Vaginal and Cervical Microbiota Composition in Patients with Endometrial Cancer
Abstract
1. Introduction
2. Results
2.1. Study Population
2.2. Qualitative Identification of Vaginal/Cervical Microbiota
2.3. Quantitative Identification of Vaginal/Cervical Microbiota
2.4. Vaginal/Cervical Microbiota Composition in Patients with Endometrial Cancer and Precancerous Lesion
2.5. Vaginal/Cervical Microbiota Composition in Regard to Menopausal Status
2.6. Relative Profiling of Vaginal/Cervical Microbiota Composition in Endometrial Cancer vs. Atypical Hyperplasia or Benign Uterine Condition
2.7. Prediction of Bacterial Communities Profile
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Sample Collection
4.3. Real-Time PCR Analysis
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Łaniewski, P.; Ilhan, Z.E.; Herbst-Kralovetz, M.M. The microbiome and gynaecological cancer development, prevention and therapy. Nat. Rev. Urol. 2020, 17, 232–250. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Chase, D.; Goulder, A.; Zenhausern, F.; Monk, B.; Herbst-Kralovetz, M. The vaginal and gastrointestinal microbiomes in gynecologic cancers: A review of applications in etiology, symptoms and treatment. Gynecol. Oncol. 2015, 138, 190–200. [Google Scholar] [CrossRef]
- Hernandes, C.; Silveira, P.; Rodrigues Sereia, A.F.; Christoff, A.P.; Mendes, H.; Valter de Oliveira, L.F.; Podgaec, S. Microbiome Profile of Deep Endometriosis Patients: Comparison of Vaginal Fluid, Endometrium and Lesion. Diagnostics 2020, 10, 163. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ko, E.Y.-L.; Wong, K.K.-W.; Chen, X.; Cheung, W.-C.; Law, T.S.-M.; Chung, J.P.-W.; Tsui, S.K.-W.; Li, T.-C.; Chim, S.S.-C. Endometrial Microbiota in Infertile Women with and without Chronic Endometritis as Diagnosed Using a Quantitative and Reference Range-Based Method. Fertil. Steril. 2019, 112, 707–717.e1. [Google Scholar] [CrossRef]
- Fang, R.L.; Chen, L.X.; Shu, W.S.; Yao, S.Z.; Wang, S.W.; Chen, Y.Q. Barcoded sequencing reveals diverse intrauterine microbiomes in patients suffering with endometrial polyps. Am. J. Transl. Res. 2016, 8, 1581–1592. [Google Scholar]
- Pelzer, E.S.; Willner, D.; Buttini, M.; Huygens, F. A Role for the Endometrial Microbiome in Dysfunctional Menstrual Bleeding. Antonie Van Leeuwenhoek 2018, 111, 933–943. [Google Scholar] [CrossRef]
- Walsh, D.M.; Hokenstad, A.N.; Chen, J.; Sung, J.; Jenkins, G.D.; Chia, N.; Nelson, H.; Mariani, A.; Walther-Antonio, M.R.S. Postmenopause as a Key Factor in the Composition of the Endometrial Cancer Microbiome (ECbiome). Sci. Rep. 2019, 9, 19213. [Google Scholar] [CrossRef]
- Wee, B.A.; Thomas, M.; Sweeney, E.L.; Frentiu, F.D.; Samios, M.; Ravel, J.; Gajer, P.; Myers, G.; Timms, P.; Allan, J.A.; et al. A Retrospective Pilot Study to Determine Whether the Reproductive Tract Microbiota Differs between Women with a History of Infertility and Fertile Women. Aust. N. Z. J. Obstet. Gynaecol. 2018, 58, 341–348. [Google Scholar] [CrossRef]
- Francescone, R.; Hou, V.; Grivennikov, S.I. Microbiome, Inflammation, and Cancer. Cancer J. 2014, 20, 181–189. [Google Scholar] [CrossRef]
- Sobstyl, M.; Brecht, P.; Sobstyl, A.; Mertowska, P.; Grywalska, E. The Role of Microbiota in the Immunopathogenesis of Endometrial Cancer. Int. J. Mol. Sci. 2022, 23, 5756. [Google Scholar] [CrossRef] [PubMed]
- Rogers, C.J.; Prabhu, K.S.; Vijay-Kumar, M. The microbiome and obesity-an established risk for certain types of cancer. Cancer J. 2014, 20, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Wessels, J.M.; Felker, A.M.; Dupont, H.A.; Kaushic, C. The relationship between sex hormones, the vaginal microbiome and immunity in HIV-1 susceptibility in women. Dis. Model. Mech. 2018, 11, dmm035147. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Hansmann, M.A.; Davis, C.C.; Suzuki, H.; Brown, C.J.; Schütte, U.; Pierson, J.D.; Forney, L.J. The vaginal bacterial communities of Japanese women resemble those of women in other racial groups. FEMS Immunol. Med. Microbiol. 2010, 58, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Braundmeier, A.G.; Lenz, K.M.; Inman, K.S.; Chia, N.; Jeraldo, P.; Walther António, M.R.; Berg Miller, M.E.; Yang, F.; Creedon, D.J.; Nelson, H.; et al. Individualized medicine and the microbiome in reproductive tract. Front Physiol. 2015, 6, 97. [Google Scholar] [CrossRef]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, R.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108, 4680–4687. [Google Scholar] [CrossRef]
- Walther-António, M.R.; Chen, J.; Multinu, F.; Hokenstad, A.; Distad, T.J.; Cheek, E.H.; Keeney, G.L.; Creedon, D.J.; Nelson, H.; Mariani, A.; et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 2016, 8, 122. [Google Scholar] [CrossRef]
- Gressel, G.M.; Usyk, M.; Frimer, M.; Kuo, D.Y.S.; Burk, R.D. Characterization of the endometrial, cervicovaginal and anorectal microbiota in post-menopausal women with endometrioid and serous endometrial cancers. PLoS ONE 2021, 16, e0259188. [Google Scholar] [CrossRef]
- Hakimjavadi, H.; George, S.H.; Taub, M.; Dodds, L.V.; Sanchez-Covarrubias, A.P.; Huang, M.; Pearson, J.M.; Slomovitz, B.M.; Kobetz, E.N.; Gharaibeh, R.; et al. The vaginal microbiome is associated with endometrial cancer grade and histology. Cancer Res. Commun. 2022, 2, 447–455. [Google Scholar] [CrossRef]
- Zhu, N.; Yang, X.; Liu, Q.; Chen, Y.; Wang, X.; Li, H.; Gao, H. “Iron triangle” of regulating the uterine microecology: Endometrial microbiota, immunity and endometrium. Front. Immunol. 2022, 13, 928475. [Google Scholar] [CrossRef]
- Baj, J.; Korona-Głowniak, I.; Forma, A.; Maani, A.; Sitarz, E.; Rahnama-Hezavah, M.; Radzikowska, E.; Portincasa, P. Mechanisms of the Epithelial-Mesenchymal Transition and Tumor Microenvironment in Helicobacter pylori-Induced Gastric Cancer. Cells 2020, 23, 1055. [Google Scholar] [CrossRef]
- Wang, N.; Fang, J.Y. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 2023, 31, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.C.; Blanchard, Z.; Maurer, K.A.; Gertz, J. Estrogen Signaling in Endometrial Cancer: A Key Oncogenic Pathway with Several Open Questions. Horm. Cancer 2019, 10, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Plottel, C.S.; Blaser, M.J. Microbiome and malignancy. Cell Host Microbe 2011, 20, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Dabek, M.; McCrae, S.I.; Stevens, V.J.; Duncan, S.H.; Louis, P. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol. Ecol. 2008, 66, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Gloux, K.; Berteau, O.; El Oumami, H.; Béguet, F.; Leclerc, M.; Doré, J. A metagenomic β-glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc. Natl. Acad. Sci. USA 2011, 15, 4539–4546. [Google Scholar] [CrossRef]
- Candeliere, F.; Raimondi, S.; Ranieri, R.; Musmeci, E.; Zambon, A.; Amaretti, A.; Rossi, M. β-Glucuronidase Pattern Predicted From Gut Metagenomes Indicates Potentially Diversified Pharmacomicrobiomics. Front. Microbiol. 2022, 13, 826994. [Google Scholar] [CrossRef]
- Elkafas, H.; Wall, M.; Al-Hendy, A.; Ismail, N. Gut and genital tract microbiomes: Dysbiosis and link to gynecological disorders. Front. Cell. Infect. Microbiol. 2022, 12, 1059825. [Google Scholar] [CrossRef]
- Dossus, L.; Rinaldi, S.; Becker, S.; Lukanova, A.; Tjonneland, A.; Olsen, A.; Stegger, J.; Overvad, K.; Chabbert-Buffet, N.; Jimenez-Corona, A.; et al. Obesity, inflammatory markers, and endometrial cancer risk: A prospective case-control study. Endocr. Relat. Cancer 2010, 17, 1007–1019. [Google Scholar] [CrossRef]
- Gagnière, J.; Raisch, J.; Veziant, J.; Barnich, N.; Bonnet, R.; Buc, E.; Bringer, M.A.; Pezet, D.; Bonnet, M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016, 22, 501–518. [Google Scholar] [CrossRef]
- Lu, W.; He, F.; Lin, Z.; Liu, S.; Tang, L.; Huang, Y.; Hu, Z. Dysbiosis of the endometrial microbiota and its association with inflammatory cytokines in endometrial cancer. Int. J. Cancer 2021, 148, 1708–1716. [Google Scholar] [CrossRef] [PubMed]
- Morselli, S.; Salvo, M.; Foschi, C.; Lazzarotto, T.; Ambretti, S.; Marangoni, A. Characterization of Gardnerella vaginalis isolates: Correlations among clades, biofilm formation and cytokine stimulation. New Microbiol. 2023, 46, 56–59. [Google Scholar] [PubMed]
- Anton, L.; Ferguson, B.; Friedman, E.S.; Gerson, K.D.; Brown, A.G.; Elovitz, M.A. Gardnerella vaginalis alters cervicovaginal epithelial cell function through microbe-specific immune responses. Microbiome 2022, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- Bertran, T.; Brachet, P.; Vareille-Delarbre, M.; Falenta, J.; Dosgilbert, A.; Vasson, M.P.; Forestier, C.; Tridon, A.; Evrard, B. Slight Pro-Inflammatory Immunomodulation Properties of Dendritic Cells by Gardnerella vaginalis: The “Invisible Man” of Bacterial Vaginosis? J. Immunol. Res. 2016, 2016, 9747480. [Google Scholar] [CrossRef]
- Santos, C.M.A.; Pires, M.C.V.; Leão, T.L.; Silva, A.K.S.; Miranda, L.S.; Martins, F.S.; Silva, A.M.; Nicoli, J.R. Anti-inflammatory effect of two Lactobacillus strains during infection with Gardnerella vaginalis and Candida albicans in a HeLa cell culture model. Microbiology 2018, 164, 349–358. [Google Scholar] [CrossRef]
- Łaniewski, P.; Herbst-Kralovetz, M.M. Bacterial vaginosis and health-associated bacteria modulate the immunometabolic landscape in 3D model of human cervix. NPJ Biofilms Microbiomes 2021, 13, 88. [Google Scholar] [CrossRef]
- Jespers, V.; Kyongo, J.; Joseph, S.; Hardy, L.; Cools, P.; Crucitti, T.; Mwaura, M.; Ndayisaba, G.; Delany-Moretlwe, S.; Buyze, J.; et al. A longitudinal analysis of the vaginal microbiota and vaginal immune mediators in women from sub-Saharan Africa. Sci. Rep. 2017, 7, 11974. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, C.Z.; Wan, J.Y.; Yao, H.; Yuan, C.S. Dissecting the Interplay Mechanism between Epigenetics and Gut Microbiota: Health Maintenance and Disease Prevention. Int. J. Mol. Sci. 2021, 22, 6933. [Google Scholar] [CrossRef]
- Inoue, F.; Sone, K.; Toyohara, Y.; Takahashi, Y.; Kukita, A.; Hara, A.; Taguchi, A.; Tanikawa, M.; Tsuruga, T.; Osuga, Y. Targeting Epigenetic Regulators for Endometrial Cancer Therapy: Its Molecular Biology and Potential Clinical Applications. Int. J. Mol. Sci. 2021, 22, 2305. [Google Scholar] [CrossRef]
- Witkin, S.S.; Nasioudis, D.; Leizer, J.; Minis, E.; Boester, A.; Forney, L.J. Epigenetics and the vaginal microbiome: Influence of the microbiota on the histone deacetylase level in vaginal epithelial cells from pregnant women. Minerva Ginecol. 2019, 71, 171–175. [Google Scholar] [CrossRef]
- Anton, L.; Sierra, L.J.; DeVine, A.; Barila, G.; Heiser, L.; Brown, A.G.; Elovitz, M.A. Common Cervicovaginal Microbial Supernatants Alter Cervical Epithelial Function: Mechanisms by Which Lactobacillus crispatus Contributes to Cervical Health. Front. Microbiol. 2018, 9, 2181. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, G.M.; Burkett, W.C.; McCoy, A.N.; Nichols, H.B.; Olshan, A.F.; Broaddus, R.; Merker, J.D.; Weissman, B.; Brewster, W.R.; Roach, J.; et al. Differences in the microbial profiles of early stage endometrial cancers between Black and White women. Gynecol. Oncol. 2022, 165, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, D.; Moore, K.; Gunderson, C.; Rowland, M.; Austin, R.; Honap, T.P.; Xu, J.; Warinner, C.; Sankaranarayanan, K.; Lewis, C.M., Jr. Shifts in gut and vaginal microbiomes are associated with cancer recurrence time in women with ovarian cancer. PeerJ. 2021, 9, e11574. [Google Scholar] [CrossRef] [PubMed]
- Chambers, L.M.; Esakov Rhoades, E.L.; Bharti, R.; Braley, C.; Tewari, S.; Trestan, L.; Alali, Z.; Bayik, D.; Lathia, J.D.; Sangwan, N.; et al. Disruption of the Gut Microbiota Confers Cisplatin Resistance in Epithelial Ovarian Cancer. Cancer Res. 2022, 82, 4654–4669. [Google Scholar] [CrossRef] [PubMed]
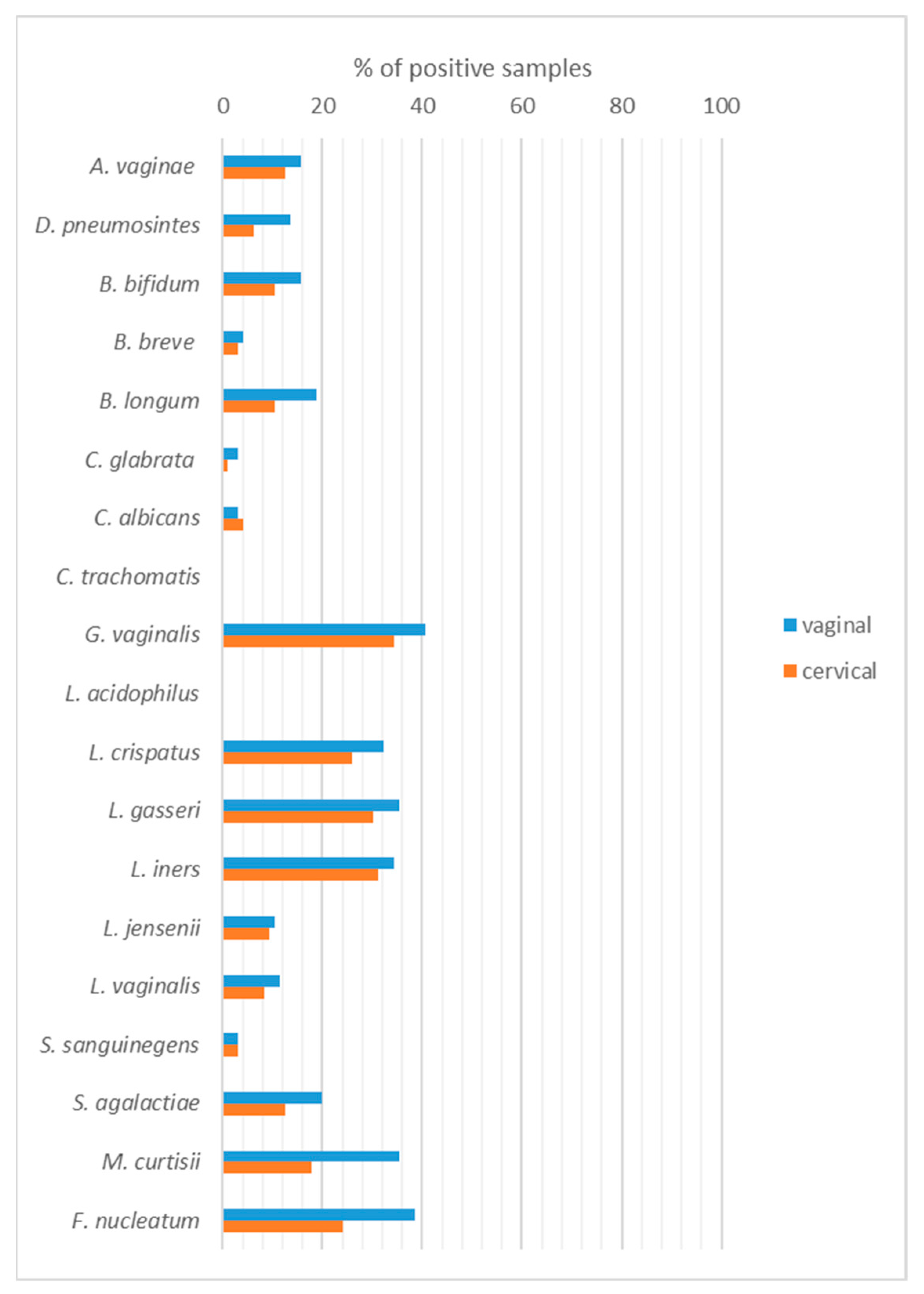
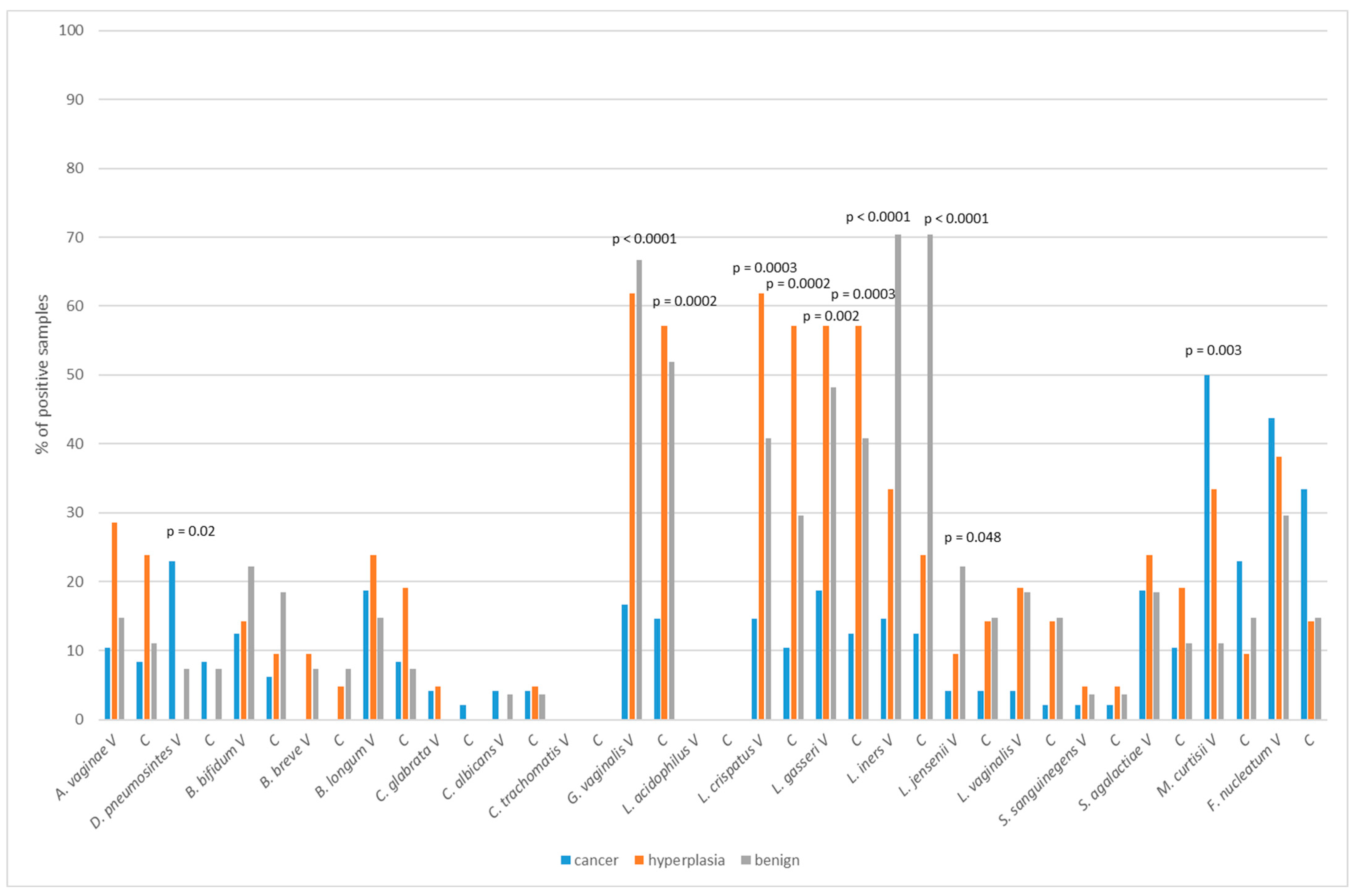
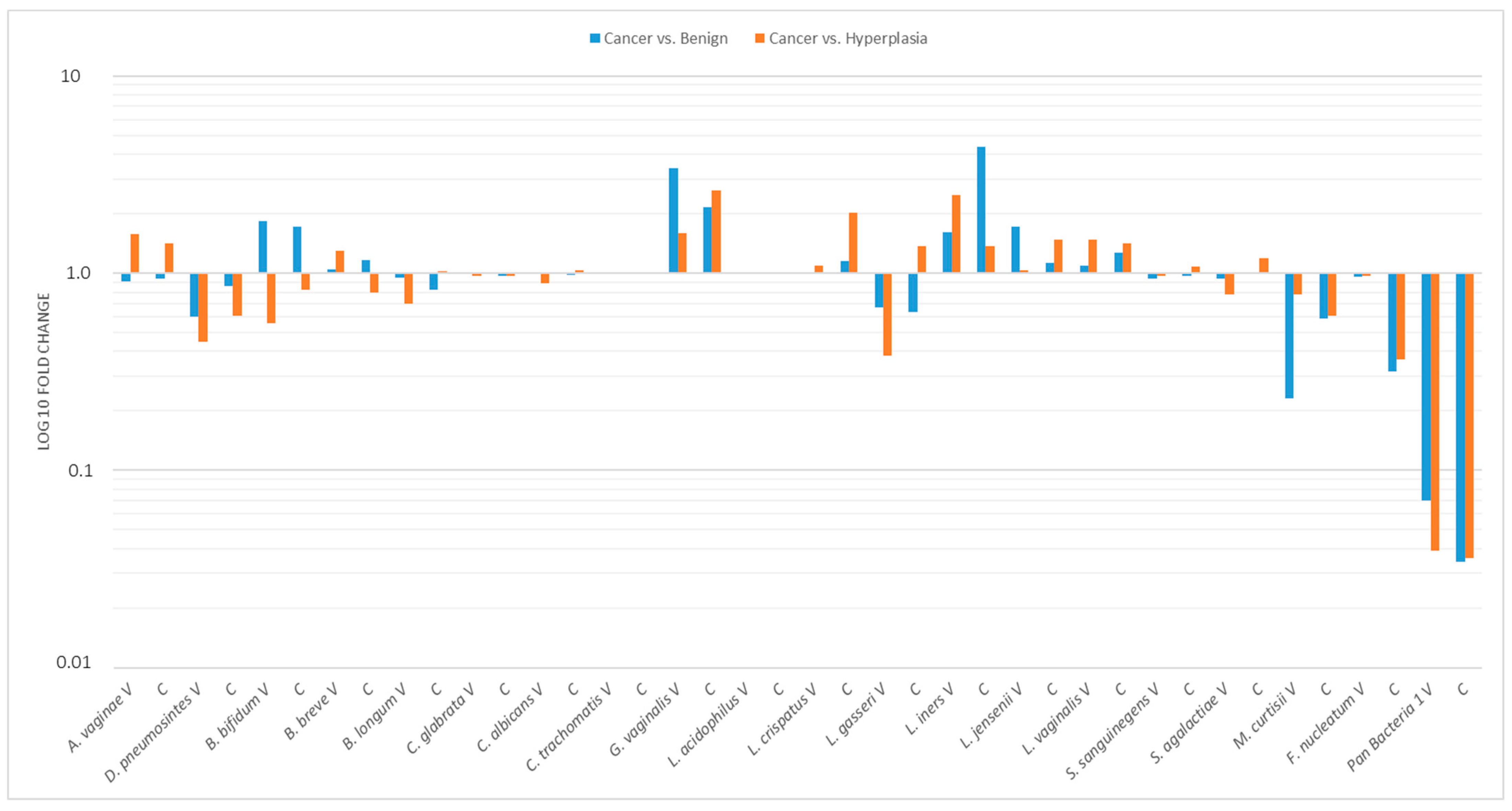
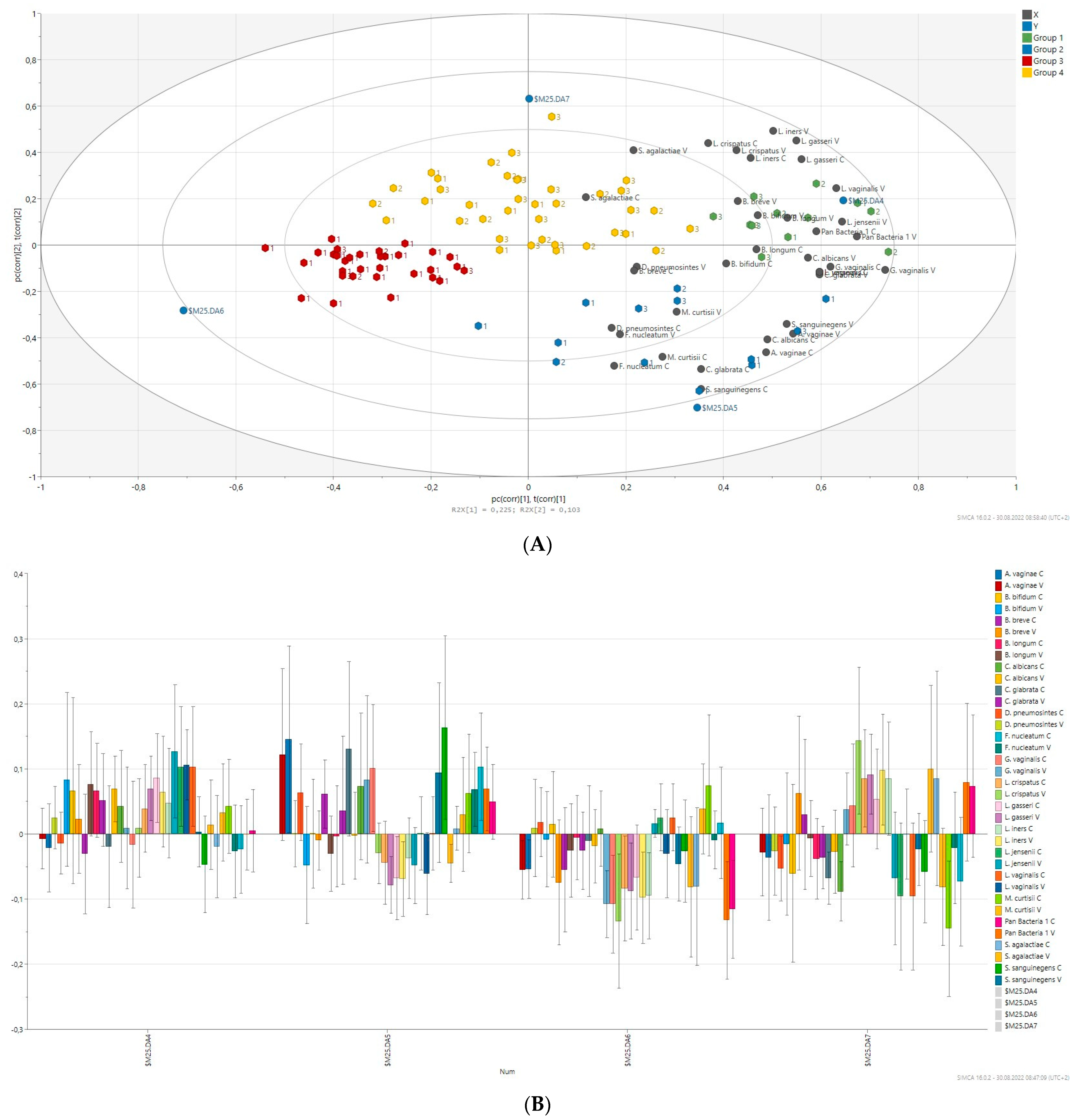
| Variables | Total (n = 96) | Benign (n = 27) | Hyperplasia (n = 21) | Cancer (n = 48) | p-Value |
|---|---|---|---|---|---|
| Age (years) mean ± SD | 60.1 ± 9.8 | 54.4 ± 9.0 | 55.6 ± 8.2 | 65.3 ± 8.2 | <0.0001 |
| BMI (mean ± SD) | 30.5 ± 6.1 | 29.7 ± 5.1 | 29.3 ± 5.8 | 31.5 ± 6.6 | 0.31 |
| Menopausal status | <0.0001 | ||||
| Pre | 26 (27.1) | 15 (55.6) | 8 (38.1) | 3 (6.3) | |
| Peri | 9 (9.4) | 3 (11.1) | 5 (23.8) | 1 (2.1) | |
| Post | 61 (63.5) | 9 (33.3) | 8 (38.1) | 44 (91.7) | |
| Histotype | NA | NA | |||
| Endometroid | 40 (41.7) | 40 (83.3) | |||
| Serous | 3 (3.1) | 3 (6.3) | |||
| Carcinosarcoma | 4 (4.2) | 4 (8.3) | |||
| Clearcell | 1 (1.0) | 1 (2.8) | |||
| Grade | NA | NA | |||
| G1 | 12 (12.5) | 12 (25.0) | |||
| G2 | 28 (29.2) | 28 (58.3) | |||
| G3 | 6 (6.3) | 6 (12.5) | |||
| unknown | 2 (2.1) | 2 (4.2) | |||
| FIGO | NA | NA | |||
| 1A | 25 (26.0) | 25 (52.8) | |||
| 1B | 16 (16.7) | 16 (33.3) | |||
| 2 | 4 (4.2) | 4 (8.3) | |||
| 3A | 1 (1.0) | 1 (2.1) | |||
| 3B | 2 (2.1) | 2 (4.2) |
| Entity | Vaginal Samples | Cervical Samples | p-Value |
|---|---|---|---|
| Cancer | 2.6 ± 1.9 | 1.6 ± 1.6 | <0.0001 |
| Hyperplasia | 4.2 ± 1.2 | 3.3 ± 1.8 | 0.034 |
| Benign | 4.0 ± 1.9 | 3.2 ± 1.8 | 0.0055 |
| Microorganisms | Pre- and Perimenopausal (n = 35) | Postmenopausal (n = 61) | p-Value | |
|---|---|---|---|---|
| A. vaginae | V | 11 (31.4%) | 4 (6.6%) | 0.002 |
| C | 8 (22.9%) | 4 (6.6%) | 0.024 | |
| D. pneumosintes | V | 3 (8.6%) | 10 (16.4%) | 0.22 |
| C | 3 (8.6%) | 3 (4.9%) | 0.38 | |
| B. bifidum | V | 9 (25.7%) | 6 (9.8%) | 0.040 |
| C | 7 (20.0%) | 3 (4.9%) | 0.026 | |
| B. breve | V | 1 (2.9%) | 3 (4.9%) | 0.54 |
| C | 1 (2.9%) | 2 (3.3%) | 0.70 | |
| B. longum | V | 7 (20.0%) | 11 (18.0%) | 0.51 |
| C | 5 (14.3%) | 5 (8.2%) | 0.27 | |
| C. glabrata | V | 1 (2.9%) | 2 (3.3%) | 0.70 |
| C | 0 (0) | 1 (1.6%) | 0.64 | |
| C. albicans | V | 2 (5.7%) | 1 (1.6%) | 0.30 |
| C | 2 (5.7%) | 2 (3.3%) | 0.46 | |
| G. vaginalis | V | 25 (71.4%) | 14 (23.0%) | <0.0001 |
| C | 22 (62.9%) | 11 (18.0%) | <0.0001 | |
| L. crispatus | V | 15 (42.8%) | 16 (26.2) | 0.074 |
| C | 13 (37.1%) | 12 (19.7%) | 0.052 | |
| L. gasseri | V | 17 (48.6%) | 17 (27.9%) | 0.035 |
| C | 17 (48.6%) | 12 (19.7) | 0.0033 | |
| L. iners | V | 21 (60.0%) | 12 (19.7%) | <0.0001 |
| C | 19 (54.3%) | 11 (18.3%) | 0.0003 | |
| L. jensenii | V | 7 (20.0%) | 3 (4.9%) | 0.026 |
| C | 7 (20.0%) | 2 (3.3%) | 0.010 | |
| L. vaginalis | V | 7 (20.0%) | 4 (6.6%) | 0.051 |
| C | 6 (17.1%) | 2 (3.3%) | 0.026 | |
| S. sanguinegens | V | 1 (2.9%) | 2 (3.3%) | 0.70 |
| C | 1 (2.9%) | 2 (3.3%) | 0.70 | |
| S. agalactiae | V | 8 (22.9%) | 11 (18.0%) | 0.38 |
| C | 7 (20.0%) | 5 (8.2%) | 0.088 | |
| M. curtisii | V | 5 (14.3%) | 29 (47.5%) | 0.0008 |
| C | 4 (11.4%) | 13 (21.3%) | 0.17 | |
| F. nucleatum | V | 12 (34.3%) | 25 (41.0%) | 0.33 |
| C | 6 (17.1%) | 17 (27.9%) | 0.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barczyński, B.; Frąszczak, K.; Grywalska, E.; Kotarski, J.; Korona-Głowniak, I. Vaginal and Cervical Microbiota Composition in Patients with Endometrial Cancer. Int. J. Mol. Sci. 2023, 24, 8266. https://doi.org/10.3390/ijms24098266
Barczyński B, Frąszczak K, Grywalska E, Kotarski J, Korona-Głowniak I. Vaginal and Cervical Microbiota Composition in Patients with Endometrial Cancer. International Journal of Molecular Sciences. 2023; 24(9):8266. https://doi.org/10.3390/ijms24098266
Chicago/Turabian StyleBarczyński, Bartłomiej, Karolina Frąszczak, Ewelina Grywalska, Jan Kotarski, and Izabela Korona-Głowniak. 2023. "Vaginal and Cervical Microbiota Composition in Patients with Endometrial Cancer" International Journal of Molecular Sciences 24, no. 9: 8266. https://doi.org/10.3390/ijms24098266
APA StyleBarczyński, B., Frąszczak, K., Grywalska, E., Kotarski, J., & Korona-Głowniak, I. (2023). Vaginal and Cervical Microbiota Composition in Patients with Endometrial Cancer. International Journal of Molecular Sciences, 24(9), 8266. https://doi.org/10.3390/ijms24098266








