Genome-Wide Expression Analysis of Long Noncoding RNAs and Their Target Genes in Metafemale Drosophila
Abstract
1. Introduction
2. Results
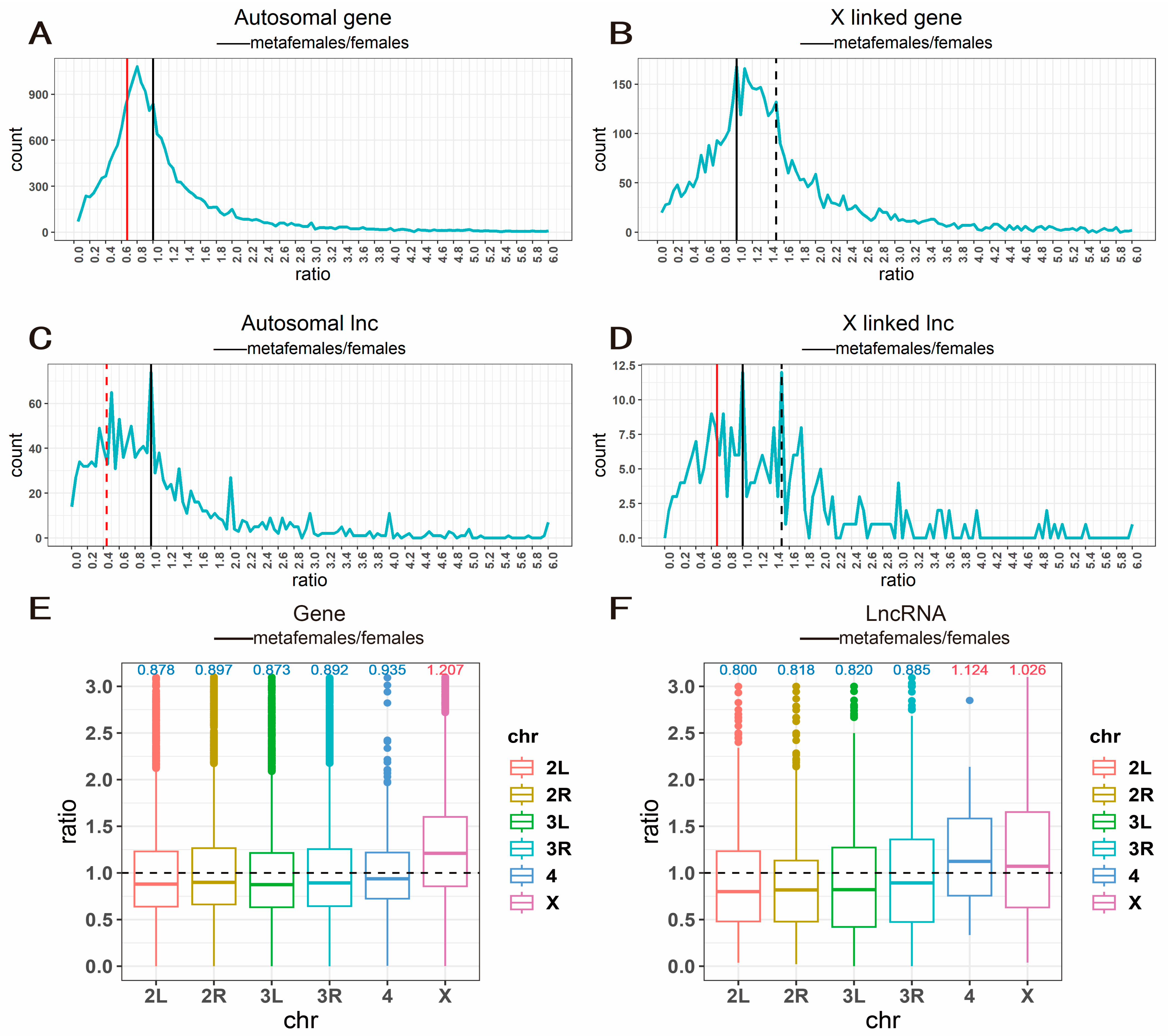
2.1. Distribution of Gene Expression Ratios in Metafemale Individuals
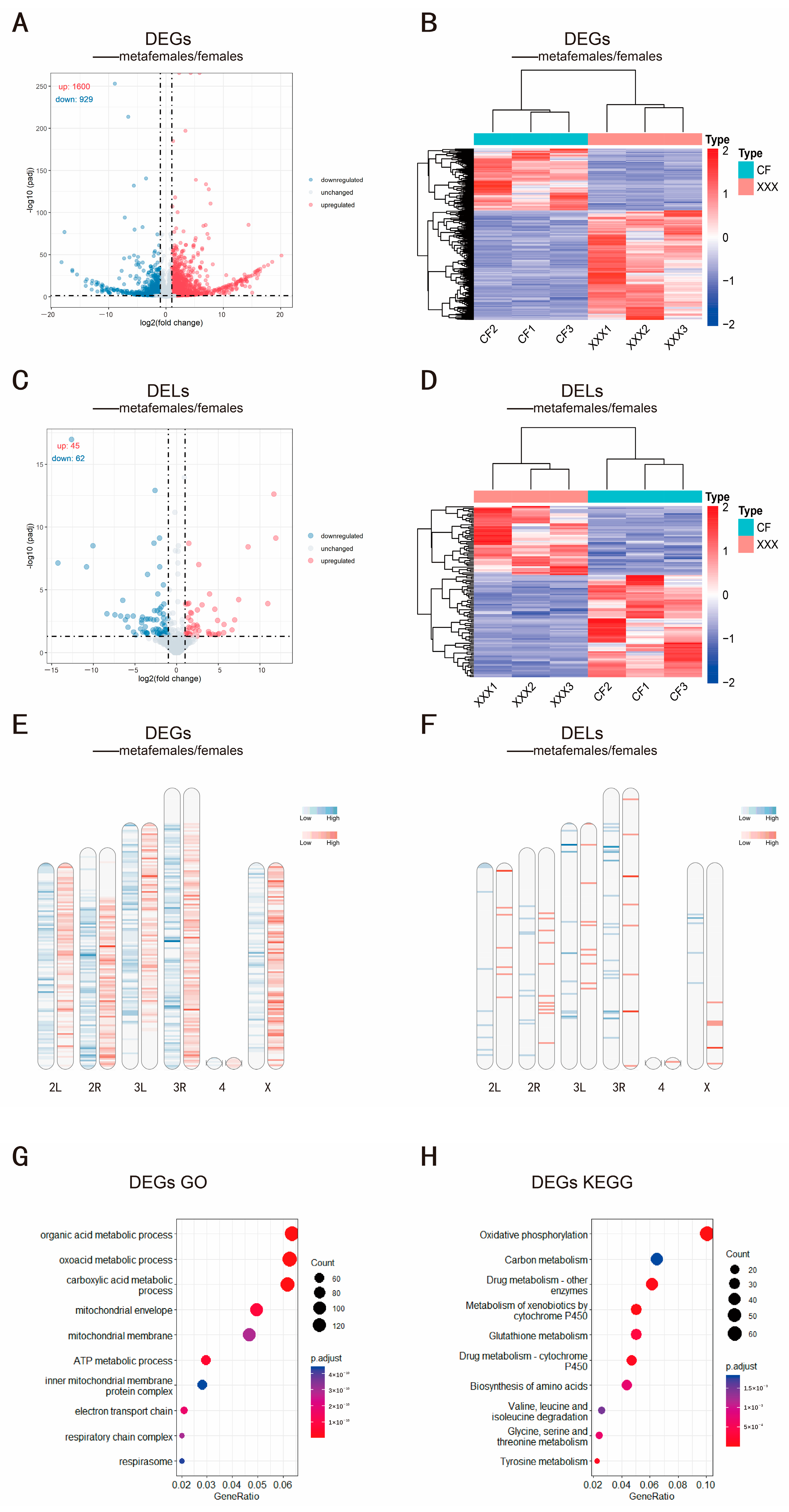
2.2. Identification of Differentially Expressed mRNAs and lncRNAs
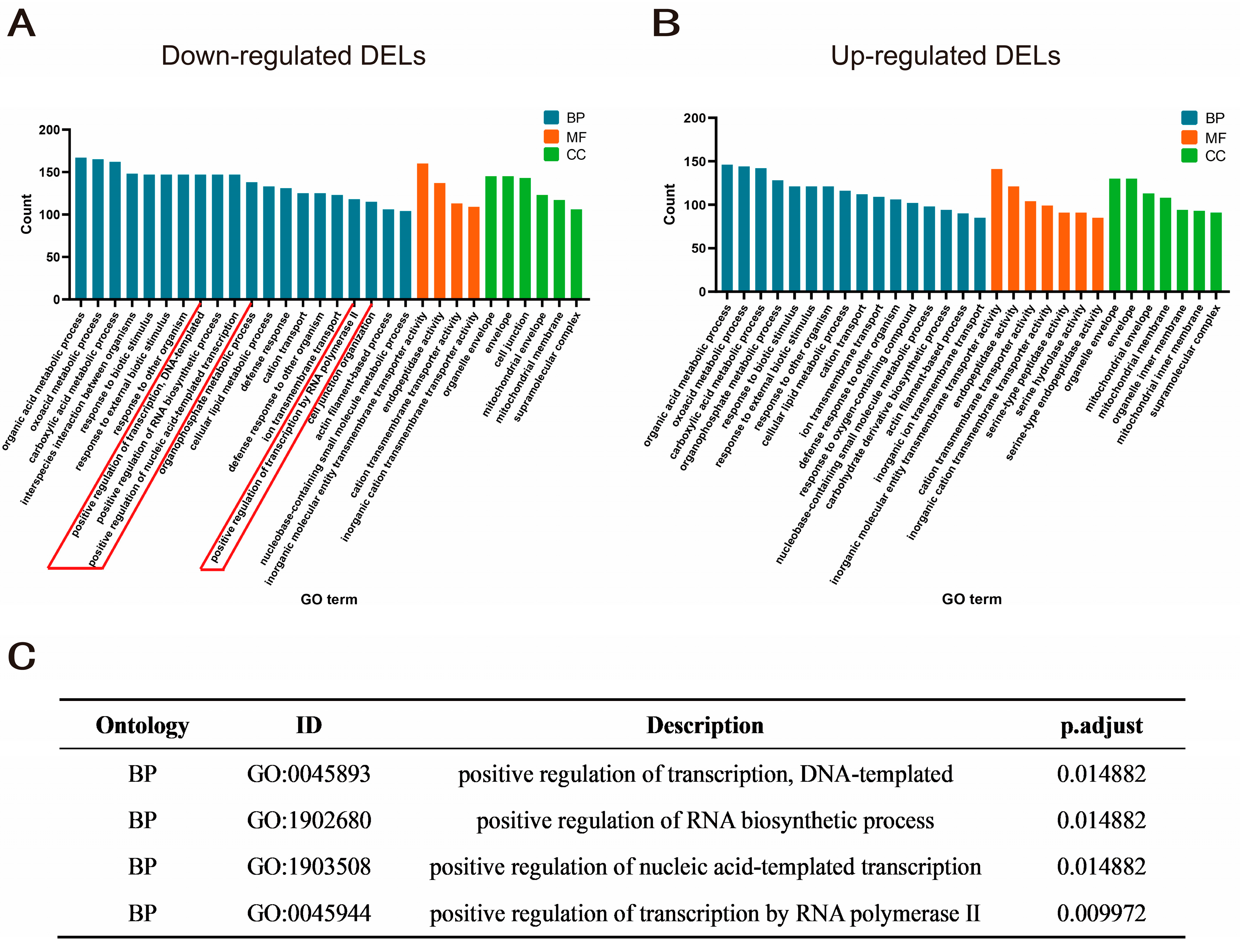
2.3. Identification and Enrichment Analysis of Differentially Expressed lncRNA Target Genes
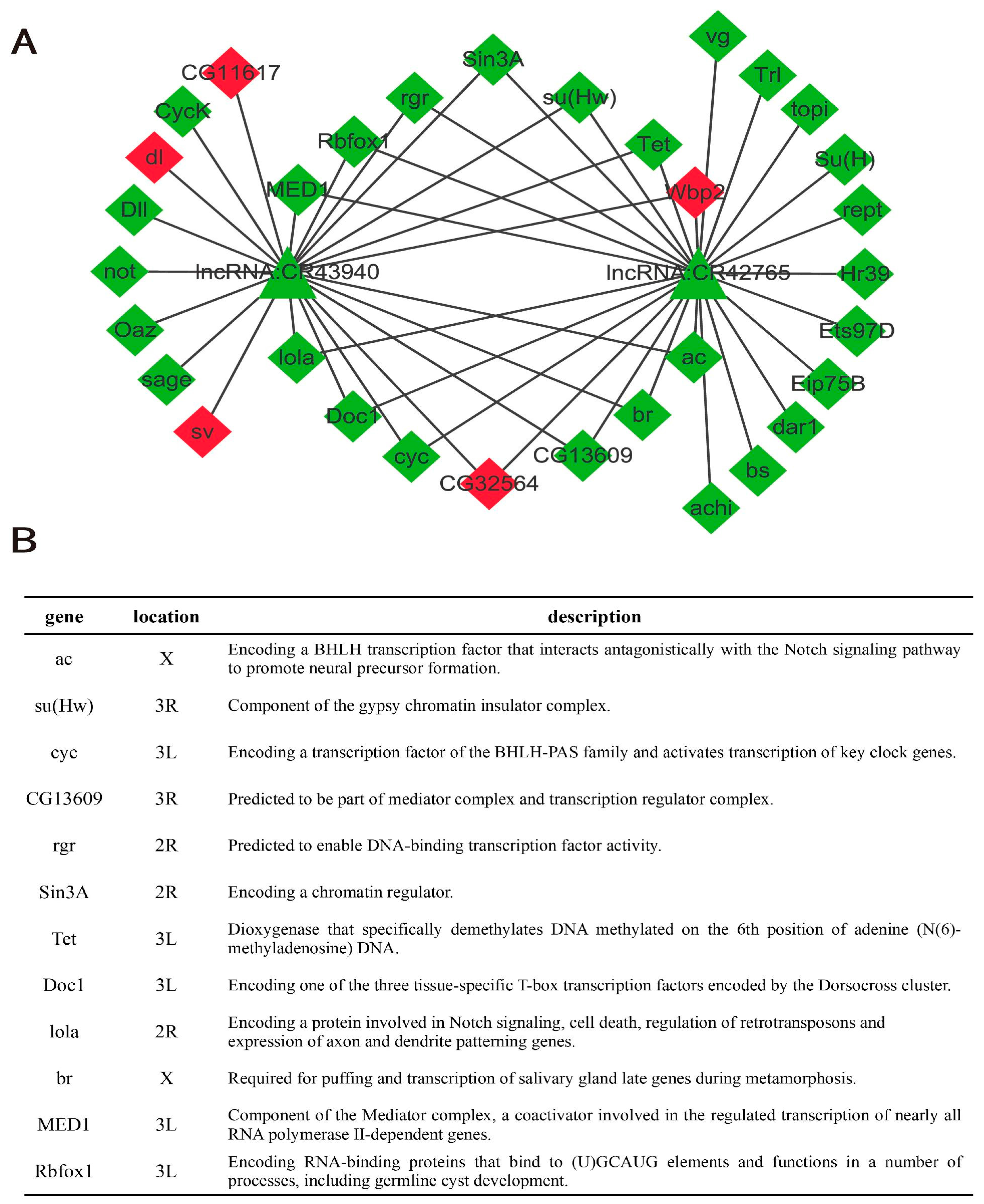
2.4. lncRNA-mRNA Interaction Network
2.5. Validation of Candidate Inverse Dosage Modulators
3. Discussion
4. Materials and Methods
4.1. Drosophila Stocks and Crosses
4.2. RNA Extraction and Sequencing
4.3. RNA Sequencing Analysis
4.4. Ratio Distribution
4.5. Differential Expression Analysis
4.6. Differential lncRNA Target Gene Prediction
4.7. Target Gene Enrichment Analysis
4.8. Transcription Factor Analysis
4.9. lncRNA-mRNA Interaction Network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Orr, B.; Godek, K.M.; Compton, D. Aneuploidy. Curr. Biol. 2015, 25, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Potapova, T.; Gorbsky, G.J. The consequences of chromosome segregation errors in mitosis and meiosis. Biology 2017, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Gogendeau, D.; Siudeja, K.; Gambarotto, D.; Pennetier, C.; Bardin, A.J.; Basto, R. Aneuploidy causes premature differentiation of neural and intestinal stem cells. Nat. Commun. 2015, 6, 8894. [Google Scholar] [CrossRef] [PubMed]
- Torres, E.M.; Sokolsky, T.; Tucker, C.M.; Chan, L.Y.; Boselli, M.; Dunham, M.J.; Amon, A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 2007, 317, 916–924. [Google Scholar] [CrossRef]
- Birchler, J.A.; Veitia, R.A. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc Natl. Acad. Sci. USA 2012, 109, 14746–14753. [Google Scholar] [CrossRef]
- Sheltzer, J.M.; Blank, H.M.; Pfau, S.J.; Tange, Y.; George, B.M.; Humpton, T.J.; Brito, I.L.; Hiraoka, Y.; Niwa, O.; Amon, A. Aneuploidy drives genomic instability in yeast. Science 2011, 333, 1026–1030. [Google Scholar] [CrossRef]
- Makarevitch, I.; Phillips, R.L.; Springer, N.M. Profiling expression changes caused by a segmental aneuploid in maize. BMC Genomics 2008, 9, 7. [Google Scholar] [CrossRef]
- Hou, J.; Shi, X.; Chen, C.; Islam, M.S.; Johnson, A.F.; Kanno, T.; Huettel, B.; Yen, M.R.; Hsu, F.M.; Ji, T.; et al. Global impacts of chromosomal imbalance on gene expression in Arabidopsis and other taxa. Proc. Natl. Acad. Sci. USA 2018, 115, E11321–E11330. [Google Scholar] [CrossRef]
- Yang, H.; Shi, X.; Chen, C.; Hou, J.; Ji, T.; Cheng, J.; Birchler, J.A. Predominantly inverse modulation of gene expression in genomically unbalanced disomic haploid maize. Plant Cell 2021, 33, 901–916. [Google Scholar] [CrossRef]
- Blakeslee, A.F.; Belling, J.; Farnham, M.E. Chromosomal duplication and mendelian phenomena in datura mutants. Science 1920, 52, 388–390. [Google Scholar] [CrossRef] [PubMed]
- Blakeslee, A.F. New Jimson weeds from old chromosomes. J. Hered. 1934, 25, 81–108. [Google Scholar] [CrossRef]
- Bridges, C.B. Triploid intersexes in drosophila melanogaster. Science 1921, 54, 252–254. [Google Scholar] [CrossRef]
- Antonarakis, S.E.; Skotko, B.G.; Rafii, M.S.; Strydom, A.; Pape, S.E.; Bianchi, D.W.; Sherman, S.L.; Reeves, R.H. Down syndrome. Nat. Rev. Dis. Primers 2020, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Reiss, A.L. Cognitive and neurological aspects of sex chromosome aneuploidies. Lancet Neurol. 2014, 13, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Boveri, T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J. Cell Sci. 2008, 121, 1–84. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.R.; Prabhu, V.R.; Hunter, K.E.; Glazier, C.M.; Whittaker, C.A.; Housman, D.E.; Amon, A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 2008, 322, 703–709. [Google Scholar] [CrossRef]
- Grell, E. The dose effect of ma-l+ and ry+ on xanthine dehydrogenase activity in Drosophila melanogaster. Z. Für Vererb. 1962, 93, 371–377. [Google Scholar]
- Birchler, J.A. A study of enzyme activities in a dosage series of the long arm of chromosome one in maize. Genetics 1979, 92, 1211–1229. [Google Scholar] [CrossRef]
- Birchler, J.A.; Newton, K.J. Modulation of protein levels in chromosomal dosage series of maize: The biochemical basis of aneuploid syndromes. Genetics 1981, 99, 247–266. [Google Scholar] [CrossRef]
- Devlin, R.H.; Holm, D.G.; Grigliatti, T.A. Autosomal dosage compensation Drosophila melanogaster strains trisomic for the left arm of chromosome 2. Proc. Natl. Acad. Sci. USA 1982, 79, 1200–1204. [Google Scholar] [CrossRef]
- Sun, L.; Johnson, A.F.; Donohue, R.C.; Li, J.; Cheng, J.; Birchler, J.A. Dosage compensation and inverse effects in triple X metafemales of Drosophila. Proc. Natl. Acad. Sci. USA 2013, 110, 7383–7388. [Google Scholar] [CrossRef]
- Zhang, Z.; Presgraves, D.C. Translational compensation of gene copy number alterations by aneuploidy in Drosophila melanogaster. Nucleic Acids Res. 2017, 45, 2986–2993. [Google Scholar] [CrossRef]
- Birchler, J.; Sun, L.; Fernandez, H.; Donohue, R.; Xie, W.; Sanyal, A. Re-evaluation of the function of the male specific lethal complex in Drosophila. J. Genet. Genomics 2011, 38, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Hiebert, J.C.; Birchler, J.A. Effects of the maleless mutation on X and autosomal gene expression in Drosophila melanogaster. Genetics 1994, 136, 913–926. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Birchler, J.A. Interaction study of the male specific lethal (MSL) complex and trans-acting dosage effects in metafemales of Drosophila melanogaster. Cytogenet. Genome. Res. 2009, 124, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Fernandez, H.R.; Donohue, R.C.; Li, J.; Cheng, J.; Birchler, J.A. Male-specific lethal complex in Drosophila counteracts histone acetylation and does not mediate dosage compensation. Proc. Natl. Acad. Sci. USA 2013, 110, E808–E817. [Google Scholar] [CrossRef] [PubMed]
- Devlin, R.H.; Holm, D.G.; Grigliatti, T.A. The influence of whole-arm trisomy on gene expression in Drosophila. Genetics 1988, 118, 87–101. [Google Scholar] [CrossRef]
- Birchler, J.A.; Hiebert, J.C.; Paigen, K. Analysis of autosomal dosage compensation involving the alcohol dehydrogenase locus in Drosophila melanogaster. Genetics 1990, 124, 679–686. [Google Scholar] [CrossRef]
- Sun, L.; Johnson, A.F.; Li, J.; Lambdin, A.S.; Cheng, J.; Birchler, J.A. Differential effect of aneuploidy on the X chromosome and genes with sex-biased expression in Drosophila. Proc. Natl. Acad. Sci. USA 2013, 110, 16514–16519. [Google Scholar] [CrossRef]
- Johnson, A.F.; Hou, J.; Yang, H.; Shi, X.; Chen, C.; Islam, M.S.; Ji, T.; Cheng, J.; Birchler, J.A. Magnitude of modulation of gene expression in aneuploid maize depends on the extent of genomic imbalance. J. Genet. Genomics 2020, 47, 93–103. [Google Scholar] [CrossRef]
- Rabinow, L.; Nguyen-Huynh, A.T.; Birchler, J.A. A trans-acting regulatory gene that inversely affects the expression of the white, brown and scarlet loci in Drosophila. Genetics 1991, 129, 463–480. [Google Scholar] [CrossRef] [PubMed]
- Sabl, J.F.; Birchler, J.A. Dosage dependent modifiers of white alleles in Drosophila melanogaster. Genet. Res. 1993, 62, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Birchler, J.A.; Bhadra, U.; Bhadra, M.P.; Auger, D.L. Dosage-dependent gene regulation in multicellular eukaryotes: Implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev. Biol. 2001, 234, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, R.; Huang, C.; Zhang, L.; Sun, L. Modulation of Global Gene Expression by Aneuploidy and CNV of Dosage Sensitive Regulatory Genes. Genes 2021, 12, 1606. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of long noncoding RNA function in development and disease. Cell Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Chen, L.L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef]
- Villamizar, O.; Chambers, C.B.; Riberdy, J.M.; Persons, D.A.; Wilber, A. Long noncoding RNA Saf and splicing factor 45 increase soluble Fas and resistance to apoptosis. Oncotarget 2016, 7, 13810–13826. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef]
- Visel, A.; Zhu, Y.; May, D.; Afzal, V.; Gong, E.; Attanasio, C.; Blow, M.J.; Cohen, J.C.; Rubin, E.M.; Pennacchio, L.A. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature 2010, 464, 409–412. [Google Scholar] [CrossRef]
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1131–1139. [Google Scholar] [CrossRef]
- Ulasov, A.V.; Rosenkranz, A.A.; Sobolev, A.S. Transcription factors: Time to deliver. J. Control. Release 2018, 269, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Weidemüller, P.; Kholmatov, M.; Petsalaki, E.; Zaugg, J.B. Transcription factors: Bridge between cell signaling and gene regulation. Proteomics 2021, 21, e2000034. [Google Scholar] [CrossRef]
- Davidson, E.H.; Rast, J.P.; Oliveri, P.; Ransick, A.; Calestani, C.; Yuh, C.H.; Minokawa, T.; Amore, G.; Hinman, V.; Arenas-Mena, C.; et al. A genomic regulatory network for development. Science 2002, 295, 1669–1678. [Google Scholar] [CrossRef]
- Zhang, X.; Miao, C.; Nan, Z.; Lyu, J.; Xi, Y.; Yang, X.; Ge, W. A positive role of Sin3A in regulating Notch signaling during Drosophila wing development. Cell Signal 2019, 53, 184–189. [Google Scholar] [CrossRef]
- Birchler, J.A. Aneuploidy in plants and flies: The origin of studies of genomic imbalance. Semin. Cell Dev. Biol. 2013, 24, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Carlson, P.S. Locating genetic loci with aneuploids. Mol. Gen. Genet. 1972, 114, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Birchler, J.A. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 1994, 266, 1999–2002. [Google Scholar] [CrossRef]
- Birchler, J.A. Parallel Universes for Models of X Chromosome Dosage Compensation in Drosophila: A Review. Cytogenet. Genome. Res. 2016, 148, 52–67. [Google Scholar] [CrossRef]
- Santaguida, S.; Vasile, E.; White, E.; Amon, A. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 2015, 29, 2010–2021. [Google Scholar] [CrossRef] [PubMed]
- Joy, J.; Barrio, L.; Santos-Tapia, C.; Romão, D.; Giakoumakis, N.N.; Clemente-Ruiz, M.; Milán, M. Proteostasis failure and mitochondrial dysfunction leads to aneuploidy-induced senescence. Dev. Cell 2021, 56, 2043–2058.e7. [Google Scholar] [CrossRef] [PubMed]
- Malone, J.H.; Cho, D.Y.; Mattiuzzo, N.R.; Artieri, C.G.; Jiang, L.; Dale, R.K.; Smith, H.E.; McDaniel, J.; Munro, S.; Salit, M.; et al. Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 2012, 13, r28. [Google Scholar] [CrossRef]
- Osborn, T.C.; Pires, J.C.; Birchler, J.A.; Auger, D.L.; Chen, Z.J.; Lee, H.S.; Comai, L.; Madlung, A.; Doerge, R.W.; Colot, V.; et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003, 19, 141–147. [Google Scholar] [CrossRef]
- Veitia, R.A.; Bottani, S.; Birchler, J.A. Cellular reactions to gene dosage imbalance: Genomic, transcriptomic and proteomic effects. Trends Genet. 2008, 24, 390–397. [Google Scholar] [CrossRef]
- Birchler, J.A.; Veitia, R.A. The gene balance hypothesis: Implications for gene regulation, quantitative traits and evolution. New Phytol. 2010, 186, 54–62. [Google Scholar] [CrossRef]
- Zhao, G.; Boekhoff-Falk, G.; Wilson, B.A.; Skeath, J.B. Linking pattern formation to cell-type specification: Dichaete and Ind directly repress achaete gene expression in the Drosophila CNS. Proc. Natl. Acad. Sci. USA 2007, 104, 3847–3852. [Google Scholar] [CrossRef]
- Marcellini, S.; Gibert, J.M.; Simpson, P. achaete, but not scute, is dispensable for the peripheral nervous system of Drosophila. Dev. Biol. 2005, 285, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, R.A.; Ekwall, K. Sin3: A flexible regulator of global gene expression and genome stability. Curr. Genet. 2005, 47, 1–17. [Google Scholar] [CrossRef]
- Zhang, T.; Sheng, Z.; Du, W. Loss of histone deacetylase HDAC1 induces cell death in Drosophila epithelial cells through JNK and Hippo signaling. Mech. Dev. 2016, 141, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Jeronimo, C.; Robert, F. The Mediator Complex: At the Nexus of RNA Polymerase II Transcription. Trends Cell Biol. 2017, 27, 765–783. [Google Scholar] [CrossRef]
- Zhang, Y.; Fu, Y.; Zhang, C.; Jia, L.; Yao, N.; Lin, Y.; Dong, Y.; Fatima, N.; Alam, N.; Wang, R.; et al. MED1 Deficiency in Macrophages Accelerates Intimal Hyperplasia via ROS Generation and Inflammation. Oxid. Med. Cell Longev. 2021, 2021, 3010577. [Google Scholar] [CrossRef]
- Birchler, J.A.; Riddle, N.C.; Auger, D.L.; Veitia, R.A. Dosage balance in gene regulation: Biological implications. Trends Genet. 2005, 21, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Aït Yahya-Graison, E.; Aubert, J.; Dauphinot, L.; Rivals, I.; Prieur, M.; Golfier, G.; Rossier, J.; Personnaz, L.; Creau, N.; Bléhaut, H.; et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: Impact on disease phenotypes. Am. J. Hum. Genet. 2007, 81, 475–491. [Google Scholar] [CrossRef]
- Raznahan, A.; Parikshak, N.N.; Chandran, V.; Blumenthal, J.D.; Clasen, L.S.; Alexander-Bloch, A.F.; Zinn, A.R.; Wangsa, D.; Wise, J.; Murphy, D.G.M.; et al. Sex-chromosome dosage effects on gene expression in humans. Proc. Natl. Acad. Sci. USA 2018, 115, 7398–7403. [Google Scholar] [CrossRef]
- Zhang, X.; Hong, D.; Ma, S.; Ward, T.; Ho, M.; Pattni, R.; Duren, Z.; Stankov, A.; Bade Shrestha, S.; Hallmayer, J.; et al. Integrated functional genomic analyses of Klinefelter and Turner syndromes reveal global network effects of altered X chromosome dosage. Proc. Natl. Acad. Sci. USA 2020, 117, 4864–4873. [Google Scholar] [CrossRef]
- Birchler, J.A.; Veitia, R.A. The gene balance hypothesis: From classical genetics to modern genomics. Plant Cell 2007, 19, 395–402. [Google Scholar] [CrossRef]
- Shi, X.; Yang, H.; Chen, C.; Hou, J.; Hanson, K.M.; Albert, P.S.; Ji, T.; Cheng, J.; Birchler, J.A. Genomic imbalance determines positive and negative modulation of gene expression in diploid maize. Plant Cell 2021, 33, 917–939. [Google Scholar] [CrossRef]
- Veitia, R.A. Exploring the etiology of haploinsufficiency. Bioessays 2002, 24, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Veitia, R.A. Nonlinear effects in macromolecular assembly and dosage sensitivity. J. Biol. 2003, 220, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Schuster-Böckler, B.; Conrad, D.; Bateman, A. Dosage sensitivity shapes the evolution of copy-number varied regions. PLoS ONE 2010, 5, e9474. [Google Scholar] [CrossRef] [PubMed]
- Desvignes, T.; Sydes, J.; Montfort, J.; Bobe, J.; Postlethwait, J.H. Evolution after Whole-Genome Duplication: Teleost MicroRNAs. Mol. Biol. Evol. 2021, 38, 3308–3331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chia, J.M.; Kumari, S.; Stein, J.C.; Liu, Z.; Narechania, A.; Maher, C.A.; Guill, K.; McMullen, M.D.; Ware, D. A genome-wide characterization of microRNA genes in maize. PLoS Genet. 2009, 5, e1000716. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Shi, X.; Chen, C.; Hou, J.; Ji, T.; Cheng, J.; Birchler, J.A. Genomic imbalance modulates transposable element expression in maize. Plant Commun. 2022, 4, 100467. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef]
- Hao, Z.; Lv, D.; Ge, Y.; Shi, J.; Weijers, D.; Yu, G.; Chen, J. RIdeogram: Drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput. Sci. 2020, 6, e251. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Aibar, S.; González-Blas, C.B.; Moerman, T.; Huynh-Thu, V.A.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.C.; Geurts, P.; Aerts, J.; et al. SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef]

| Node_Name | MCC | DMNC | MNC | Degree | EPC | Bottleneck | Eccentricity | Closeness | Radiality | Betweenness | Stress |
|---|---|---|---|---|---|---|---|---|---|---|---|
| lncRNA:CR43940 | 31 | 0 | 1 | 32 | 50.165 | 4 | 0.19721 | 96.48333 | 6.27612 | 3619.788 | 202,948 |
| lncRNA:CR42765 | 30 | 0 | 1 | 30 | 49.031 | 4 | 0.19721 | 94.9 | 6.21069 | 3721.355 | 213,714 |
| lncRNA:CR46258 | 29 | 0 | 1 | 29 | 49.566 | 3 | 0.19721 | 94.11667 | 6.20134 | 2571.495 | 165,596 |
| br | 25 | 0 | 1 | 25 | 44.288 | 4 | 0.16434 | 96.13333 | 6.24808 | 5976.691 | 263,602 |
| lncRNA:CR45232 | 25 | 0 | 1 | 25 | 46.97 | 6 | 0.19721 | 90.03333 | 6.08919 | 2361.111 | 173,502 |
| lncRNA:CR44948 | 23 | 0 | 1 | 23 | 45.544 | 6 | 0.19721 | 86.81667 | 5.99572 | 1835.18 | 130,014 |
| lncRNA:CR45170 | 23 | 0 | 1 | 23 | 45.698 | 1 | 0.19721 | 90.51667 | 6.17331 | 2923.464 | 174,554 |
| lncRNA:CR43651 | 22 | 0 | 1 | 22 | 46.481 | 2 | 0.19721 | 88.55 | 6.08919 | 1555.737 | 120,006 |
| lncRNA:CR45972 | 21 | 0 | 1 | 21 | 44.079 | 17 | 0.19721 | 87.28333 | 6.07049 | 2236.299 | 136,518 |
| Wbp2 | 20 | 0 | 1 | 20 | 44.254 | 52 | 0.16434 | 88.28333 | 6.07049 | 2891.457 | 144,714 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Yan, R.; Liu, H.; Zhang, S.; Wang, R.; Zhang, B.; Sun, L. Genome-Wide Expression Analysis of Long Noncoding RNAs and Their Target Genes in Metafemale Drosophila. Int. J. Mol. Sci. 2023, 24, 8381. https://doi.org/10.3390/ijms24098381
Liu X, Yan R, Liu H, Zhang S, Wang R, Zhang B, Sun L. Genome-Wide Expression Analysis of Long Noncoding RNAs and Their Target Genes in Metafemale Drosophila. International Journal of Molecular Sciences. 2023; 24(9):8381. https://doi.org/10.3390/ijms24098381
Chicago/Turabian StyleLiu, Xinyu, Ran Yan, Haosheng Liu, Shuai Zhang, Ruixue Wang, Bowen Zhang, and Lin Sun. 2023. "Genome-Wide Expression Analysis of Long Noncoding RNAs and Their Target Genes in Metafemale Drosophila" International Journal of Molecular Sciences 24, no. 9: 8381. https://doi.org/10.3390/ijms24098381
APA StyleLiu, X., Yan, R., Liu, H., Zhang, S., Wang, R., Zhang, B., & Sun, L. (2023). Genome-Wide Expression Analysis of Long Noncoding RNAs and Their Target Genes in Metafemale Drosophila. International Journal of Molecular Sciences, 24(9), 8381. https://doi.org/10.3390/ijms24098381






