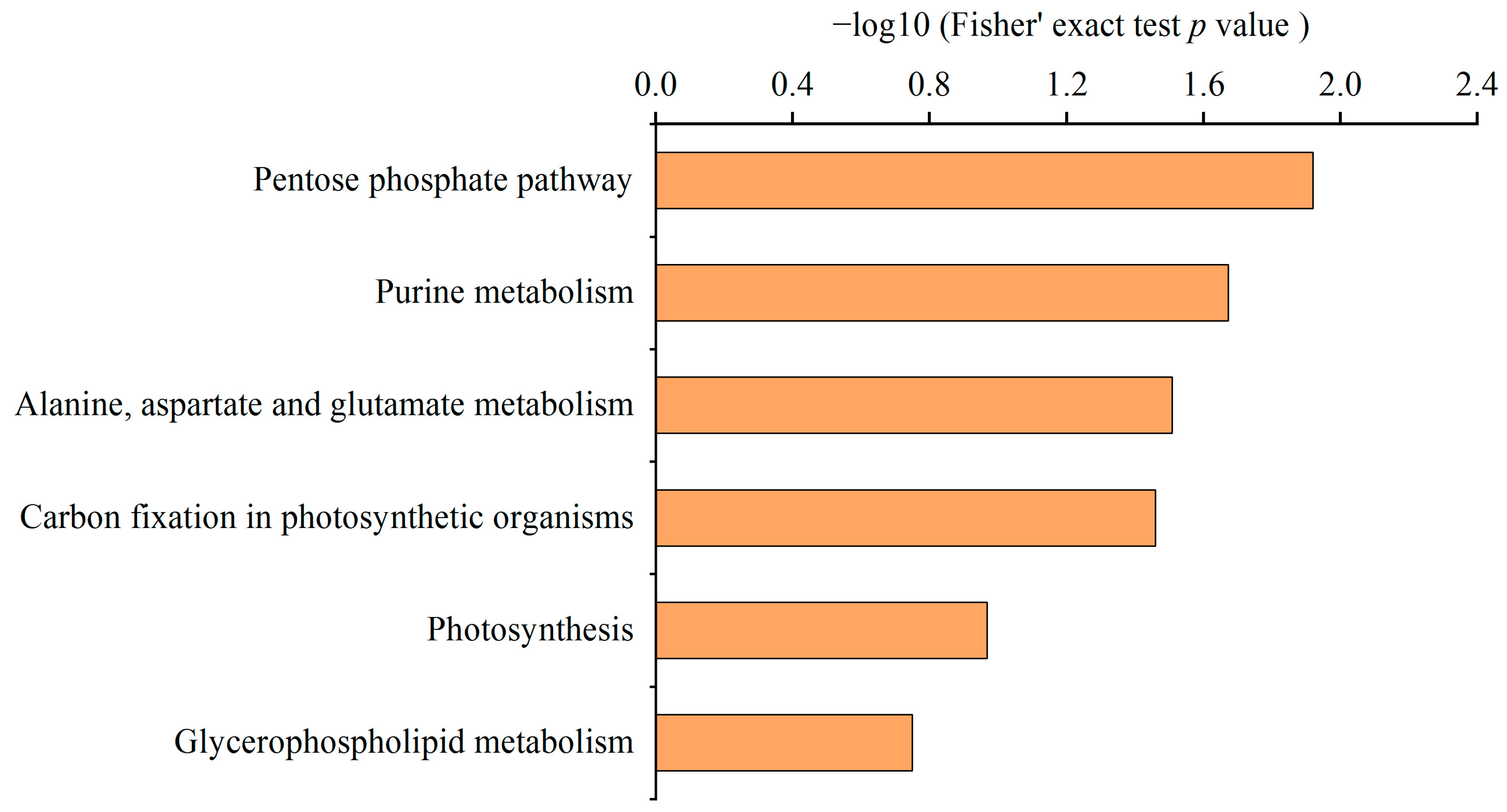
2.3. Functional Analysis of the Malonylated Proteins in N. flagelliforme during Drought Stress
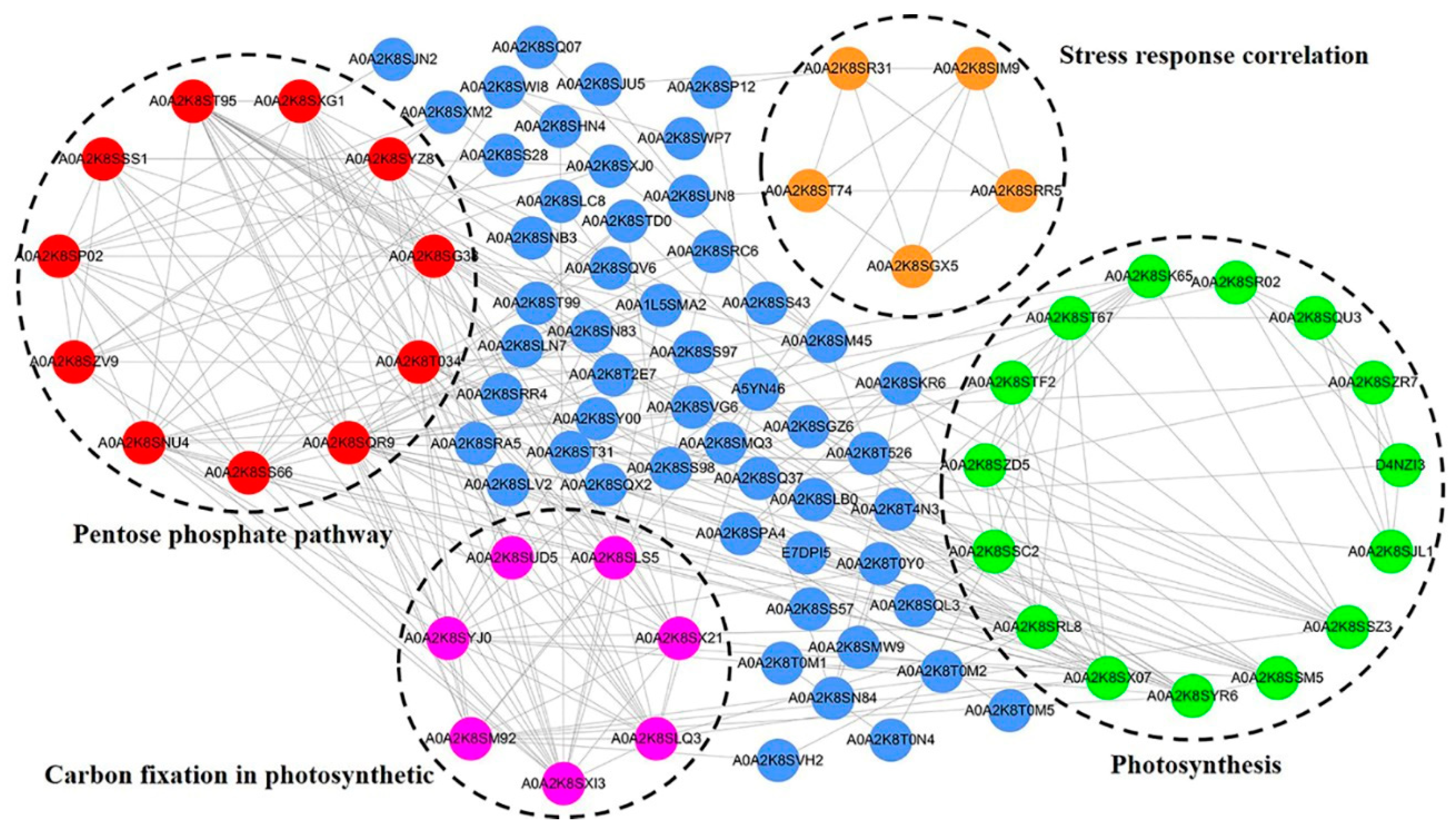
2.4. Protein–Protein Interaction Network
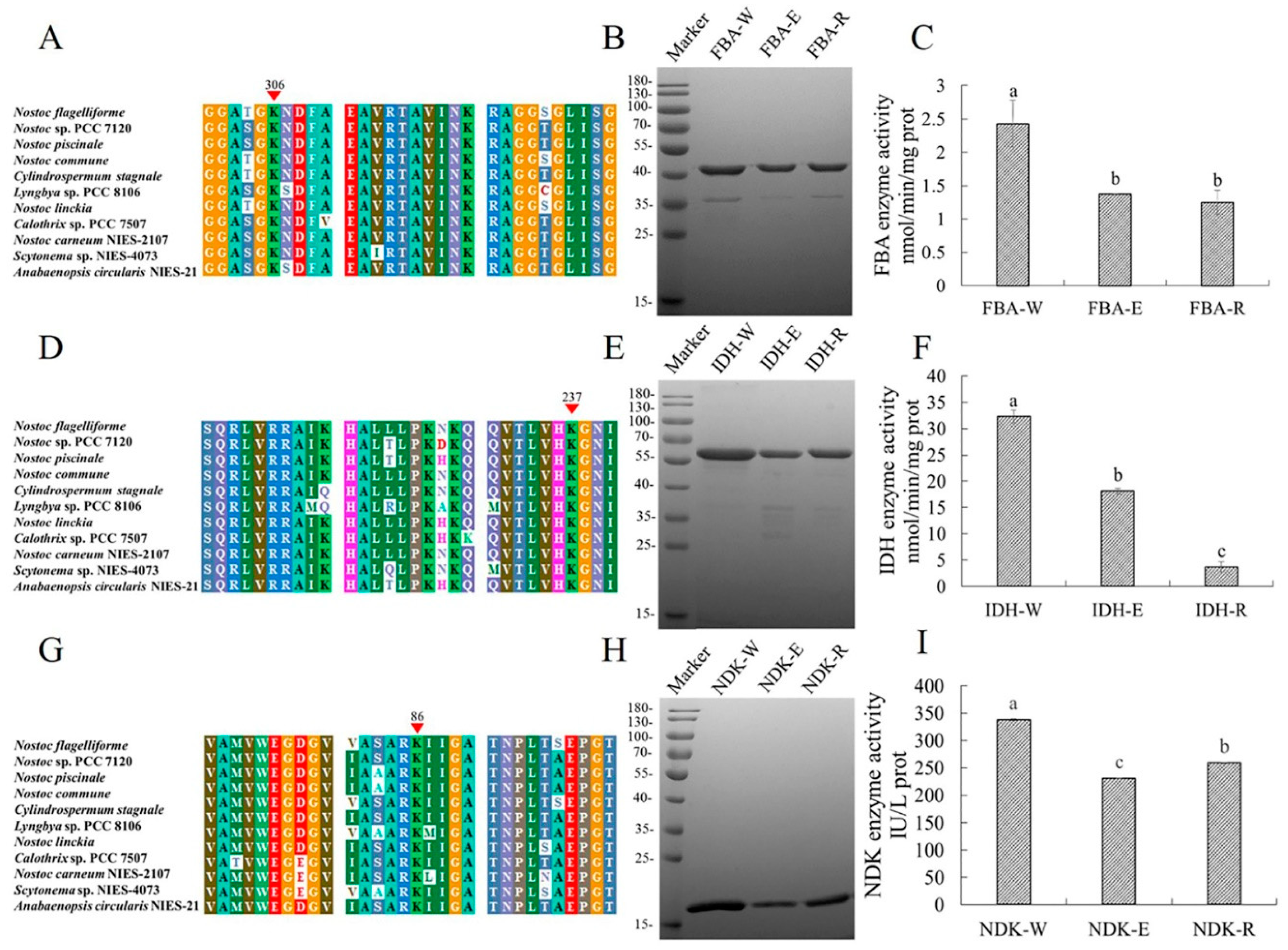
2.5. Conservative Analysis of Lysine Malonylated Proteins in N. flagelliforme
2.6. Impact of Lysine Malonylation on Enzymatic Activity
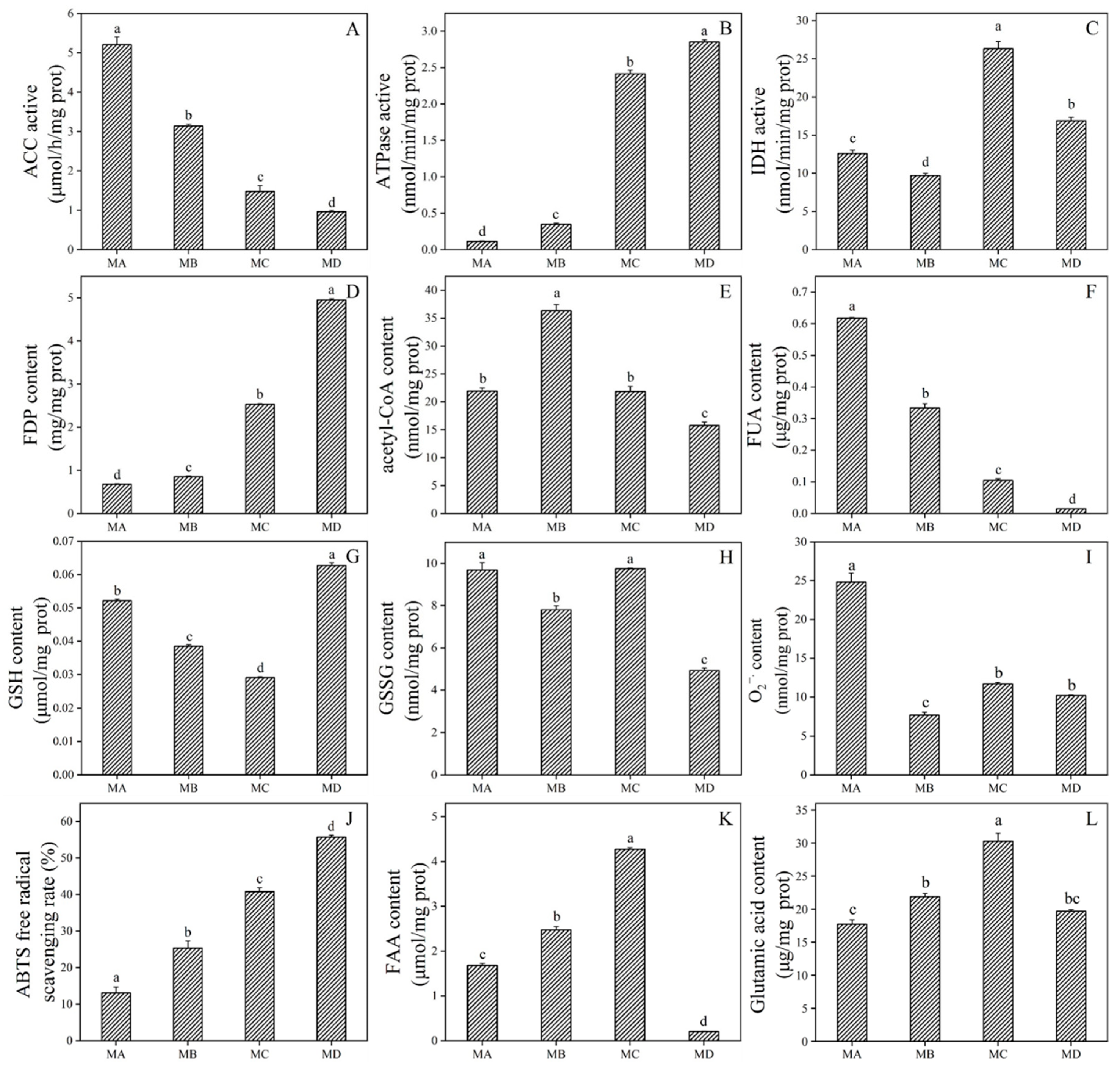
2.7. Physiological Response to Drought Stress
3. Discussion
3.1. Photosynthesis
3.2. Central Carbon Metabolism
3.3. Antioxidant Defense
3.4. Lipid Metabolism and Amino Acid Biosynthesis
4. Materials and Methods
4.1. Materials and Stress Treatments
4.2. Protein Extraction
4.3. SDS-PAGE and Western Blot Analysis
4.4. Trypsin Digestion and Affinity Enrichment
4.5. LC-MS/MS Analysis
4.6. Data Analysis
4.7. Bioinformatics Analysis
4.8. Cloning, Mutagenesis, and Purification of FBA, IDH, and NDK Proteins
4.9. Enzymatic Activities and Metabolite Assays
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Borrell, A.; Wong, A.; George-Jaeggli, B.; van Oosterom, E.; Mace, E.; Godwin, I.; Liu, G.; Mullet, J.; Klein, P.; Hammer, G.; et al. Genetic modification of PIN genes induces causal mechanisms of stay-green drought adaptation phenotype. J. Exp. Bot. 2022, 73, 6711–6726. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, M. Arbuscular mycorrhizal fungi mediated enhanced biomass, root morphological traits and nutrient uptake under drought stress: A meta-analysis. J. Fungi 2022, 8, 660. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Guo, H.X.; Wu, X.; Wang, J.R.; Li, H.J.; Zhang, R.H. Transcriptomic and physiological responses of contrasting maize genotypes to drought stress. Front. Plant Sci. 2022, 13, 928897. [Google Scholar] [CrossRef]
- Haque, M.; Mosharaf, M.; Khatun, M.; Haque, M.; Biswas, M.; Islam, M.; Islam, M.; Shozib, H.; Miah, M.; Molla, A.; et al. Biofilm producing rhizobacteria with multiple plant growth-promoting traits promote growth of tomato under water-deficit stress. Front. Microbiol. 2020, 11, 542053. [Google Scholar] [CrossRef]
- Saharan, B.S.; Brar, B.; Duhan, J.S.; Kumar, R.; Marwaha, S.; Rajput, V.D.; Minkina, T. Molecular and physiological mechanisms to mitigate abiotic stress conditions in plants. Life 2022, 12, 1634. [Google Scholar] [CrossRef]
- Kissoudis, C.; van de Wiel, C.; Visser, R.G.; van der Linden, G. Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Front. Plant Sci. 2014, 5, 207. [Google Scholar] [CrossRef] [PubMed]
- Jonwal, S.; Verma, N.; Sinha, A. Regulation of photosynthetic light reaction proteins via reversible phosphorylation. Plant Sci. 2022, 321, 111312. [Google Scholar] [CrossRef]
- Philp, A.; Rowland, T.; Perez-Schindler, J.; Schenk, S. Understanding the acetylome: Translating targeted proteomics into meaningful physiology. Am. J. Physiol. Cell Physiol. 2014, 307, C763–C773. [Google Scholar] [CrossRef]
- Peng, C.; Lu, Z.; Xie, Z.Y.; Cheng, Z.; Chen, Y.; Tan, M.J.; Luo, H.; Zhang, Y.; He, W.; Yang, K.; et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteom. 2011, 10, M111.012658. [Google Scholar] [CrossRef]
- Shi, Y.; Zhu, J.; Xu, Y.; Tang, X.; Yang, Z.; Huang, A. Malonyl-proteome profiles of Staphylococcus aureus reveal lysine malonylation modification in enzymes involved in energy metabolism. Proteome Sci. 2021, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.B.; Wang, G.Y.; Lin, Q.; Liang, W.X.; Gao, Z.Q.; Mu, P.; Li, G.Q.; Song, L.M. Systematic analysis of the lysine malonylome in common wheat. BMC Genom. 2018, 19, 209. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Y.; Yang, M.K.; Lin, X.H.; Liu, X.; Huang, H.; Ge, F. Malonylome analysis reveals the involvement of lysine malonylation in metabolism and photosynthesis in cyanobacteria. J. Proteome Res. 2017, 16, 2030–2043. [Google Scholar] [CrossRef]
- Xu, M.; Tian, X.M.; Ku, T.T.; Wang, G.Y.; Zhang, E.Y. Global identification and systematic analysis of lysine malonylation in maize (Zea mays L.). Front. Plant Sci. 2021, 12, 728338. [Google Scholar] [CrossRef]
- Mujahid, H.; Meng, X.X.; Xing, S.H.; Peng, X.J.; Wang, C.L.; Peng, Z.H. Malonylome analysis in developing rice (Oryza sativa) seeds suggesting that protein lysine malonylation is well-conserved and overlaps with acetylation and succinylation substantially. J. Proteom. 2018, 170, 88–98. [Google Scholar] [CrossRef]
- Xu, J.Y.; Xu, Z.; Zhou, Y.; Ye, B.C. Lysine malonylome may affect the central metabolism and erythromycin biosynthesis pathway in Saccharopolyspora erythraea. J. Proteome Res. 2016, 15, 1685–1701. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.P.; Cai, T.X.; Li, T.T.; Xue, P.; Zhou, B.; He, X.L.; Wei, P.; Liu, P.S.; Yang, F.Q.; Wei, T.T. Lysine malonylation is elevated in type 2 diabetic mouse models and enriched in metabolic associated proteins. Mol. Cell. Proteom. 2015, 14, 227–236. [Google Scholar] [CrossRef]
- Qian, L.L.; Nie, L.T.; Chen, M.; Liu, P.; Zhu, J.; Zhai, L.H.; Tao, S.C.; Cheng, Z.Y.; Zhao, Y.M.; Tan, M.J. Global profiling of protein lysine malonylation in Escherichia coli reveals its role in energy metabolism. J. Proteome Res. 2016, 15, 2060–2071. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, Q.; Li, X.X.; Hu, J.H.; Song, F.; Liang, W.L.; Ma, X.R.; Wang, L.X.; Liang, W.Y. Effect of drought stress on degradation and remodeling of membrane lipids in Nostoc flagelliforme. Foods 2022, 11, 1798. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Liu, B.; Ji, B.Y. Profiling of small molecular metabolites in Nostoc flagelliforme during periodic desiccation. Mar. Drugs 2019, 17, 298. [Google Scholar] [CrossRef]
- Wang, B.; Yang, J.J.; Xu, C.; Yi, L.X.; Wan, C.H. Dynamic expression of intra- and extra-cellular proteome and the influence of epiphytic bacteria for Nostoc flagelliforme in response to rehydration. Environ. Microbiol. 2020, 22, 1251–1264. [Google Scholar] [CrossRef]
- Gao, X.; Zhu, Z.X.; Xu, H.Y.; Liu, L.T.; An, J.; Ji, B.Y.; Ye, S.F. Cold adaptation in drylands: Transcriptomic insights into cold-stressed Nostoc flagelliforme and characterization of a hypothetical gene with cold and nitrogen stress tolerance. Environ. Microbiol. 2021, 23, 713–727. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.Y.; Yan, F.K.; Wang, M.; Li, X.X.; Zhang, Z.; Ma, X.R.; Hu, J.H.; Wang, J.; Wang, L.X. Comprehensive phosphoproteomic analysis of Nostoc flagelliforme in response to dehydration provides insights into plant ROS signaling transduction. ACS Omega 2021, 6, 13554–13566. [Google Scholar] [CrossRef]
- Wang, L.X.; Li, X.X.; Wang, M.; Ma, X.R.; Song, F.; Hu, J.H.; Liang, W.L.; Liang, W.Y. Carbon metabolism and the ROS scavenging system participate in Nostoc flagelliforme’s adaptive response to dehydration conditions through protein acetylation. J. Proteome Res. 2022, 21, 482–493. [Google Scholar] [CrossRef]
- Li, X.X.; Wang, L.X.; Wang, M.; Zhang, Z.; Ma, C.X.; Ma, X.R.; Na, X.F.; Liang, W.Y. Global analysis of protein succinylation modification of Nostoc flagelliforme in response to dehydration. J. Proteom. 2021, 237, 104149. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.K.; Huang, H.; Ge, F. Lysine propionylation is a widespread post-translational modification involved in regulation of photosynthesis and metabolism in cyanobacteria. Int. J. Mol. Sci. 2019, 20, 4792. [Google Scholar] [CrossRef]
- Ye, J.; Ding, W.N.; Chen, Y.J.; Zhu, X.N.; Sun, J.T.; Zheng, W.J.; Zhang, B.T.; Zhu, S.H. A nucleoside diphosphate kinase gene OsNDPK4 is involved in root development and defense responses in rice (Oryza sativa L.). Planta 2020, 251, 77. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, Q.P.; Mao, P.S. Ultrastructural and photosynthetic responses of pod walls in alfalfa to drought stress. Int. J. Mol. Sci. 2020, 21, 4457. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Xia, H.Q.; Li, Q.; Li, Z.Z.; Zhai, C.; Weng, L.; Mi, H.L.; Yan, S.; Datla, R.; Wang, H.; et al. PALE-GREEN LEAF 1, a rice cpSRP54 protein, is essential for the assembly of the PSI-LHCI supercomplex. Plant Direct 2022, 6, e436. [Google Scholar] [CrossRef]
- Wang, X.P.; Liu, H.L.; Zhang, D.; Zou, D.T.; Wang, J.G.; Zheng, H.L.; Jia, Y.; Qu, Z.J.; Sun, B.; Zhao, H.W. Photosynthetic carbon fixation and sucrose metabolism supplemented by weighted gene co-expression network analysis in response to water stress in rice with overlapping growth stages. Front. Plant Sci. 2022, 13, 864605. [Google Scholar] [CrossRef]
- Miyagawa, Y.; Tamoi, M.; Shigeoka, S. Overexpression of a cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat. Biotechnol. 2001, 19, 965–969. [Google Scholar] [CrossRef]
- Ichikawa, Y.; Tamoi, M.; Sakuyama, H.; Maruta, T.; Ashida, H.; Yokota, A.; Shigeoka, S. Generation of transplastomic lettuce with enhanced growth and high yield. GM Crops 2010, 1, 322–326. [Google Scholar] [CrossRef]
- Nishida, Y.; Rardin, M.J.; Carrico, C.; He, W.; Sahu, A.K.; Gut, P.; Najjar, R.; Fitch, M.; Hellerstein, M.; Gibson, B.W.; et al. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol. Cell 2015, 59, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Škerlová, J.; Berndtsson, J.; Nolte, H.; Ott, M.; Stenmark, P. Structure of the native pyruvate dehydrogenase complex reveals the mechanism of substrate insertion. Nat. Commun. 2021, 12, 5277. [Google Scholar] [CrossRef] [PubMed]
- Soo, P.C.; Horng, Y.T.; Lai, M.J.; Wei, J.R.; Hsieh, S.C.; Chang, Y.L.; Tsai, Y.H.; Lai, H.C. Pirin regulates pyruvate catabolism by interacting with the pyruvate dehydrogenase E1 subunit and modulating pyruvate dehydrogenase activity. J. Bacteriol. 2007, 189, 109–118. [Google Scholar] [CrossRef]
- Xiao, X.; Wang, Q.L.; Ma, X.; Lang, D.Y.; Guo, Z.G.; Zhang, X.H. Physiological biochemistry-combined transcriptomic analysis reveals mechanism of Bacillus cereus G2 improved salt-stress tolerance of Glycyrrhiza uralensis Fisch. seedlings by balancing carbohydrate metabolism. Front. Plant Sci. 2021, 12, 712363. [Google Scholar] [CrossRef]
- Wang, B.; Yang, L.; Zhang, Y.Q.; Chen, S.L.; Gao, X.; Wan, C.H. Investigation of the dynamical expression of Nostoc flagelliforme proteome in response to rehydration. J. Proteom. 2018, 192, 160–168. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Colak, G.; Pougovkina, O.; Dai, L.Z.; Tan, M.J.; Te Brinke, H.; Huang, H.; Cheng, Z.Y.; Park, J.; Wan, X.L.; Liu, X.J.; et al. Proteomic and biochemical studies of lysine malonylation suggest its malonic aciduria-associated regulatory role in mitochondrial function and fatty acid oxidation. Mol. Cell. Proteom. 2015, 14, 3056–3071. [Google Scholar]
- Moradi, P.; Ford-Lloyd, B.; Pritchard, J. Metabolomic approach reveals the biochemical mechanisms underlying drought stress tolerance in Thyme. Anal. Biochem. 2017, 527, 49–62. [Google Scholar] [CrossRef]
- Ashrafi, M.; Azimi-Moqadam, M.R.; Moradi, P.; MohseniFard, E.; Shekari, F.; Kompany-Zareh, M. Effect of drought stress on metabolite adjustments in drought tolerant and sensitive thyme. Plant Physiol. Biochem. 2018, 132, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Li, X.X.; Ding, M.M.; Wang, M.; Yang, S.J.; Ma, X.R.; Hu, J.; Song, F.; Wang, L.X.; Liang, W.Y. Proteome profiling reveals changes in energy metabolism, transport and antioxidation during drought stress in Nostoc flagelliforme. BMC Plant Biol. 2022, 22, 162. [Google Scholar] [CrossRef]
- Petersen, B.; Petersen, T.N.; Andersen, P.; Nielsen, M.; Lundegaard, C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 2009, 9, 51. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.P.; Xin, Y.; Sheng, Z.L.; Liu, H.; Jiang, A.L.; Wang, F.; Yang, J.; Xi, X.J.; Zha, Q.; Zhang, L.Q.; et al. Systematic identification and analysis of lysine succinylation in strawberry stigmata. J. Agric. Food Chem. 2018, 66, 13310–13320. [Google Scholar]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.F.; Fan, R.; Wang, X.; Wang, D.; Zhang, X. TaRAR1 and TaSGT1 associate with TaHsp90 to function in bread wheat (Triticum aestivum L.) seedling growth and stripe rust resistance. Plant Mol. Biol. 2015, 87, 577–589. [Google Scholar] [CrossRef] [PubMed]



| Comparisons | Significant Changes in Abundance | Consistent Presence/Absence Expression Profile | ||
|---|---|---|---|---|
| Increased | Decreased | Increased | Decreased | |
| MB vs. MA | 11 | 2 | 118 | 26 |
| MC vs. MA | 0 | 18 | 1 | 37 |
| MD vs. MA | 0 | 10 | 7 | 33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Zhu, Q.; Yao, N.; Liang, W.; Ma, X.; Li, J.; Li, X.; Wang, L.; Liang, W. The Enzyme Lysine Malonylation of Calvin Cycle and Gluconeogenesis Regulated Glycometabolism in Nostoc flagelliforme to Adapt to Drought Stress. Int. J. Mol. Sci. 2023, 24, 8446. https://doi.org/10.3390/ijms24098446
Wang M, Zhu Q, Yao N, Liang W, Ma X, Li J, Li X, Wang L, Liang W. The Enzyme Lysine Malonylation of Calvin Cycle and Gluconeogenesis Regulated Glycometabolism in Nostoc flagelliforme to Adapt to Drought Stress. International Journal of Molecular Sciences. 2023; 24(9):8446. https://doi.org/10.3390/ijms24098446
Chicago/Turabian StyleWang, Meng, Qiang Zhu, Ning Yao, Wangli Liang, Xiaoxia Ma, Jingjing Li, Xiaoxu Li, Lingxia Wang, and Wenyu Liang. 2023. "The Enzyme Lysine Malonylation of Calvin Cycle and Gluconeogenesis Regulated Glycometabolism in Nostoc flagelliforme to Adapt to Drought Stress" International Journal of Molecular Sciences 24, no. 9: 8446. https://doi.org/10.3390/ijms24098446
APA StyleWang, M., Zhu, Q., Yao, N., Liang, W., Ma, X., Li, J., Li, X., Wang, L., & Liang, W. (2023). The Enzyme Lysine Malonylation of Calvin Cycle and Gluconeogenesis Regulated Glycometabolism in Nostoc flagelliforme to Adapt to Drought Stress. International Journal of Molecular Sciences, 24(9), 8446. https://doi.org/10.3390/ijms24098446