Role of cGAS–Sting Signaling in Alzheimer’s Disease
Abstract
1. Introduction
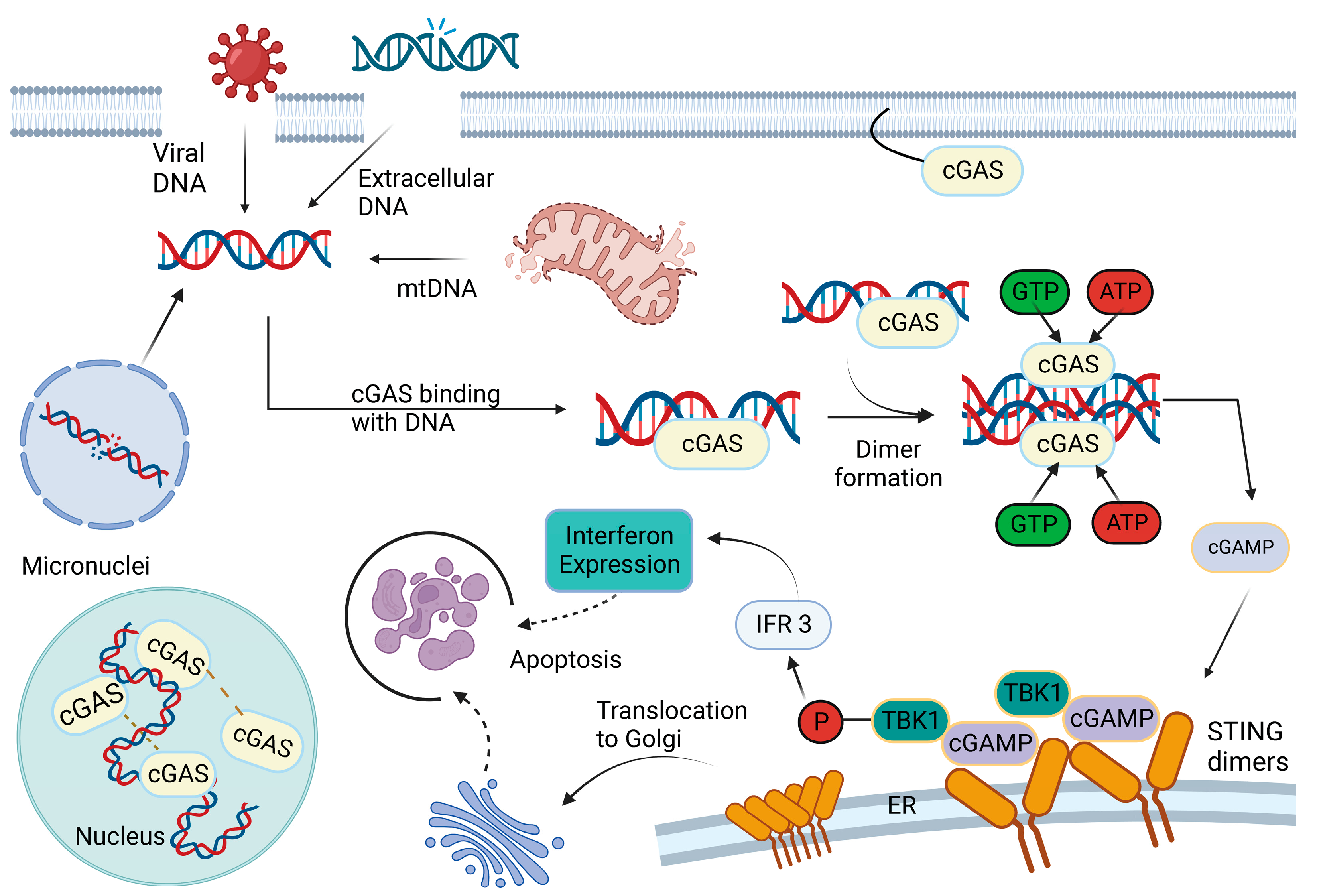
2. cGAS–STING Pathway
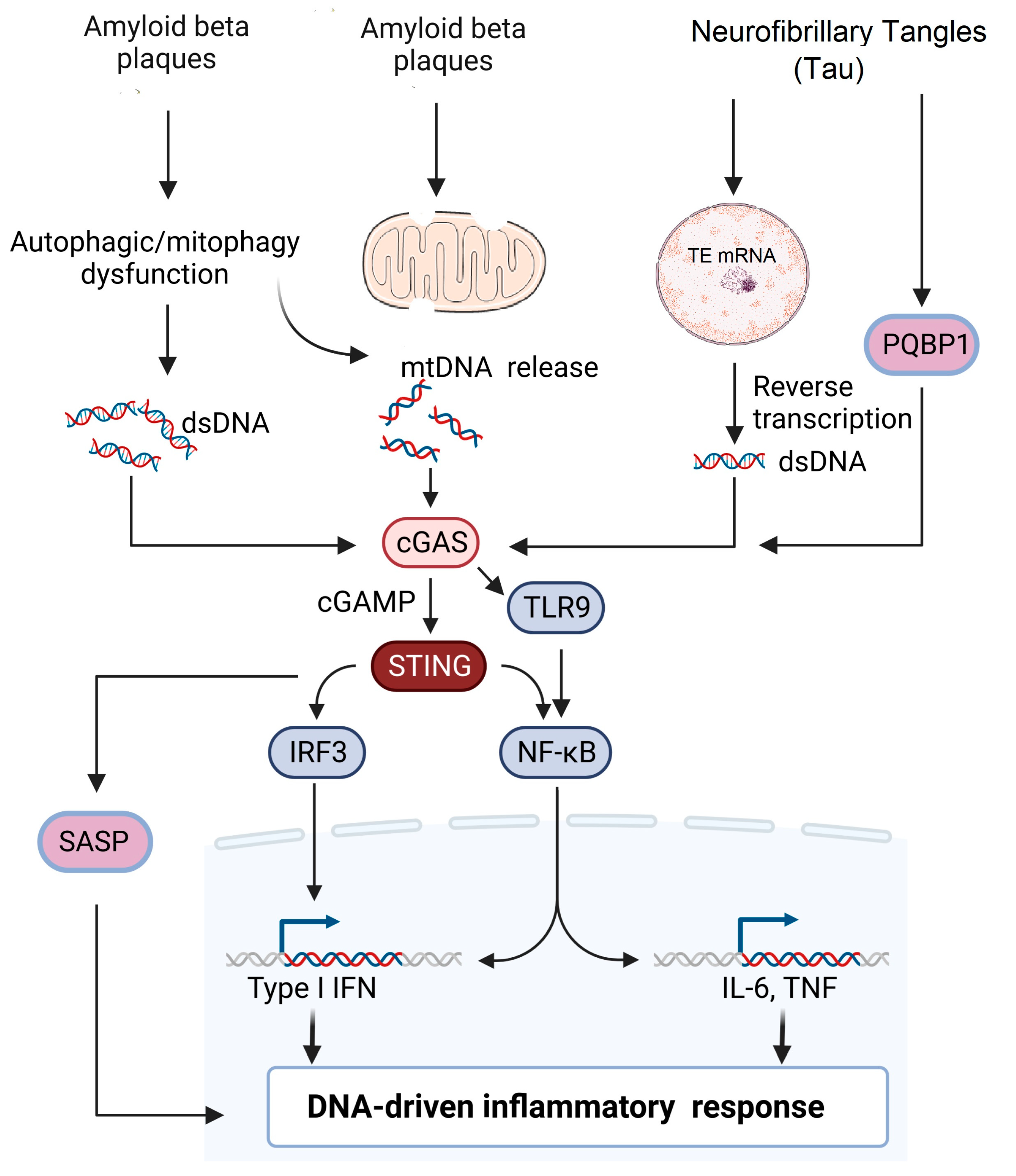
3. Activation of cGAS: Sources and Mechanisms
4. Role of cGAS–STING Pathway in Alzheimer’s Disease
5. Current Therapeutics to Target STING in AD
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, V. The Trinity of cGAS, TLR9, and ALRs Guardians of the Cellular Galaxy against Host-Derived Self-DNA. Front. Immunol. 2020, 11, 624597. [Google Scholar] [CrossRef]
- Santa, P.; Garreau, A.; Serpas, L.; Ferriere, A.; Blanco, P.; Soni, C.; Sisirak, V. The Role of Nucleases and Nucleic Acid Editing Enzymes in the Regulation of Self-Nucleic Acid Sensing. Front. Immunol. 2021, 12, 629922. [Google Scholar] [CrossRef]
- Stetson, D.B.; Medzhitov, R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 2006, 24, 93–103. [Google Scholar] [CrossRef]
- Zierhut, C.; Yamaguchi, N.; Paredes, M.; Luo, J.D.; Carroll, T.; Funabiki, H. The Cytoplasmic DNA Sensor cGAS Promotes Mitotic Cell Death. Cell 2019, 178, 302–315.e323. [Google Scholar] [CrossRef] [PubMed]
- Ablasser, A.; Chen, Z.J. cGAS in action: Expanding roles in immunity and inflammation. Science 2019, 363, eaat8657. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Mei, J.; Guo, H.; Zhu, J.; Wang, C. Intervention of cGAS‒STING signaling in sterile inflammatory diseases. J. Mol. Cell Biol. 2022, 14, mjac005. [Google Scholar] [CrossRef]
- Rezabakhsh, A.; Montazersaheb, S.; Nabat, E.; Hassanpour, M.; Montaseri, A.; Malekinejad, H.; Movassaghpour, A.A.; Rahbarghazi, R.; Garjani, A. Effect of hydroxychloroquine on oxidative/nitrosative status and angiogenesis in endothelial cells under high glucose condition. Bioimpacts 2017, 7, 219–226. [Google Scholar] [CrossRef]
- Bai, J.; Liu, F. Nuclear cGAS: Sequestration and beyond. Protein Cell 2022, 13, 90–101. [Google Scholar] [CrossRef]
- Hopfner, K.-P.; Hornung, V. Molecular mechanisms and cellular functions of cGAS–STING signalling. Nat. Rev. Mol. Cell Biol. 2020, 21, 501–521. [Google Scholar] [CrossRef]
- Gui, X.; Yang, H.; Li, T.; Tan, X.; Shi, P.; Li, M.; Du, F.; Chen, Z.J. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 2019, 567, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shang, G.; Gui, X.; Zhang, X.; Bai, X.-c.; Chen, Z.J. Structural basis of STING binding with and phosphorylation by TBK1. Nature 2019, 567, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cai, X.; Wu, J.; Cong, Q.; Chen, X.; Li, T.; Du, F.; Ren, J.; Wu, Y.T.; Grishin, N.V.; et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015, 347, aaa2630. [Google Scholar] [CrossRef]
- Wu, J.; Dobbs, N.; Yang, K.; Yan, N. Interferon-Independent Activities of Mammalian STING Mediate Antiviral Response and Tumor Immune Evasion. Immunity 2020, 53, 115–126.e115. [Google Scholar] [CrossRef]
- Ghosh, M.; Saha, S.; Li, J.; Montrose, D.C.; Martinez, L.A. p53 engages the cGAS/STING cytosolic DNA sensing pathway for tumor suppression. Mol. Cell 2023, 83, 266–280.e266. [Google Scholar] [CrossRef]
- Pei, J.; Zhang, Y.; Luo, Q.; Zheng, W.; Li, W.; Zeng, X.; Li, Q.; Quan, J. STAT3 inhibition enhances CDN-induced STING signaling and antitumor immunity. Cancer Lett. 2019, 450, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Seo, G.J.; Choi, Y.J.; Kwak, M.J.; Ge, J.; Rodgers, M.A.; Shi, M.; Leslie, B.J.; Hopfner, K.P.; Ha, T.; et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe. 2014, 15, 228–238. [Google Scholar] [CrossRef]
- Chen, M.; Meng, Q.; Qin, Y.; Liang, P.; Tan, P.; He, L.; Zhou, Y.; Chen, Y.; Huang, J.; Wang, R.F.; et al. TRIM14 Inhibits cGAS Degradation Mediated by Selective Autophagy Receptor p62 to Promote Innate Immune Responses. Mol. Cell 2016, 64, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, T.; Fujita, N.; Hayashi, T.; Takahara, K.; Satoh, T.; Lee, H.; Matsunaga, K.; Kageyama, S.; Omori, H.; Noda, T.; et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. USA 2009, 106, 20842–20846. [Google Scholar] [CrossRef] [PubMed]
- Konno, H.; Konno, K.; Barber, G.N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 2013, 155, 688–698. [Google Scholar] [CrossRef]
- Liu, D.; Wu, H.; Wang, C.; Li, Y.; Tian, H.; Siraj, S.; Sehgal, S.A.; Wang, X.; Wang, J.; Shang, Y.; et al. STING directly activates autophagy to tune the innate immune response. Cell Death Differ. 2019, 26, 1735–1749. [Google Scholar] [CrossRef]
- Prabakaran, T.; Bodda, C.; Krapp, C.; Zhang, B.-c.; Christensen, M.H.; Sun, C.; Reinert, L.; Cai, Y.; Jensen, S.B.; Skouboe, M.K.; et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018, 37, e97858. [Google Scholar] [CrossRef] [PubMed]
- Nassour, J.; Radford, R.; Correia, A.; Fusté, J.M.; Schoell, B.; Jauch, A.; Shaw, R.J.; Karlseder, J. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature 2019, 565, 659–663. [Google Scholar] [CrossRef] [PubMed]
- Gonugunta, V.K.; Sakai, T.; Pokatayev, V.; Yang, K.; Wu, J.; Dobbs, N.; Yan, N. Trafficking-Mediated STING Degradation Requires Sorting to Acidified Endolysosomes and Can Be Targeted to Enhance Anti-tumor Response. Cell Rep. 2017, 21, 3234–3242. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, H.; Ren, J.; Chen, Q.; Chen, Z.J. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA 2017, 114, E4612–E4620. [Google Scholar] [CrossRef] [PubMed]
- Glück, S.; Guey, B.; Gulen, M.F.; Wolter, K.; Kang, T.W.; Schmacke, N.A.; Bridgeman, A.; Rehwinkel, J.; Zender, L.; Ablasser, A. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 2017, 19, 1061–1070. [Google Scholar] [CrossRef]
- Gulen, M.F.; Koch, U.; Haag, S.M.; Schuler, F.; Apetoh, L.; Villunger, A.; Radtke, F.; Ablasser, A. Signalling strength determines proapoptotic functions of STING. Nat. Commun. 2017, 8, 427. [Google Scholar] [CrossRef]
- Petrasek, J.; Iracheta-Vellve, A.; Csak, T.; Satishchandran, A.; Kodys, K.; Kurt-Jones, E.A.; Fitzgerald, K.A.; Szabo, G. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc. Natl. Acad. Sci. USA 2013, 110, 16544–16549. [Google Scholar] [CrossRef]
- Selvey, S.; Thompson, E.W.; Matthaei, K.; Lea, R.A.; Irving, M.G.; Griffiths, L.R. Beta-actin—An unsuitable internal control for RT-PCR. Mol. Cell Probes. 2001, 15, 307–311. [Google Scholar] [CrossRef]
- West, A.P.; Khoury-Hanold, W.; Staron, M.; Tal, M.C.; Pineda, C.M.; Lang, S.M.; Bestwick, M.; Duguay, B.A.; Raimundo, N.; MacDuff, D.A.; et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 2015, 520, 553–557. [Google Scholar] [CrossRef]
- Bai, J.; Cervantes, C.; Liu, J.; He, S.; Zhou, H.; Zhang, B.; Cai, H.; Yin, D.; Hu, D.; Li, Z.; et al. DsbA-L prevents obesity-induced inflammation and insulin resistance by suppressing the mtDNA release-activated cGAS-cGAMP-STING pathway. Proc. Natl. Acad. Sci. USA 2017, 114, 12196–12201. [Google Scholar] [CrossRef]
- Bai, J.; Cervantes, C.; He, S.; He, J.; Plasko, G.R.; Wen, J.; Li, Z.; Yin, D.; Zhang, C.; Liu, M.; et al. Mitochondrial stress-activated cGAS-STING pathway inhibits thermogenic program and contributes to overnutrition-induced obesity in mice. Commun. Biol. 2020, 3, 257. [Google Scholar] [CrossRef]
- Wu, Z.; Oeck, S.; West, A.P.; Mangalhara, K.C.; Sainz, A.G.; Newman, L.E.; Zhang, X.-O.; Wu, L.; Yan, Q.; Bosenberg, M.; et al. Mitochondrial DNA stress signalling protects the nuclear genome. Nat. Metab. 2019, 1, 1209–1218. [Google Scholar] [CrossRef] [PubMed]
- Gehrke, N.; Mertens, C.; Zillinger, T.; Wenzel, J.; Bald, T.; Zahn, S.; Tüting, T.; Hartmann, G.; Barchet, W. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity 2013, 39, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Sliter, D.A.; Martinez, J.; Hao, L.; Chen, X.; Sun, N.; Fischer, T.D.; Burman, J.L.; Li, Y.; Zhang, Z.; Narendra, D.P.; et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature 2018, 561, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Nath, P.; Chauhan, N.R.; Jena, K.K.; Datey, A.; Kumar, N.D.; Mehto, S.; De, S.; Nayak, T.K.; Priyadarsini, S.; Rout, K.; et al. Inhibition of IRGM establishes a robust antiviral immune state to restrict pathogenic viruses. EMBO Rep. 2021, 22, e52948. [Google Scholar] [CrossRef]
- Lou, H.; Pickering, M.C. Extracellular DNA and autoimmune diseases. Cell Mol. Immunol. 2018, 15, 746–755. [Google Scholar] [CrossRef]
- King, K.R.; Aguirre, A.D.; Ye, Y.X.; Sun, Y.; Roh, J.D.; Ng, R.P., Jr.; Kohler, R.H.; Arlauckas, S.P.; Iwamoto, Y.; Savol, A.; et al. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat. Med. 2017, 23, 1481–1487. [Google Scholar] [CrossRef]
- Luo, W.; Wang, Y.; Zhang, L.; Ren, P.; Zhang, C.; Li, Y.; Azares, A.R.; Zhang, M.; Guo, J.; Ghaghada, K.B.; et al. Critical Role of Cytosolic DNA and Its Sensing Adaptor STING in Aortic Degeneration, Dissection, and Rupture. Circulation 2020, 141, 42–66. [Google Scholar] [CrossRef]
- Lood, C.; Blanco, L.P.; Purmalek, M.M.; Carmona-Rivera, C.; De Ravin, S.S.; Smith, C.K.; Malech, H.L.; Ledbetter, J.A.; Elkon, K.B.; Kaplan, M.J. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med. 2016, 22, 146–153. [Google Scholar] [CrossRef]
- Wang, R.; Zhu, Y.; Liu, Z.; Chang, L.; Bai, X.; Kang, L.; Cao, Y.; Yang, X.; Yu, H.; Shi, M.J.; et al. Neutrophil extracellular traps promote tPA-induced brain hemorrhage via cGAS in mice with stroke. Blood 2021, 138, 91–103. [Google Scholar] [CrossRef]
- Deng, L.; Liang, H.; Xu, M.; Yang, X.; Burnette, B.; Arina, A.; Li, X.D.; Mauceri, H.; Beckett, M.; Darga, T.; et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity 2014, 41, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Kawane, K.; Ohtani, M.; Miwa, K.; Kizawa, T.; Kanbara, Y.; Yoshioka, Y.; Yoshikawa, H.; Nagata, S. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 2006, 443, 998–1002. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Gutman, D.; Saijo, S.; Barber, G.N. STING manifests self DNA-dependent inflammatory disease. Proc. Natl. Acad. Sci. USA 2012, 109, 19386–19391. [Google Scholar] [CrossRef] [PubMed]
- Guey, B.; Wischnewski, M.; Decout, A.; Makasheva, K.; Kaynak, M.; Sakar, M.S.; Fierz, B.; Ablasser, A. BAF restricts cGAS on nuclear DNA to prevent innate immune activation. Science 2020, 369, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.A.; Spangler, C.J.; Strauss, J.D.; Cesmat, A.P.; Liu, P.; McGinty, R.K.; Zhang, Q. Structural basis of nucleosome-dependent cGAS inhibition. Science 2020, 370, 450–454. [Google Scholar] [CrossRef]
- Cao, D.; Han, X.; Fan, X.; Xu, R.M.; Zhang, X. Structural basis for nucleosome-mediated inhibition of cGAS activity. Cell Res. 2020, 30, 1088–1097. [Google Scholar] [CrossRef]
- Kujirai, T.; Zierhut, C.; Takizawa, Y.; Kim, R.; Negishi, L.; Uruma, N.; Hirai, S.; Funabiki, H.; Kurumizaka, H. Structural basis for the inhibition of cGAS by nucleosomes. Science 2020, 370, 455–458. [Google Scholar] [CrossRef]
- Michalski, S.; de Oliveira Mann, C.C.; Stafford, C.A.; Witte, G.; Bartho, J.; Lammens, K.; Hornung, V.; Hopfner, K.P. Structural basis for sequestration and autoinhibition of cGAS by chromatin. Nature 2020, 587, 678–682. [Google Scholar] [CrossRef]
- Ahn, J.; Xia, T.; Konno, H.; Konno, K.; Ruiz, P.; Barber, G.N. Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 2014, 5, 5166. [Google Scholar] [CrossRef]
- Song, X.; Ma, F.; Herrup, K. Accumulation of Cytoplasmic DNA Due to ATM Deficiency Activates the Microglial Viral Response System with Neurotoxic Consequences. J. Neurosci. 2019, 39, 6378–6394. [Google Scholar] [CrossRef] [PubMed]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; Elias, A.E.; Skvir, N.J.; Criscione, S.W.; Caligiana, A.; Brocculi, G.; Adney, E.M.; Boeke, J.D.; et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 2019, 566, 73–78. [Google Scholar] [CrossRef]
- Harding, S.M.; Benci, J.L.; Irianto, J.; Discher, D.E.; Minn, A.J.; Greenberg, R.A. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 2017, 548, 466–470. [Google Scholar] [CrossRef]
- Mackenzie, K.J.; Carroll, P.; Martin, C.A.; Murina, O.; Fluteau, A.; Simpson, D.J.; Olova, N.; Sutcliffe, H.; Rainger, J.K.; Leitch, A.; et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 2017, 548, 461–465. [Google Scholar] [CrossRef]
- Ivanov, A.; Pawlikowski, J.; Manoharan, I.; van Tuyn, J.; Nelson, D.M.; Rai, T.S.; Shah, P.P.; Hewitt, G.; Korolchuk, V.I.; Passos, J.F.; et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 2013, 202, 129–143. [Google Scholar] [CrossRef]
- Dou, Z.; Ghosh, K.; Vizioli, M.G.; Zhu, J.; Sen, P.; Wangensteen, K.J.; Simithy, J.; Lan, Y.; Lin, Y.; Zhou, Z.; et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 2017, 550, 402–406. [Google Scholar] [CrossRef]
- Uggenti, C.; Lepelley, A.; Depp, M.; Badrock, A.P.; Rodero, M.P.; El-Daher, M.T.; Rice, G.I.; Dhir, S.; Wheeler, A.P.; Dhir, A.; et al. cGAS-mediated induction of type I interferon due to inborn errors of histone pre-mRNA processing. Nat. Genet. 2020, 52, 1364–1372. [Google Scholar] [CrossRef]
- Ramesh, S.; Govindarajulu, M.; Jones, E.; Suppiramaniam, V.; Moore, T.; Dhanasekaran, M. Mitochondrial dysfunction and the role of Mitophagy in Alzheimer’ s Disease. Alzheimer’s Disease & Treatment; MedDocs Publishers LLC: Reno, NV, USA, 2018. [Google Scholar]
- Cai, Q.; Jeong, Y.Y. Mitophagy in Alzheimer’s Disease and Other Age-Related Neurodegenerative Diseases. Cells 2020, 9, 150. [Google Scholar] [CrossRef]
- Chakravorty, A.; Jetto, C.T.; Manjithaya, R. Dysfunctional Mitochondria and Mitophagy as Drivers of Alzheimer’s Disease Pathogenesis. Front. Aging Neurosci. 2019, 11, 311. [Google Scholar] [CrossRef]
- Jauhari, A.; Baranov, S.V.; Suofu, Y.; Kim, J.; Singh, T.; Yablonska, S.; Li, F.; Wang, X.; Oberly, P.; Minnigh, M.B.; et al. Melatonin inhibits cytosolic mitochondrial DNA-induced neuroinflammatory signaling in accelerated aging and neurodegeneration. J. Clin. Investig. 2020, 130, 3124–3136. [Google Scholar] [CrossRef]
- Moya, G.E.; Rivera, P.D.; Dittenhafer-Reed, K.E. Evidence for the Role of Mitochondrial DNA Release in the Inflammatory Response in Neurological Disorders. Int. J. Mol. Sci. 2021, 22, 7030. [Google Scholar] [CrossRef]
- Murthy, A.M.V.; Robinson, N.; Kumar, S. Crosstalk between cGAS-STING signaling and cell death. Cell Death Differ. 2020, 27, 2989–3003. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Ma, G.; Li, X.; Zhao, J.; Zhao, Z.; Zeng, J. Activation of innate immune cGAS-STING pathway contributes to Alzheimer’s pathogenesis in 5×FAD mice. Nat. Aging 2023, 3, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhang, H.; Zhang, Q.; Yao, X.; Ni, W.; Zhou, K. Emerging role of STING signalling in CNS injury: Inflammation, autophagy, necroptosis, ferroptosis and pyroptosis. J. Neuroinflamm. 2022, 19, 242. [Google Scholar] [CrossRef]
- Taylor, J.M.; Moore, Z.; Minter, M.R.; Crack, P.J. Type-I interferon pathway in neuroinflammation and neurodegeneration: Focus on Alzheimer's disease. J. Neural. Transm. 2018, 125, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Wirths, O.; Breyhan, H.; Marcello, A.; Cotel, M.C.; Brück, W.; Bayer, T.A. Inflammatory changes are tightly associated with neurodegeneration in the brain and spinal cord of the APP/PS1KI mouse model of Alzheimer's disease. Neurobiol. Aging 2010, 31, 747–757. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Wiste, H.J.; Lesnick, T.G.; Weigand, S.D.; Knopman, D.S.; Vemuri, P.; Pankratz, V.S.; Senjem, M.L.; Gunter, J.L.; Mielke, M.M.; et al. Brain β-amyloid load approaches a plateau. Neurology 2013, 80, 890–896. [Google Scholar] [CrossRef]
- Reinert, L.S.; Lopušná, K.; Winther, H.; Sun, C.; Thomsen, M.K.; Nandakumar, R.; Mogensen, T.H.; Meyer, M.; Vægter, C.; Nyengaard, J.R.; et al. Sensing of HSV-1 by the cGAS–STING pathway in microglia orchestrates antiviral defence in the CNS. Nat. Commun. 2016, 7, 13348. [Google Scholar] [CrossRef] [PubMed]
- Reinert, L.S.; Rashidi, A.S.; Tran, D.N.; Katzilieris-Petras, G.; Hvidt, A.K.; Gohr, M.; Fruhwürth, S.; Bodda, C.; Thomsen, M.K.; Vendelbo, M.H.; et al. Brain immune cells undergo cGAS/STING-dependent apoptosis during herpes simplex virus type 1 infection to limit type I IFN production. J. Clin. Investig. 2021, 131, e136824. [Google Scholar] [CrossRef]
- Cox, D.J.; Field, R.H.; Williams, D.G.; Baran, M.; Bowie, A.G.; Cunningham, C.; Dunne, A. DNA sensors are expressed in astrocytes and microglia in vitro and are upregulated during gliosis in neurodegenerative disease. Glia 2015, 63, 812–825. [Google Scholar] [CrossRef]
- Jin, M.; Shiwaku, H.; Tanaka, H.; Obita, T.; Ohuchi, S.; Yoshioka, Y.; Jin, X.; Kondo, K.; Fujita, K.; Homma, H.; et al. Tau activates microglia via the PQBP1-cGAS-STING pathway to promote brain inflammation. Nat. Commun. 2021, 12, 6565. [Google Scholar] [CrossRef] [PubMed]
- Elzinga, S.E.; Henn, R.; Murdock, B.J.; Kim, B.; Hayes, J.M.; Mendelson, F.; Webber-Davis, I.; Teener, S.; Pacut, C.; Lentz, S.I.; et al. cGAS/STING and innate brain inflammation following acute high-fat feeding. Front. Immunol. 2022, 13, 1012594. [Google Scholar] [CrossRef] [PubMed]
- Hickman, S.; Izzy, S.; Sen, P.; Morsett, L.; El Khoury, J. Microglia in neurodegeneration. Nat. Neurosci. 2018, 21, 1359–1369. [Google Scholar] [CrossRef] [PubMed]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e1217. [Google Scholar] [CrossRef]
- Paul, B.D.; Snyder, S.H.; Bohr, V.A. Signaling by cGAS-STING in Neurodegeneration, Neuroinflammation, and Aging. Trends Neurosci. 2021, 44, 83–96. [Google Scholar] [CrossRef]
- Lasry, A.; Ben-Neriah, Y. Senescence-associated inflammatory responses: Aging and cancer perspectives. Trends Immunol. 2015, 36, 217–228. [Google Scholar] [CrossRef]
- Zhang, P.; Kishimoto, Y.; Grammatikakis, I.; Gottimukkala, K.; Cutler, R.G.; Zhang, S.; Abdelmohsen, K.; Bohr, V.A.; Misra Sen, J.; Gorospe, M.; et al. Senolytic therapy alleviates Aβ-associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer's disease model. Nat. Neurosci. 2019, 22, 719–728. [Google Scholar] [CrossRef]
- Tan, M.S.; Yu, J.T.; Jiang, T.; Zhu, X.C.; Tan, L. The NLRP3 inflammasome in Alzheimer’s disease. Mol. Neurobiol. 2013, 48, 875–882. [Google Scholar] [CrossRef]
- Li, T.; Chen, Z.J. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 2018, 215, 1287–1299. [Google Scholar] [CrossRef]
- Hou, Y.; Wei, Y.; Lautrup, S.; Yang, B.; Wang, Y.; Cordonnier, S.; Mattson, M.P.; Croteau, D.L.; Bohr, V.A. NAD(+) supplementation reduces neuroinflammation and cell senescence in a transgenic mouse model of Alzheimer’s disease via cGAS-STING. Proc. Natl. Acad. Sci. USA 2021, 118, e2011226118. [Google Scholar] [CrossRef]
- Yeh, F.L.; Wang, Y.; Tom, I.; Gonzalez, L.C.; Sheng, M. TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron 2016, 91, 328–340. [Google Scholar] [CrossRef]
- N'Diaye, E.N.; Branda, C.S.; Branda, S.S.; Nevarez, L.; Colonna, M.; Lowell, C.; Hamerman, J.A.; Seaman, W.E. TREM-2 (triggering receptor expressed on myeloid cells 2) is a phagocytic receptor for bacteria. J. Cell Biol. 2009, 184, 215–223. [Google Scholar] [CrossRef]
- Takahashi, K.; Rochford, C.D.; Neumann, H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 2005, 201, 647–657. [Google Scholar] [CrossRef]
- Jiang, T.; Zhang, Y.D.; Chen, Q.; Gao, Q.; Zhu, X.C.; Zhou, J.S.; Shi, J.Q.; Lu, H.; Tan, L.; Yu, J.T. TREM2 modifies microglial phenotype and provides neuroprotection in P301S tau transgenic mice. Neuropharmacology 2016, 105, 196–206. [Google Scholar] [CrossRef]
- Jiang, T.; Yu, J.T.; Zhu, X.C.; Tan, M.S.; Gu, L.Z.; Zhang, Y.D.; Tan, L. Triggering receptor expressed on myeloid cells 2 knockdown exacerbates aging-related neuroinflammation and cognitive deficiency in senescence-accelerated mouse prone 8 mice. Neurobiol. Aging 2014, 35, 1243–1251. [Google Scholar] [CrossRef]
- Jiang, T.; Tan, L.; Zhu, X.C.; Zhang, Q.Q.; Cao, L.; Tan, M.S.; Gu, L.Z.; Wang, H.F.; Ding, Z.Z.; Zhang, Y.D.; et al. Upregulation of TREM2 ameliorates neuropathology and rescues spatial cognitive impairment in a transgenic mouse model of Alzheimer's disease. Neuropsychopharmacology 2014, 39, 2949–2962. [Google Scholar] [CrossRef]
- Xu, Q.; Xu, W.; Cheng, H.; Yuan, H.; Tan, X. Efficacy and mechanism of cGAMP to suppress Alzheimer’s disease by elevating TREM2. Brain Behav. Immun. 2019, 81, 495–508. [Google Scholar] [CrossRef]
- Ahmad, S.; Orellana, A.; Kohler, I.; Frölich, L.; de Rojas, I.; Gil, S.; Boada, M.; Hernández, I.; Hausner, L.; Bakker, M.H.M.; et al. Association of lysophosphatidic acids with cerebrospinal fluid biomarkers and progression to Alzheimer’s disease. Alzheimer’s Res. Ther. 2020, 12, 124. [Google Scholar] [CrossRef]
- Sun, Y.; Kim, N.-H.; Yang, H.; Kim, S.-H.; Huh, S.-O. Lysophosphatidic acid induces neurite retraction in differentiated neuroblastoma cells via GSK-3β activation. Mol. Cells 2011, 31, 483–489. [Google Scholar] [CrossRef]
- Ramesh, S.; Govindarajulu, M.; Suppiramaniam, V.; Moore, T.; Dhanasekaran, M. Autotaxin-Lysophosphatidic Acid Signaling in Alzheimer’s Disease. Int. J. Mol. Sci. 2018, 19, 1827. [Google Scholar] [CrossRef]
- Guo, C.; Jeong, H.H.; Hsieh, Y.C.; Klein, H.U.; Bennett, D.A.; De Jager, P.L.; Liu, Z.; Shulman, J.M. Tau Activates Transposable Elements in Alzheimer's Disease. Cell Rep. 2018, 23, 2874–2880. [Google Scholar] [CrossRef]
- Ramirez, P.; Zuniga, G.; Sun, W.; Beckmann, A.; Ochoa, E.; DeVos, S.L.; Hyman, B.; Chiu, G.; Roy, E.R.; Cao, W.; et al. Pathogenic tau accelerates aging-associated activation of transposable elements in the mouse central nervous system. Prog. Neurobiol. 2022, 208, 102181. [Google Scholar] [CrossRef]
- Li, Q.; Cao, Y.; Dang, C.; Han, B.; Han, R.; Ma, H.; Hao, J.; Wang, L. Inhibition of double-strand DNA-sensing cGAS ameliorates brain injury after ischemic stroke. EMBO Mol. Med. 2020, 12, e11002. [Google Scholar] [CrossRef]
- Nazmi, A.; Mukhopadhyay, R.; Dutta, K.; Basu, A. STING Mediates Neuronal Innate Immune Response Following Japanese Encephalitis Virus Infection. Sci. Rep. 2012, 2, 347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, C.; Li, H.; Jiao, J. Deficiency of STING Signaling in Embryonic Cerebral Cortex Leads to Neurogenic Abnormalities and Autistic-Like Behaviors. Adv. Sci. 2020, 7, 2002117. [Google Scholar] [CrossRef]
- Ma, F.; Li, B.; Liu, S.Y.; Iyer, S.S.; Yu, Y.; Wu, A.; Cheng, G. Positive feedback regulation of type I IFN production by the IFN-inducible DNA sensor cGAS. J. Immunol. 2015, 194, 1545–1554. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.P.; Henry, R.J.; Shirey, K.A.; Doran, S.J.; Makarevich, O.D.; Ritzel, R.M.; Meadows, V.A.; Vogel, S.N.; Faden, A.I.; Stoica, B.A.; et al. Interferon-β Plays a Detrimental Role in Experimental Traumatic Brain Injury by Enhancing Neuroinflammation That Drives Chronic Neurodegeneration. J. Neurosci. 2020, 40, 2357–2370. [Google Scholar] [CrossRef]
- Shih, A.Y.; Damm-Ganamet, K.L.; Mirzadegan, T. Dynamic Structural Differences between Human and Mouse STING Lead to Differing Sensitivity to DMXAA. Biophys. J. 2018, 114, 32–39. [Google Scholar] [CrossRef]
- Zhang, H.; Han, M.-J.; Tao, J.; Ye, Z.-Y.; Du, X.-X.; Deng, M.-J.; Zhang, X.-Y.; Li, L.-F.; Jiang, Z.-F.; Su, X.-D. Rat and human STINGs profile similarly towards anticancer/antiviral compounds. Sci. Rep. 2015, 5, 18035. [Google Scholar] [CrossRef]
- Sheridan, C. Drug developers switch gears to inhibit STING. Nat. Biotechnol. 2019, 37, 199–201. [Google Scholar] [CrossRef] [PubMed]
| Model | Intervention | Outcome Measures | Results | References |
|---|---|---|---|---|
| 5xFAD mice | H-151 (STING inhibitor) | -Neuroinflammatory genes (Hippocampus) -Aβ42 fractions (cortex) -Aβ (DG and cortex) -Iba1 and GFAP (DG) -Microglial phagocytosis activity | -Reduced -Reduced -Reduced -Reduced -Enhanced | [63] |
| -HMC3 human microglial cells -APP/PS1 mice | H-151 (STING inhibitor) NAD+ precursor nicotinamide riboside (NR) | - Aβ42 induced IL6 production -Neuroinflammatory markers [Iba1 and GFAP, -Proinflammatory cytokines and chemokines, NLRP3, -γ-H2AX (DNA damage marker) and Cleaved-caspase-3 (apoptosis marker] -cGAS and STING protein -Cellular senescence -Learning and Memory -Synaptic Plasticity (LTP) | -Inhibition -Reduced -Reduced -Reduced -Improved -Improved | [81] |
| -AANAT-KO mice (accelerated aging model) -AANAT-KO Primary cerebrocortical neurons | Melatonin Melatonin | - mtDNA release -cGAS/STING/IRF3 protein expression -Caspase-1 -Proinflammatory cytokines -MMP -Mitochondrial ROS -Cytosolic mtDNA -Caspase-1 -Proinflammatory cytokines | -Decreased -Decreased -Decreased -Decreased -Increased -Decreased -Decreased -Decreased -Decreased | [60] |
| -APP/PS1 mice | cGAMP | -Spatial memory - Aβ pathology -Proinflammatory cytokines -Neuronal apoptosis | -Improved -Reduced -Reduced -Reduced | [88] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Govindarajulu, M.; Ramesh, S.; Beasley, M.; Lynn, G.; Wallace, C.; Labeau, S.; Pathak, S.; Nadar, R.; Moore, T.; Dhanasekaran, M. Role of cGAS–Sting Signaling in Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 8151. https://doi.org/10.3390/ijms24098151
Govindarajulu M, Ramesh S, Beasley M, Lynn G, Wallace C, Labeau S, Pathak S, Nadar R, Moore T, Dhanasekaran M. Role of cGAS–Sting Signaling in Alzheimer’s Disease. International Journal of Molecular Sciences. 2023; 24(9):8151. https://doi.org/10.3390/ijms24098151
Chicago/Turabian StyleGovindarajulu, Manoj, Sindhu Ramesh, McNeil Beasley, Graham Lynn, Caleigh Wallace, Sammie Labeau, Suhrud Pathak, Rishi Nadar, Timothy Moore, and Muralikrishnan Dhanasekaran. 2023. "Role of cGAS–Sting Signaling in Alzheimer’s Disease" International Journal of Molecular Sciences 24, no. 9: 8151. https://doi.org/10.3390/ijms24098151
APA StyleGovindarajulu, M., Ramesh, S., Beasley, M., Lynn, G., Wallace, C., Labeau, S., Pathak, S., Nadar, R., Moore, T., & Dhanasekaran, M. (2023). Role of cGAS–Sting Signaling in Alzheimer’s Disease. International Journal of Molecular Sciences, 24(9), 8151. https://doi.org/10.3390/ijms24098151







