Chicken Erythrocyte: Epigenomic Regulation of Gene Activity
Abstract
1. Introduction
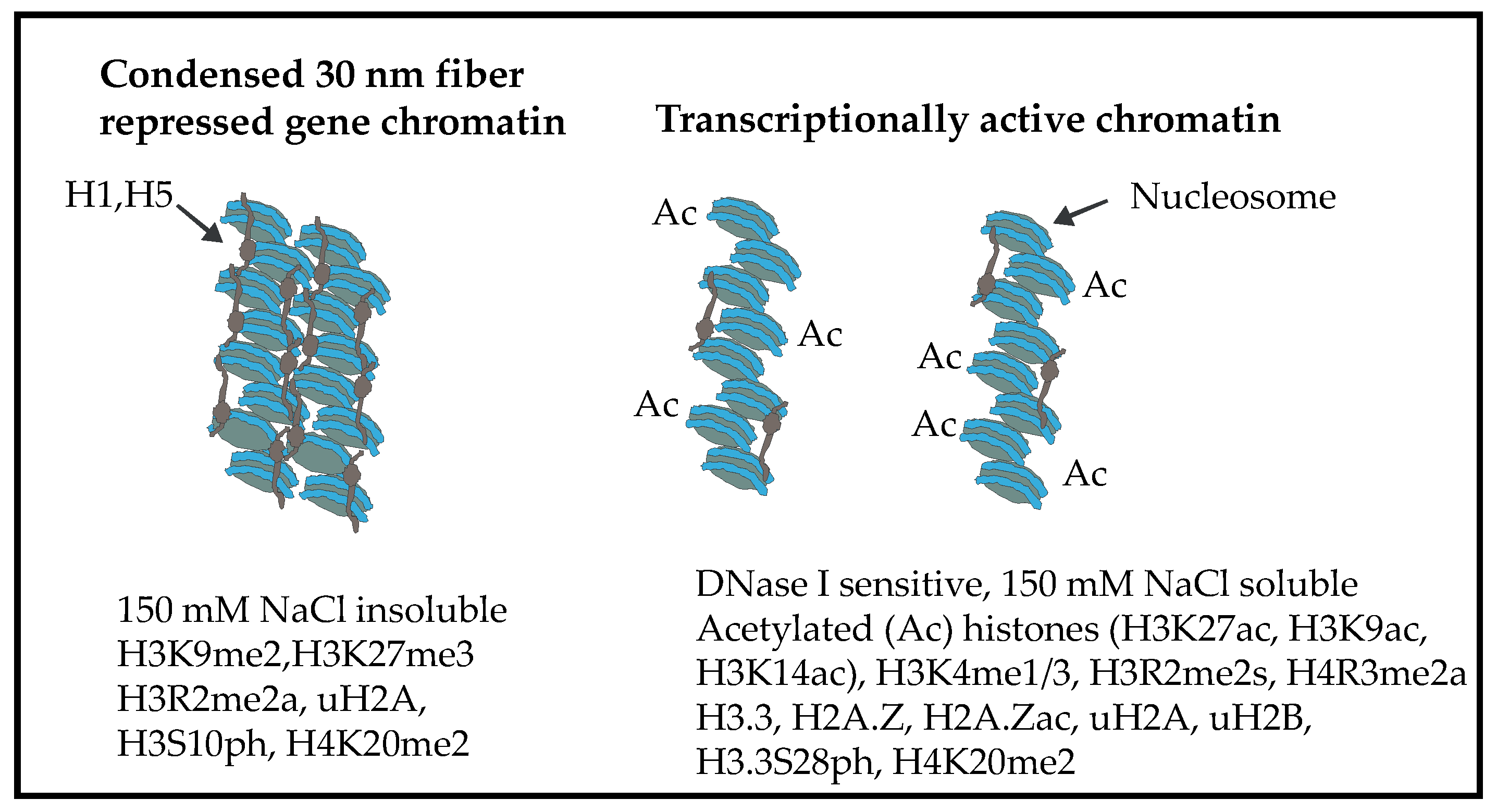
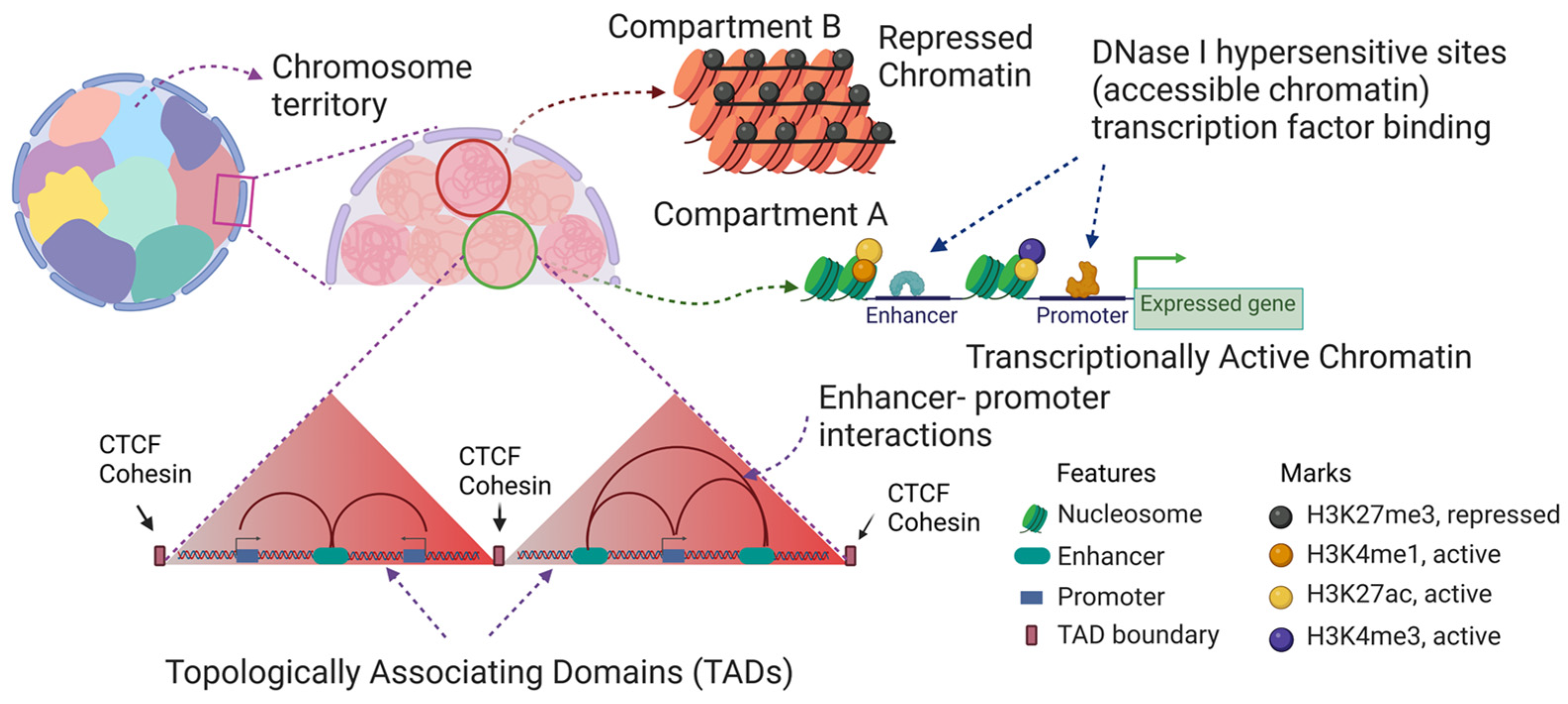
2. Chromatin Structure of the Chicken Erythrocyte
3. Chicken Erythrocyte Promoters
4. DNA Methylation
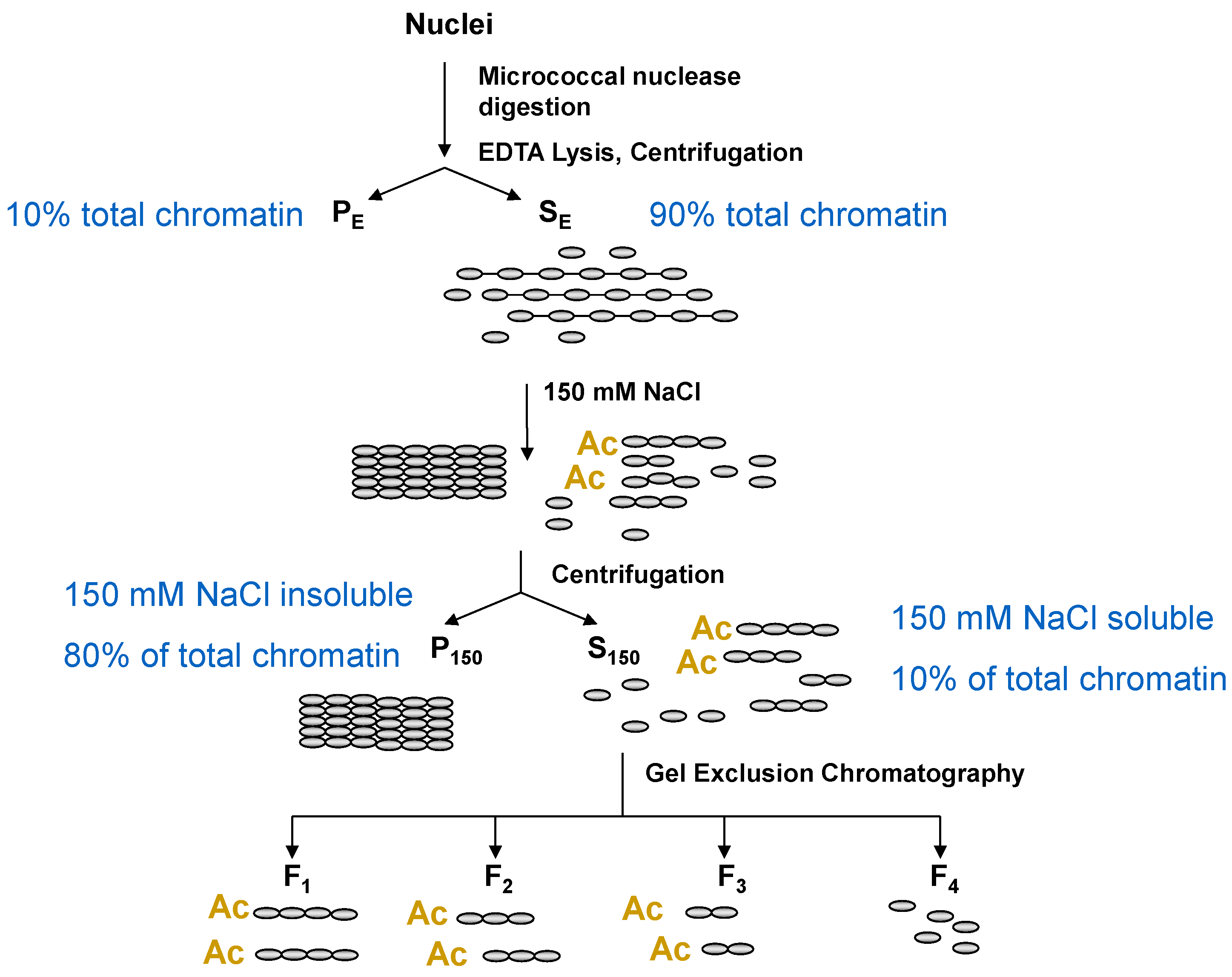
5. Chromatin Fractionation: A Powerful Method to Study Chromatin Composition of Expressed and Repressed Chromatin
6. Chicken Erythrocyte Core Histone PTMs
6.1. Metabolism
6.2. Histone H2A and H2B PTMs
6.3. Histone H3 PTMs
6.4. Histone H4 PTMs
7. Histone Acetylation, Chromatin Solubility, and the DNase I Sensitive Chromatin Domains
8. Chicken Erythrocyte Histone Genes and Variants
9. Chicken Histone PTMs, Nucleosome-Free Regions, and Genomic Mapping
10. Broad Histone PTM Domains and Chromatin Remodeling by CHD1
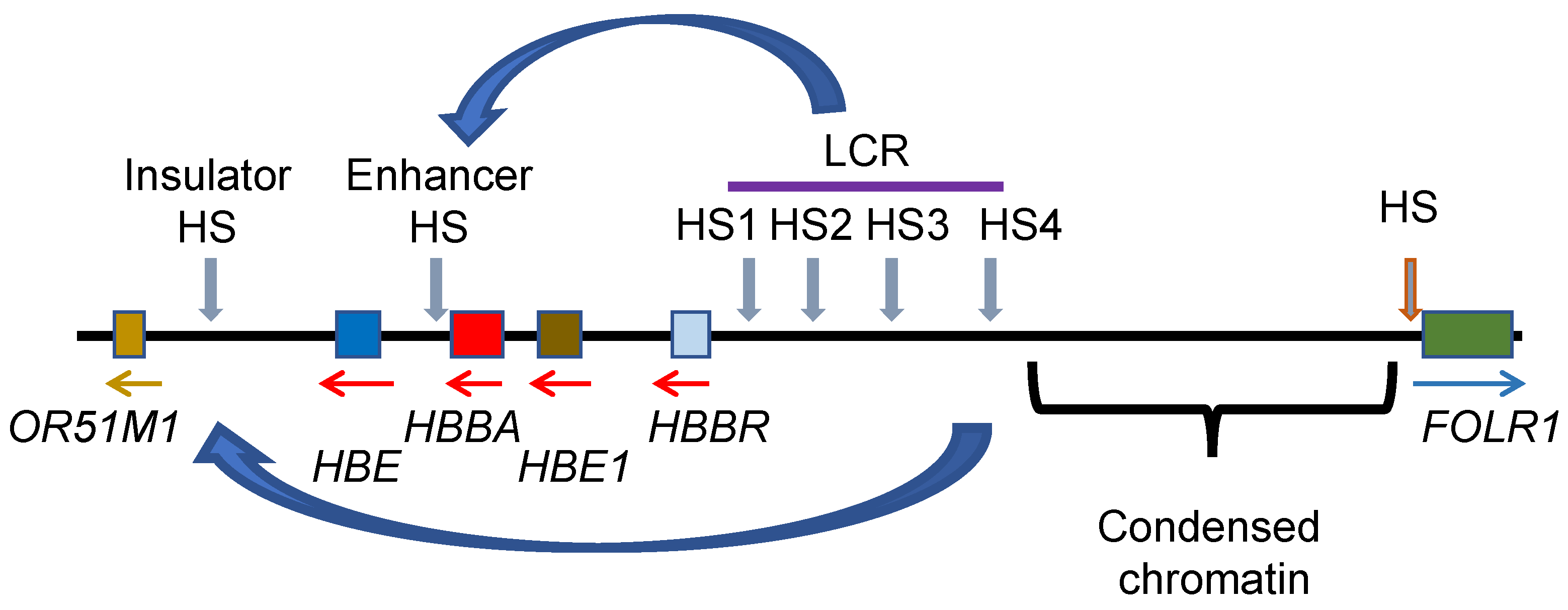
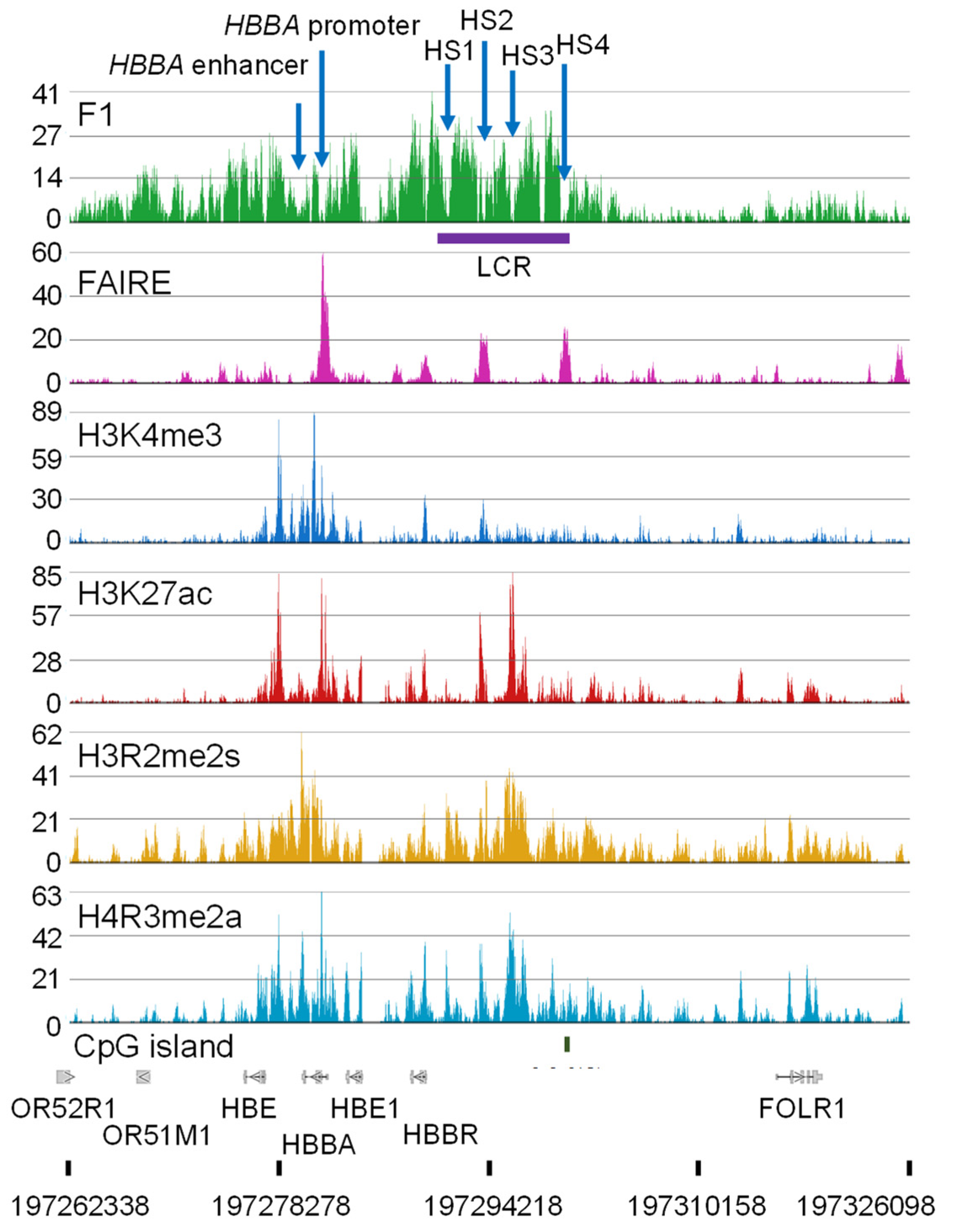
11. Genome Organization of the Genes Involved in Oxygen Transport
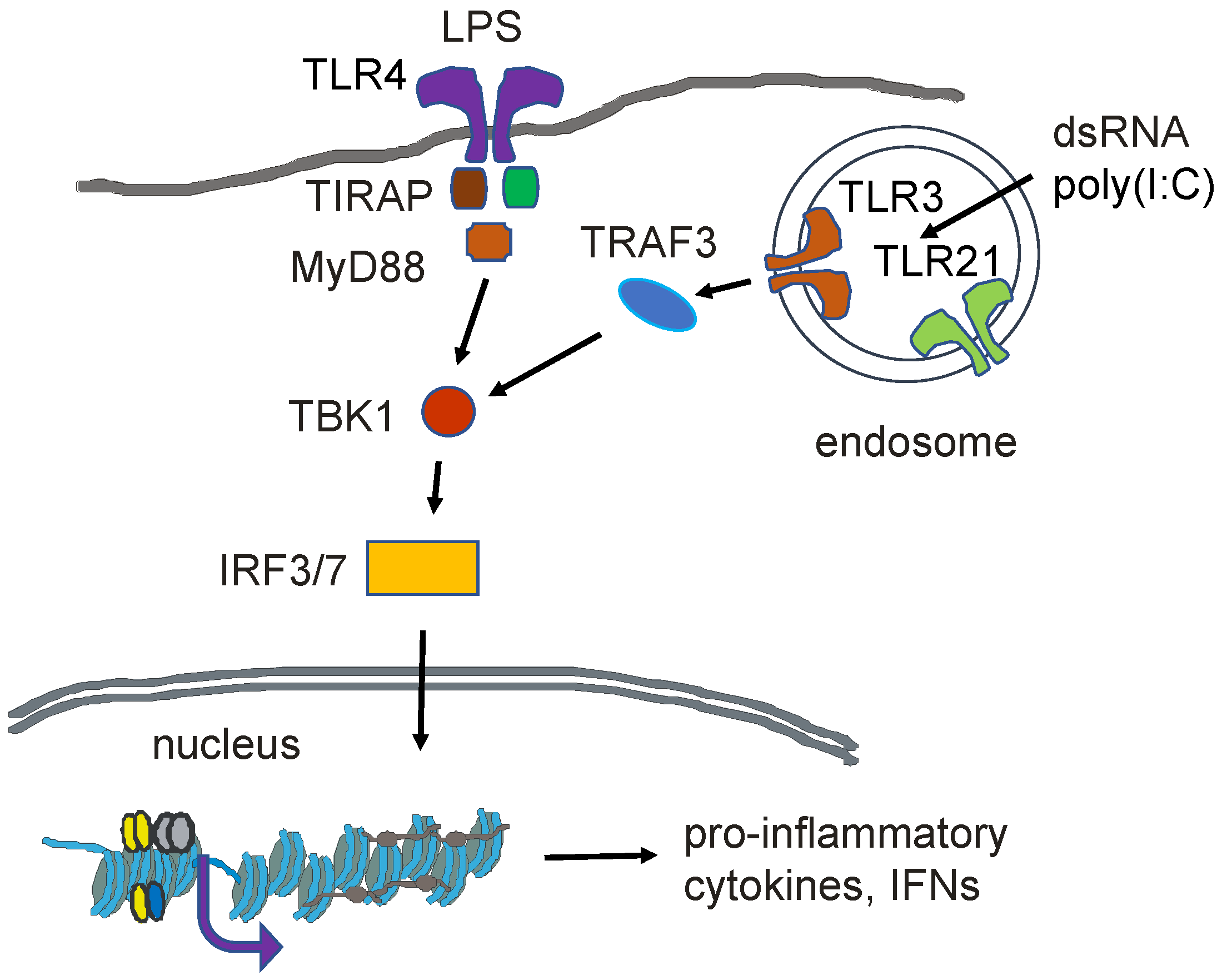
12. Innate Immunity
13. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Eda, M. Origin of the domestic chicken from modern biological and zooarchaeological approaches. Anim. Front. Rev. Mag. Anim. Agric. 2021, 11, 52. [Google Scholar] [CrossRef]
- Carelli-Alinovi, C.; Pirolli, D.; Giardina, B.; Misiti, F. Protein kinase C mediates caspase 3 activation: A role for erythrocyte morphology changes. Clin. Hemorheol. Microcirc. 2015, 59, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Recillas-Targa, F.; Bell, A.C.; Felsenfeld, G. Positional enhancer-blocking activity of the chicken β-globin insulator in transiently transfected cells. Proc. Natl. Acad. Sci. USA 1999, 96, 14354–14359. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Mahamid, J.; Dombrowski, M.; Baumeister, W.; Olins, A.L.; Olins, D.E. Interphase epichromatin: Last refuge for the 30-nm chromatin fiber? Chromosoma 2021, 130, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Morera, D.; Roher, N.; Ribas, L.; Balasch, J.C.; Donate, C.; Callol, A.; Boltana, S.; Roberts, S.; Goetz, G.; Goetz, F.W.; et al. RNA-Seq reveals an integrated immune response in nucleated erythrocytes. PLoS ONE 2011, 6, e26998. [Google Scholar] [CrossRef]
- Affolter, M.; Cote, J.; Renaud, J.; Ruiz-Carrillo, A.; Côté, J.; Renaud, J.; Ruiz-Carrillo, A. Regulation of histone and beta A-globin gene expression during differentiation of chicken erythroid cells. Mol. Cell. Biol. 1987, 7, 3663–3672. [Google Scholar] [CrossRef]
- Kurumizaka, H.; Kujirai, T.; Takizawa, Y. Contributions of histone variants in nucleosome structure and function. J. Mol. Biol. 2021, 433, 166678. [Google Scholar] [CrossRef]
- Ridsdale, J.A.; Hendzel, M.J.; Delcuve, G.P.; Davie, J.R. Histone acetylation alters the capacity of the H1 histones to condense transcriptionally active/competent chromatin. J. Biol. Chem. 1990, 265, 5150–5156. [Google Scholar] [CrossRef]
- Delcuve, G.P.; Davie, J.R. Chromatin structure of erythroid-specific genes of immature and mature chicken erythrocytes. Biochem. J. 1989, 263, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Ridsdale, J.A.; Rattner, J.B.; Davie, J.R. Erythroid-specific gene chromatin has an altered association with linker histones. Nucleic Acids Res. 1988, 16, 5915–5926. [Google Scholar] [CrossRef]
- Jahan, S.; Xu, W.; He, S.; Gonzalez, C.; Delcuve, G.P.; Davie, J.R. The chicken erythrocyte epigenome. Epigenetics Chromatin 2016, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Beacon, T.H.; Davie, J.R. Transcriptionally active chromatin-lessons learned from the chicken erythrocyte chromatin fractionation. Cells 2021, 10, 1354. [Google Scholar] [CrossRef]
- Misteli, T. The Self-organizing genome: Principles of genome architecture and function. Cell 2020, 183, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Fishman, V.; Battulin, N.; Nuriddinov, M.; Maslova, A.; Zlotina, A.; Strunov, A.; Chervyakova, D.; Korablev, A.; Serov, O.; Krasikova, A. 3D organization of chicken genome demonstrates evolutionary conservation of topologically associated domains and highlights unique architecture of erythrocytes’ chromatin. Nucleic Acids Res. 2018, 47, 648–665. [Google Scholar] [CrossRef]
- Ryzhkova, A.; Taskina, A.; Khabarova, A.; Fishman, V.; Battulin, N. Erythrocytes 3D genome organization in vertebrates. Sci. Rep. 2021, 11, 4414. [Google Scholar] [CrossRef] [PubMed]
- Kantidze, O.L.; Iarovaia, O.V.; Philonenko, E.S.; Yakutenko, I.I.; Razin, S.V. Unusual compartmentalization of CTCF and other transcription factors in the course of terminal erythroid differentiation. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2007, 1773, 924–933. [Google Scholar] [CrossRef]
- Gardiner-Garden, M.; Frommer, M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987, 196, 261–282. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Gemmell, N.J. Abundance, arrangement, and function of sequence motifs in the chicken promoters. BMC Genom. 2014, 15, 900. [Google Scholar] [CrossRef]
- Smale, S.T.; Tarakhovsky, A.; Natoli, G. Chromatin contributions to the regulation of innate immunity. Annu. Rev. Immunol. 2014, 32, 489–511. [Google Scholar] [CrossRef]
- Ramirez-Carrozzi, V.R.; Braas, D.; Bhatt, D.M.; Cheng, C.S.; Hong, C.; Doty, K.R.; Black, J.C.; Hoffmann, A.; Carey, M.; Smale, S.T. A Unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 2009, 138, 114–128. [Google Scholar] [CrossRef]
- Davie, J.R.; Xu, W.; Delcuve, G.P. Histone H3K4 trimethylation: Dynamic interplay with pre-mRNA splicing. Biochem. Cell Biol. 2015, 94, 1–11. [Google Scholar] [CrossRef]
- Migliori, V.; Müller, J.; Phalke, S.; Low, D.; Bezzi, M.; Mok, W.C.; Sahu, S.K.; Gunaratne, J.; Capasso, P.; Bassi, C.; et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat. Struct. Mol. Biol. 2012, 19, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Jahan, S.; Beacon, T.H.; He, S.; Gonzalez, C.; Xu, W.; Delcuve, G.P.; Jia, S.; Hu, P.; Davie, J.R. Chromatin organization of transcribed genes in chicken polychromatic erythrocytes. Gene 2019, 699, 80–87. [Google Scholar] [CrossRef]
- Ndlovu, M.N.; Denis, H.; Fuks, F. Exposing the DNA methylome iceberg. Trends Biochem. Sci. 2011, 36, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Haigh, L.S.; Owens, B.B.; Hellewell, S.; Ingram, V.M. DNA methylation in chicken alpha-globin gene expression. Proc. Natl. Acad. Sci. USA 1982, 79, 5332–5336. [Google Scholar] [CrossRef] [PubMed]
- Cheung, P.; Lau, P. Epigenetic regulation by histone methylation and histone variants. Mol. Endocrinol. 2005, 19, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Tada, M.; Hayashi, A.; Asano, Y.; Kubiura-Ichimaru, M.; Ito, T.; Yoshii, M.; Kimura, H.; Matsuda, Y.; Oshimura, M. Evidence for divergence of DNA methylation maintenance and a conserved inhibitory mechanism from DNA demethylation in chickens and mammals. Genes Genom. 2021, 43, 269–280. [Google Scholar] [CrossRef]
- Okuzaki, Y.; Kaneoka, H.; Nishijima, K.I.; Murakami, S.; Ozawa, Y.; Iijima, S. Molecular cloning of chicken TET family genes and role of chicken TET1 in erythropoiesis. Biochem. Biophys. Res. Commun. 2017, 490, 753–759. [Google Scholar] [CrossRef]
- Pértille, F.; Brantsæter, M.; Nordgreen, J.; Coutinho, L.L.; Janczak, A.M.; Jensen, P.; Guerrero-Bosagna, C.; Pertille, F.; Brantsaeter, M.; Nordgreen, J.; et al. DNA methylation profiles in red blood cells of adult hens correlate with their rearing conditions. J. Exp. Biol. 2017, 220, 3579–3587. [Google Scholar] [CrossRef]
- Pértille, F.; Ibelli, A.M.G.; Sharif, M.E.; Poleti, M.D.; Fröhlich, A.S.; Rezaei, S.; Ledur, M.C.; Jensen, P.; Guerrero-Bosagna, C.; Coutinho, L.L. Putative epigenetic biomarkers of stress in red blood cells of chickens reared across different biomes. Front. Genet. 2020, 11, 508809. [Google Scholar] [CrossRef]
- Rezaei, S.; Uffenorde, J.; Gimm, O.; Hosseinpour Feizi, M.A.; Miemczyk, S.; Coutinho, L.L.; Jensen, P.; Guerrero-Bosagna, C.; Pértille, F. GBS-MeDIP: A protocol for parallel identification of genetic and epigenetic variation in the same reduced fraction of genomes across individuals. STAR Protoc. 2022, 3, 101202. [Google Scholar] [CrossRef] [PubMed]
- Ridsdale, J.A.; Davie, J.R. Chicken erythrocyte polynucleosomes which are soluble at physiological ionic strength and contain linker histones are highly enriched in β-globin gene sequences. Nucleic Acids Res. 1987, 15, 1081–1096. [Google Scholar] [CrossRef] [PubMed]
- Locklear, L.; Ridsdale, A.J.; Bazett-Jones, D.P.; Davie, J.R. Ultrastructure of transcriptionally competent chromatin. Nucleic Acids Res. 1990, 18, 7015–7024. [Google Scholar] [CrossRef]
- Czarnota, G.J.; Bazett-Jones, D.P.; Mendez, E.; Allfrey, V.G.; Ottensmeyer, F.P. High resolution microanalysis and three-dimensional nucleosome structure associated with transcribing chromatin. Micron 1997, 28, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Walia, H.; Chen, H.Y.; Sun, J.M.; Holth, L.T.; Davie, J.R. Histone acetylation is required to maintain the unfolded nucleosome structure associated with transcribing DNA. J. Biol. Chem. 1998, 273, 14516–14522. [Google Scholar] [CrossRef] [PubMed]
- Allegra, P.; Sterner, R.; Clayton, D.F.; Allfrey, V.G. Affinity chromatographic purification of nucleosomes containing transcriptionally active DNA sequences. J. Mol. Biol. 1987, 196, 379–388. [Google Scholar] [CrossRef]
- Beacon, T.H.; Delcuve, G.P.; López, C.; Nardocci, G.; Kovalchuk, I.; van Wijnen, A.J.; Davie, J.R. The dynamic broad epigenetic (H3K4me3, H3K27ac) domain as a mark of essential genes. Clin. Epigenetics 2021, 13, 138. [Google Scholar] [CrossRef]
- Boon, R.; Silveira, G.G.; Mostoslavsky, R. Nuclear metabolism and the regulation of the epigenome. Nat. Metab. 2020, 2, 1190–1203. [Google Scholar] [CrossRef]
- Reid, M.A.; Dai, Z.; Locasale, J.W. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol. 2017, 19, 1298–1306. [Google Scholar] [CrossRef]
- Zhang, D.E.; Nelson, D.A. Histone acetylation in chicken erythrocytes. Rates of acetylation and evidence that histones in both active and potentially active chromatin are rapidly modified. Biochem. J. 1988, 250, 233–240. [Google Scholar] [CrossRef]
- Li, W.; Nagaraja, S.; Delcuve, G.P.; Hendzel, M.J.; Davie, J.R. Effects of histone acetylation, ubiquitination and variants on nucleosome stability. Biochem. J. 1993, 296, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Hendzel, M.J.; Davie, J.R. Nucleosomal histones of transcriptionally active/competent chromatin preferentially exchange with newly synthesized histones in quiescent chicken erythrocytes. Biochem. J. 1990, 271, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Davie, J.R. Two-dimensional gel systems for rapid histone analysis for use in minislab polyacrylamide gel electrophoresis. Anal. Biochem. 1982, 120, 276–281. [Google Scholar] [CrossRef]
- Jahan, S. Characterization of Transcriptionally Active Chicken Erythrocyte Chromatin. Available online: https://mspace.lib.umanitoba.ca/handle/1993/32700 (accessed on 19 June 2020).
- Beacon, T.H.; Xu, W.; Davie, J.R. Genomic landscape of transcriptionally active histone arginine methylation marks, H3R2me2s and H4R3me2a, relative to nucleosome depleted regions. Gene 2020, 742, 144593. [Google Scholar] [CrossRef]
- Spencer, V.A.; Davie, J.R. Dynamically acetylated histone association with transcriptionally active and competent genes in the avian Adult β-globin gene domain. J. Biol. Chem. 2001, 276, 34810–34815. [Google Scholar] [CrossRef]
- Hendzel, M.J.; Davie, J.R. Dynamically acetylated histones of chicken erythrocytes are selectively methylated. Biochem. J. 1991, 273, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Tang, H. Analysis of core histones by liquid chromatography-mass spectrometry and peptide mapping. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003, 783, 173–179. [Google Scholar] [CrossRef]
- Zhang, D.E.; Nelson, D.A. Histone acetylation in chicken erythrocytes. Rates of deacetylation in immature and mature red blood cells. Biochem. J. 1988, 250, 241–245. [Google Scholar] [CrossRef]
- Weinert, B.T.; Narita, T.; Satpathy, S.; Srinivasan, B.; Hansen, B.K.; Schölz, C.; Hamilton, W.B.; Zucconi, B.E.; Wang, W.W.; Liu, W.R.; et al. Time-resolved analysis reveals rapid dynamics and broad scope of the CBP/p300 acetylome. Cell 2018, 174, 231–244.e12. [Google Scholar] [CrossRef]
- Jahan, S.; Sun, J.M.; He, S.; Davie, J.R. Transcription-dependent association of HDAC2 with active chromatin. J. Cell. Physiol. 2018, 233, 1650–1657. [Google Scholar] [CrossRef]
- Sun, J.-M.; Chen, H.Y.; Moniwa, M.; Samuel, S.; Davie, J.R. Purification and characterization of chicken erythrocyte histone deacetylase 1. Biochemistry 1999, 38, 5939–5947. [Google Scholar] [CrossRef] [PubMed]
- Hebbes, T.R.; Allen, S.C.H. Multiple histone acetyltransferases are associated with a chicken erythrocyte chromatin fraction enriched in active genes. J. Biol. Chem. 2000, 275, 31347–31352. [Google Scholar] [CrossRef] [PubMed]
- Hendzel, M.J.; Delcuve, G.P.; Davie, J.R. Histone deacetylase is a component of the internal nuclear matrix. J. Biol. Chem. 1991, 266, 21936–21942. [Google Scholar] [CrossRef] [PubMed]
- Hendzel, M.J.; Sun, J.-M.; Chen, H.Y.; Rattner, J.B.; Davie, J.R. Histone acetyltransferase Is associated with the nuclear matrix. J. Biol. Chem. 1994, 269, 22894–22901. [Google Scholar] [CrossRef]
- Beacon, T.H.; Davie, J.R. The chicken model organism for epigenomic research. Genome 2021, 64, 476–489. [Google Scholar] [CrossRef]
- Stalder, J.; Larsen, A.; Engel, J.D.; Dolan, M.; Groudine, M.; Weintraub, H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase, I. Cell 1980, 20, 451–460. [Google Scholar] [CrossRef]
- Hebbes, T.R.; Clayton, A.L.; Thorne, A.W.; Crane-Robinson, C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 1994, 13, 1823–1830. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, N.; Weintraub, H. Localization of DNAase I-sensitive sequences to specific regions of interphase nuclei. Cell 1985, 43, 471–482. [Google Scholar] [CrossRef]
- Sun, J.M.; Ferraiuolo, R.; Davie, J.R. In situ footprinting of chicken histone H5 gene in mature and immature erythrocytes reveals common factor-binding sites. Chromosoma 1996, 104, 504–510. [Google Scholar] [CrossRef]
- Jin, C.; Zang, C.; Wei, G.; Cui, K.; Peng, W.; Zhao, K.; Felsenfeld, G. H3.3/H2A.Z double variant-containing nucleosomes mark “nucleosome-free regions” of active promoters and other regulatory regions. Nat. Genet. 2009, 41, 941–945. [Google Scholar] [CrossRef]
- Sun, J.-M.; Chen, H.Y.; Espino, P.S.; Davie, J.R. Phosphorylated serine 28 of histone H3 is associated with destabilized nucleosomes in transcribed chromatin. Nucleic Acids Res. 2007, 35, 6640–6647. [Google Scholar] [CrossRef]
- Jahan, S.; Beacon, T.H.; Xu, W.; Davie, J.R. Atypical chromatin structure of immune-related genes expressed in chicken erythrocytes. Biochem. Cell Biol. 2020, 98, 171–177. [Google Scholar] [CrossRef]
- Radman-Livaja, M.; Quan, T.K.; Valenzuela, L.; Armstrong, J.A.; van Welsem, T.; Kim, T.S.; Lee, L.J.; Buratowski, S.; van Leeuwen, F.; Rando, O.J.; et al. A key role for Chd1 in histone H3 dynamics at the 3′ ends of long genes in yeast. PLoS Genet. 2012, 8, e1002811. [Google Scholar] [CrossRef] [PubMed]
- Alendar, A.; Berns, A. Sentinels of chromatin: Chromodomain helicase DNA-binding proteins in development and disease. Genes Dev. 2021, 35, 1403–1430. [Google Scholar] [CrossRef] [PubMed]
- Zubek, J.; Stitzel, M.L.; Ucar, D.; Plewczynski, D.M. Computational inference of H3K4me3 and H3K27ac domain length. PeerJ 2016, 2016, e1750. [Google Scholar] [CrossRef]
- Nodelman, I.M.; Das, S.; Faustino, A.M.; Fried, S.D.; Bowman, G.D.; Armache, J.P. Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state. Nat. Struct. Mol. Biol. 2022, 29, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Farnung, L.; Vos, S.M.; Wigge, C.; Cramer, P. Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature 2017, 550, 539–542. [Google Scholar] [CrossRef]
- Lee, Y.; Park, D.; Iyer, V.R. The ATP-dependent chromatin remodeler Chd1 is recruited by transcription elongation factors and maintains H3K4me3/H3K36me3 domains at actively transcribed and spliced genes. Nucleic Acids Res. 2017, 45, 8646. [Google Scholar] [CrossRef]
- Baumgart, S.J.; Najafova, Z.; Hossan, T.; Xie, W.; Nagarajan, S.; Kari, V.; Ditzel, N.; Kassem, M.; Johnsen, S.A. CHD1 regulates cell fate determination by activation of differentiation-induced genes. Nucleic Acids Res. 2017, 45, 7722–7735. [Google Scholar] [CrossRef]
- Fuchs, S.M.; Krajewski, K.; Baker, R.W.; Miller, V.L.; Strahl, B.D. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr. Biol. 2011, 21, 53–58. [Google Scholar] [CrossRef]
- Schoberleitner, I.; Bauer, I.; Huang, A.; Andreyeva, E.N.; Sebald, J.; Pascher, K.; Rieder, D.; Brunner, M.; Podhraski, V.; Oemer, G.; et al. CHD1 controls H3.3 incorporation in adult brain chromatin to maintain metabolic homeostasis and normal lifespan. Cell Rep. 2021, 37, 109769. [Google Scholar] [CrossRef]
- Jin, C.; Felsenfeld, G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007, 21, 1519–1529. [Google Scholar] [CrossRef]
- Harada, A.; Okada, S.; Konno, D.; Odawara, J.; Yoshimi, T.; Yoshimura, S.; Kumamaru, H.; Saiwai, H.; Tsubota, T.; Kurumizaka, H.; et al. Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J. 2012, 31, 2994–3007. [Google Scholar] [CrossRef]
- Wang, L.C.; Chen, C.T.; Lee, H.Y.; Li, S.H.; Lir, J.T.; Chin, S.C.; Pu, C.E.; Wang, C.H. Sexing a wider range of avian species based on two CHD1 introns with a unified reaction condition. Zoo Biol. 2007, 26, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Dierks, C.; Altgilbers, S.; Weigend, A.; Preisinger, R.; Weigend, S. Sexing assay for chickens and other birds for large-scale application based on a conserved sequence variant in CHD1 genes on W and Z chromosomes. Anim. Genet. 2022, 53, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Howell, R.M.; Woodford, K.J.; Weitzmann, M.N.; Usdin, K. The chicken beta-globin gene promoter forms a novel “cinched” tetrahelical structure. J. Biol. Chem. 1996, 271, 5208–5214. [Google Scholar] [CrossRef] [PubMed]
- Ulianov, S.V.; Gavrilov, A.A.; Razin, S.V. Spatial organization of the chicken beta-globin gene domain in erythroid cells of embryonic and adult lineages. Epigenetics Chromatin 2012, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.H.; Bell, A.C.; Felsenfeld, G. Characterization of the chicken beta-globin insulator. Proc. Natl. Acad. Sci. USA 1997, 94, 575–580. [Google Scholar] [CrossRef]
- Dickson, J.; Gowher, H.; Strogantsev, R.; Gaszner, M.; Hair, A.; Felsenfeld, G.; West, A.G. VEZF1 elements mediate protection from DNA methylation. PLoS Genet. 2010, 6, e1000804. [Google Scholar] [CrossRef]
- Aker, M.; Bomsztyk, K.; Emery, D.W. Poly(ADP-ribose) polymerase-1 (PARP-1) contributes to the barrier function of a vertebrate chromatin insulator. J. Biol. Chem. 2010, 285, 37589–37597. [Google Scholar] [CrossRef]
- Benabdallah, N.S.; Williamson, I.; Illingworth, R.S.; Kane, L.; Boyle, S.; Sengupta, D.; Grimes, G.R.; Therizols, P.; Bickmore, W.A. Decreased enhancer-promoter proximity accompanying enhancer activation. Mol. Cell 2019, 76, 473–484.e7. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.S.; Rehman, S.; Yousaf, W.; Hassan, F.U.; Ahmad, W.; Liu, Q.; Pan, H. The potential of toll-like receptors to modulate avian immune system: Exploring the effects of genetic variants and phytonutrients. Front. Genet. 2021, 12, 671235. [Google Scholar] [CrossRef] [PubMed]
- Sick, C.; Schultz, U.; Münster, U.; Meier, J.; Kaspers, B.; Staeheli, P. Promoter structures and differential responses to viral and nonviral inducers of chicken type I interferon genes. J. Biol. Chem. 1998, 273, 9749–9754. [Google Scholar] [CrossRef] [PubMed]
- Besant, P.G.; Attwood, P.V. Histone H4 histidine phosphorylation: Kinases, phosphatases, liver regeneration and cancer. Biochem. Soc. Trans. 2012, 40, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Shi, X. H3.3S31 phosphorylation: Linking transcription elongation to stimulation responses. Signal Transduct. Target. Ther. 2020, 5, 176. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.A.; Ng, M.K.; Cheung, P. The use of mononucleosome immunoprecipitation for analysis of combinatorial histone post-translational modifications and purification of nucleosome-interacting proteins. Front. Cell Dev. Biol. 2020, 8, 331. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beacon, T.H.; Davie, J.R. Chicken Erythrocyte: Epigenomic Regulation of Gene Activity. Int. J. Mol. Sci. 2023, 24, 8287. https://doi.org/10.3390/ijms24098287
Beacon TH, Davie JR. Chicken Erythrocyte: Epigenomic Regulation of Gene Activity. International Journal of Molecular Sciences. 2023; 24(9):8287. https://doi.org/10.3390/ijms24098287
Chicago/Turabian StyleBeacon, Tasnim H., and James R. Davie. 2023. "Chicken Erythrocyte: Epigenomic Regulation of Gene Activity" International Journal of Molecular Sciences 24, no. 9: 8287. https://doi.org/10.3390/ijms24098287
APA StyleBeacon, T. H., & Davie, J. R. (2023). Chicken Erythrocyte: Epigenomic Regulation of Gene Activity. International Journal of Molecular Sciences, 24(9), 8287. https://doi.org/10.3390/ijms24098287




