PIMT Controls Insulin Synthesis and Secretion through PDX1
Abstract
1. Introduction
2. Results
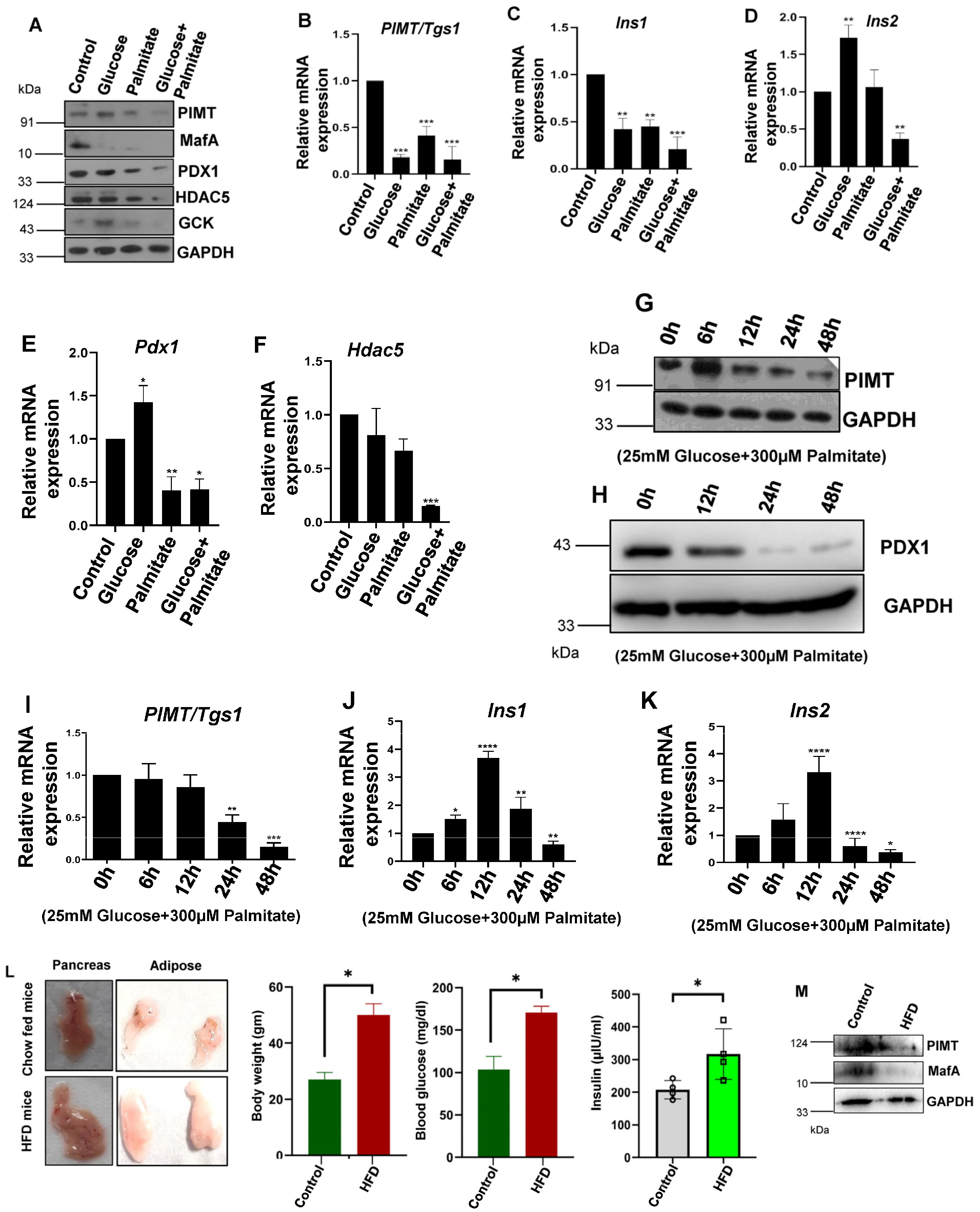
2.1. PIMT (PRIP-Interacting Protein with Methyl Transferase Activity) Expression Is Reduced in Cells Treated with Glucolipotoxic Conditions and HFD-Fed Mice Islets
2.2. RNA Sequencing Analysis of PIMT Knockdown Beta Cells Revealed Impairment of Insulin Secretory Pathway
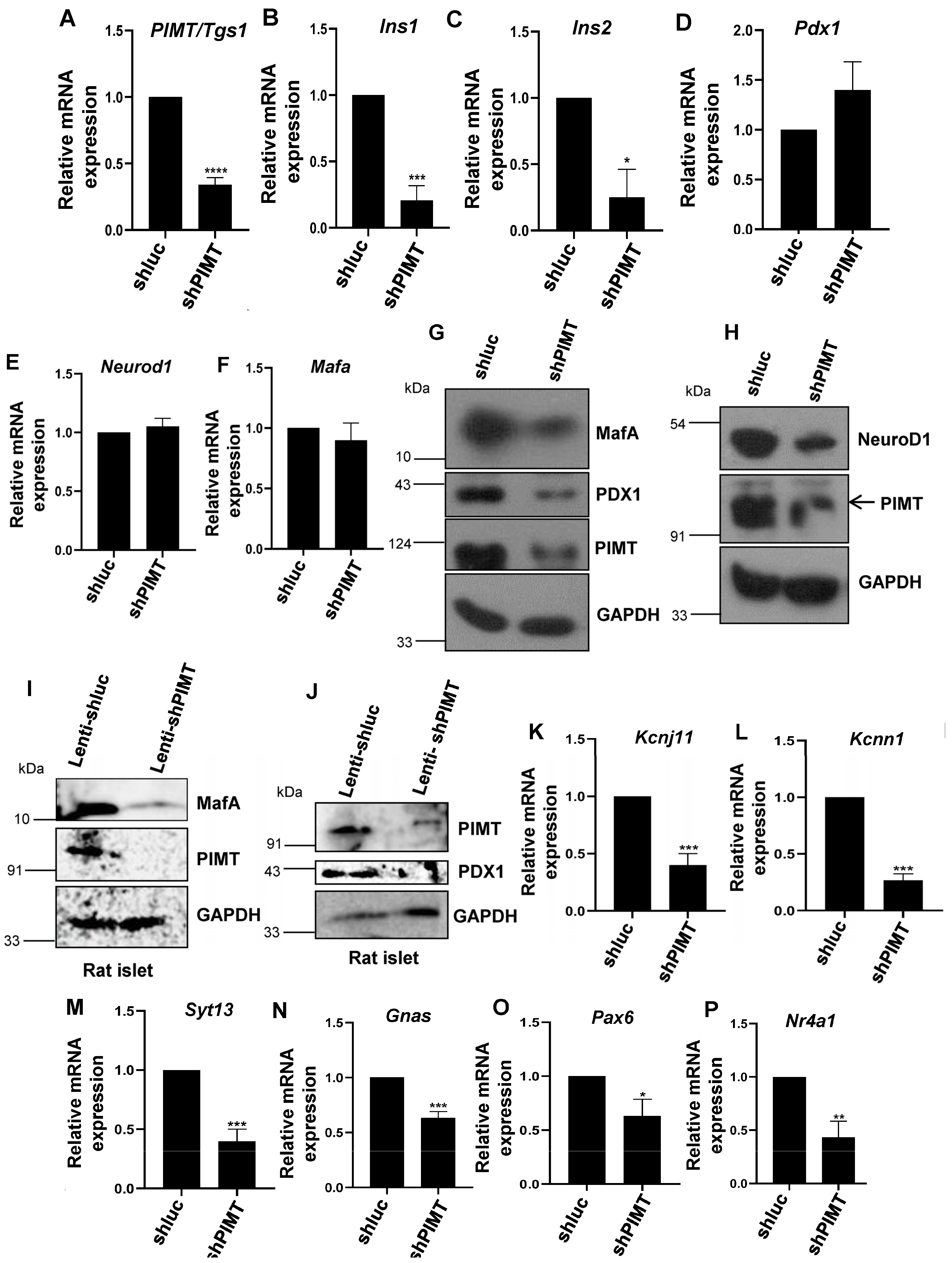
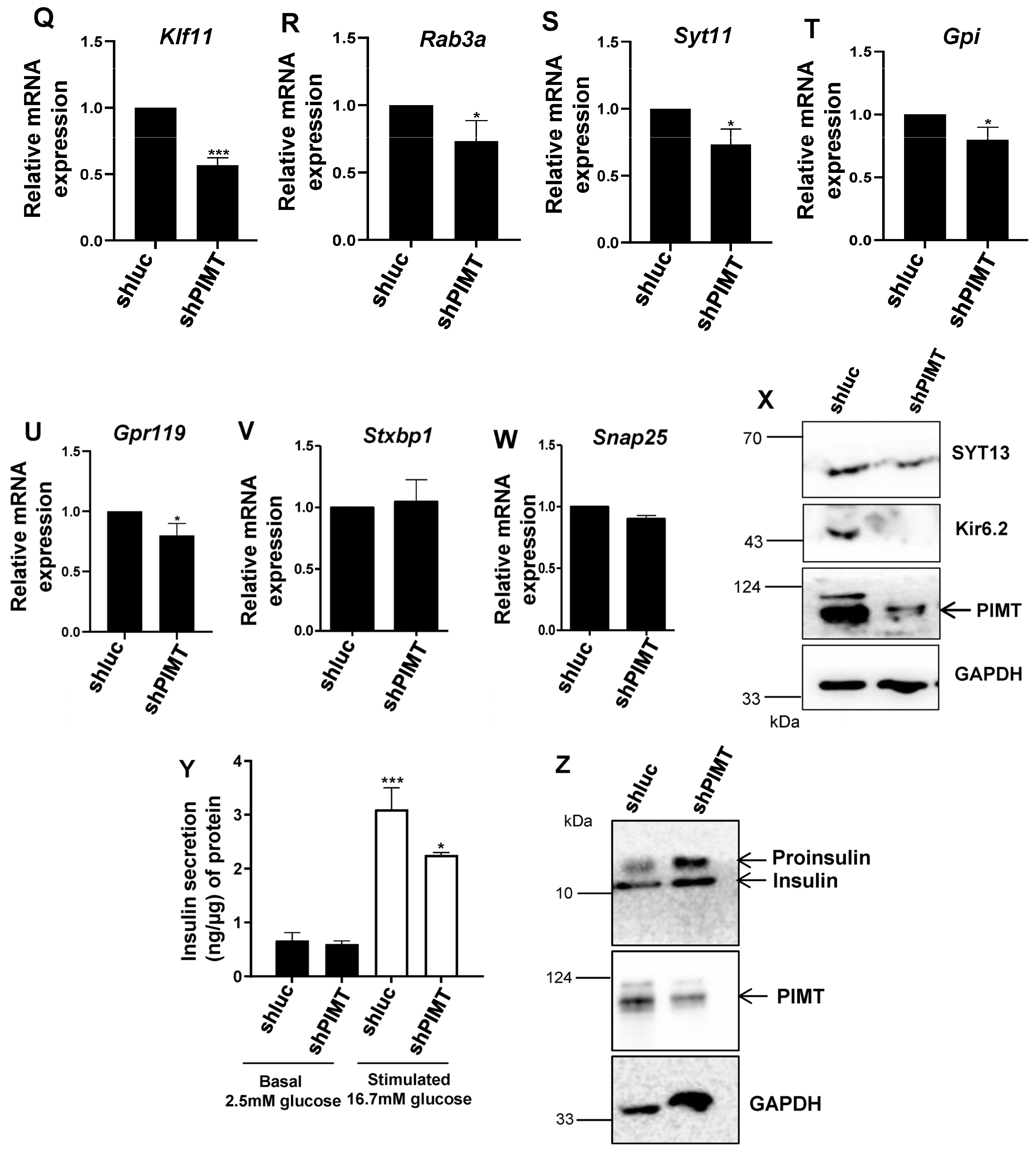
2.3. PIMT Knockdown Attenuates Insulin Secretion and the Associated Transcriptional Machinery
2.4. PIMT Interacts with PDX1 and MafA and Is Recruited to Ins1 and Ins2 in Beta Cells
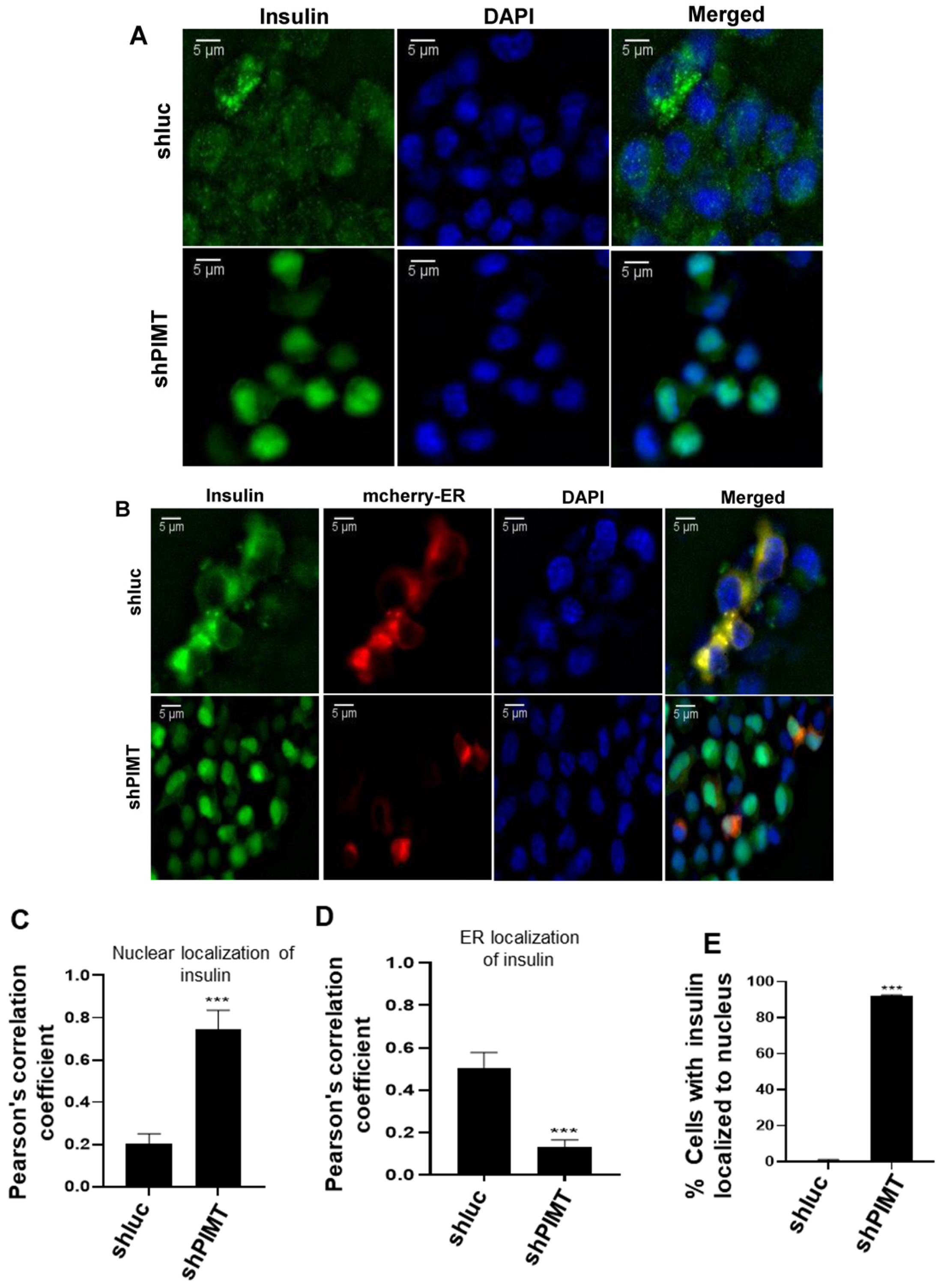
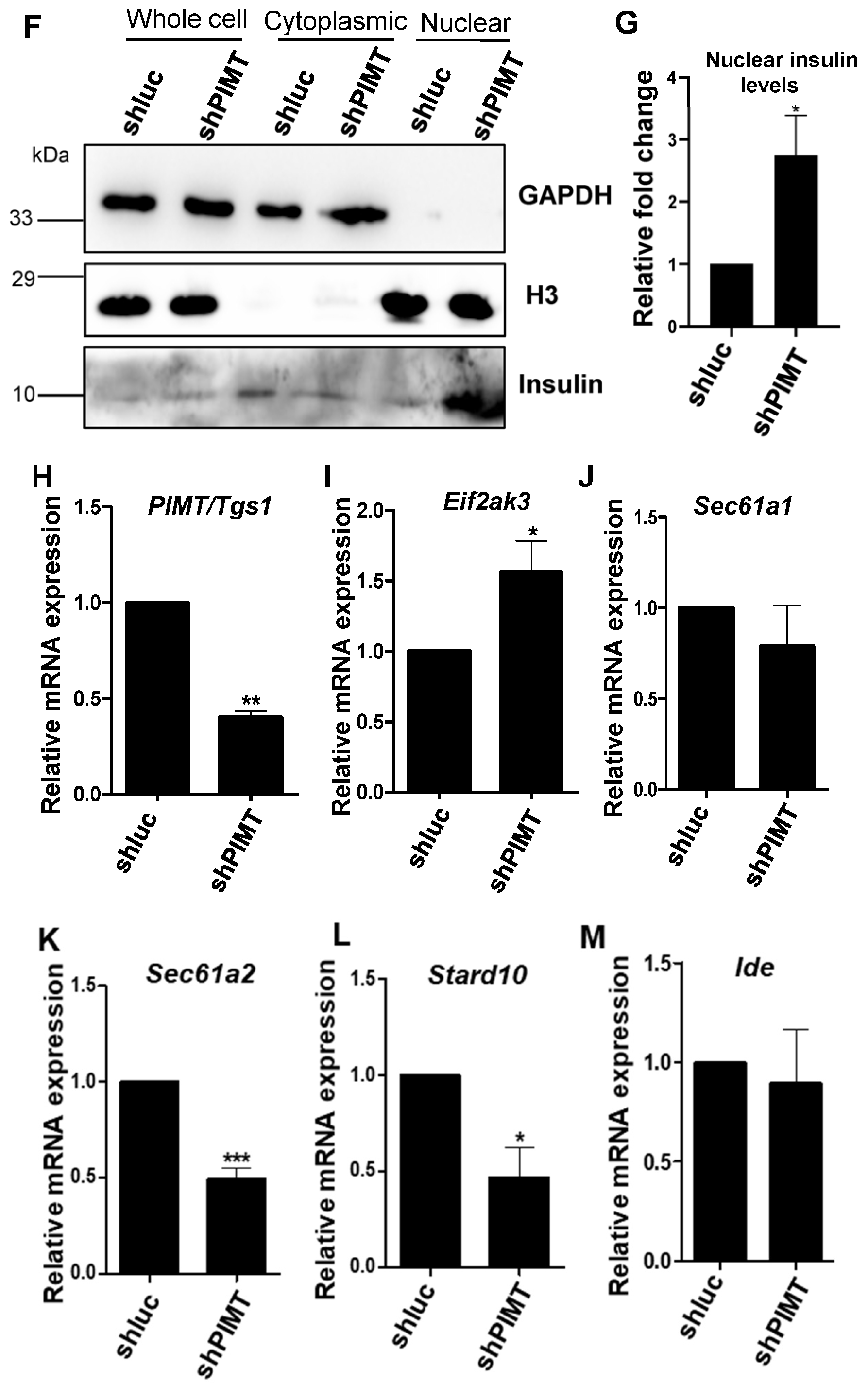
2.5. PIMT Knockdown Prompted Localization of Insulin to the Nucleus in the Beta Cells
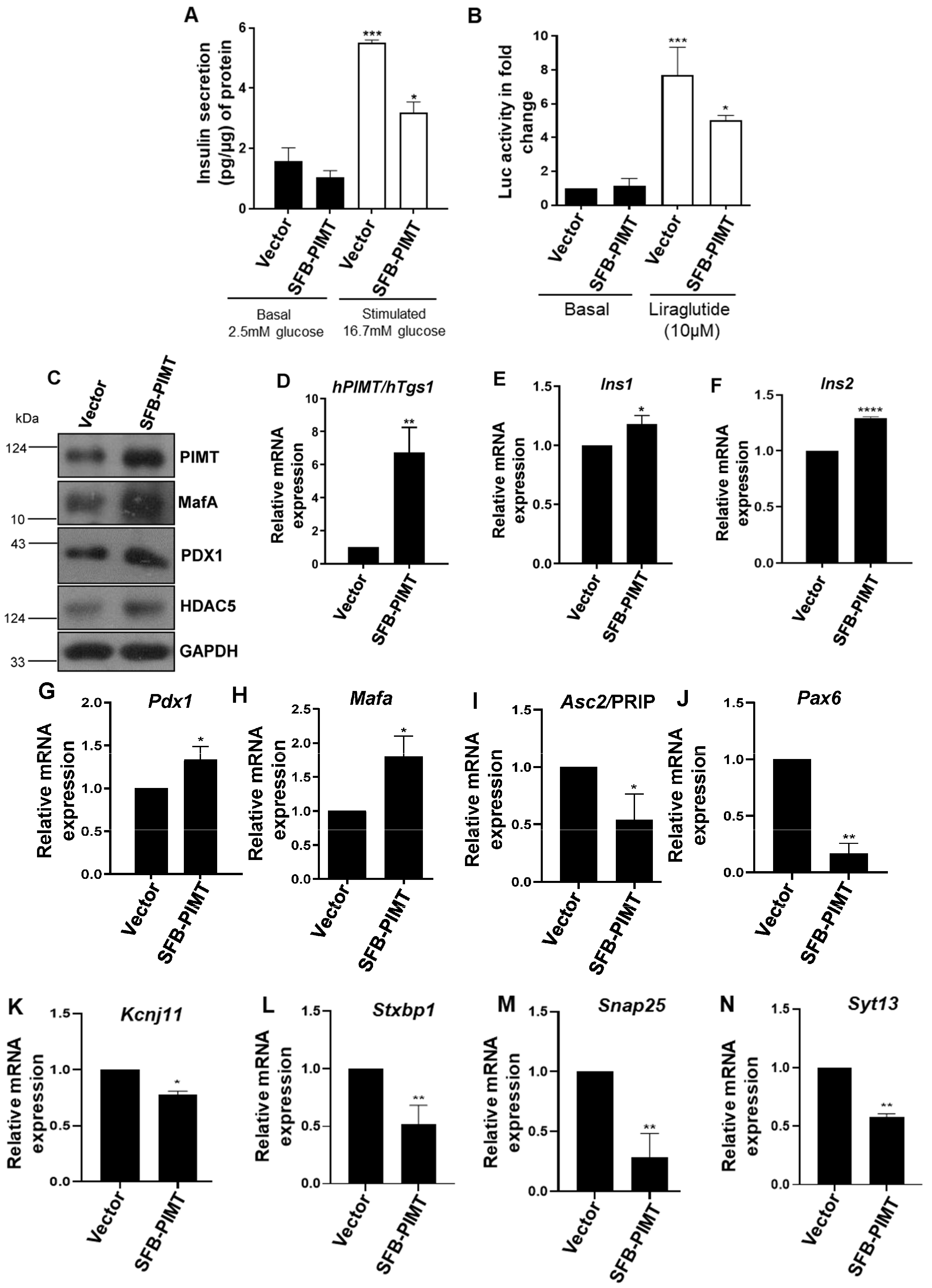
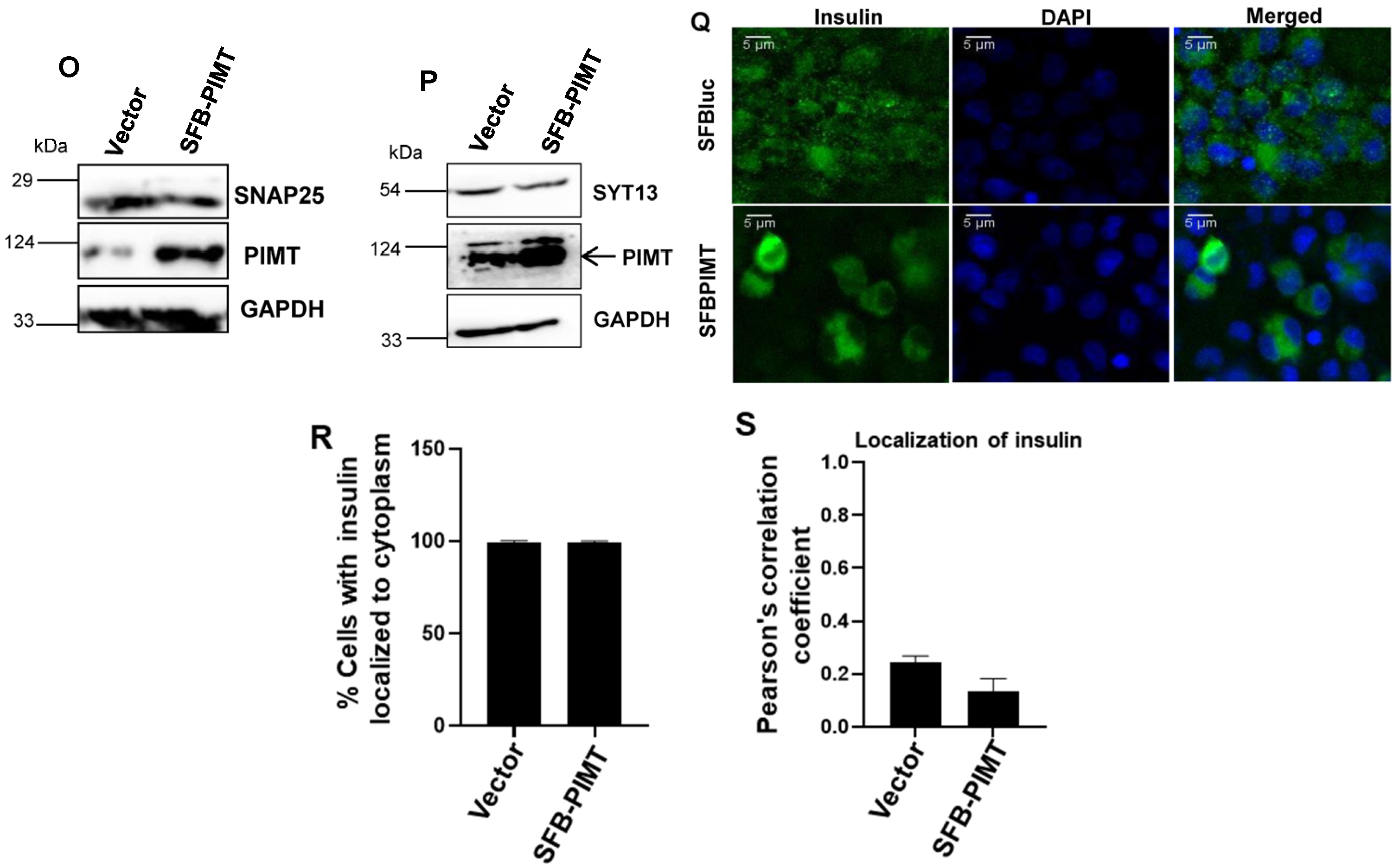
2.6. PIMT Overexpression Impairs Glucose-Stimulated Insulin Secretion and the Associated Gene Expression
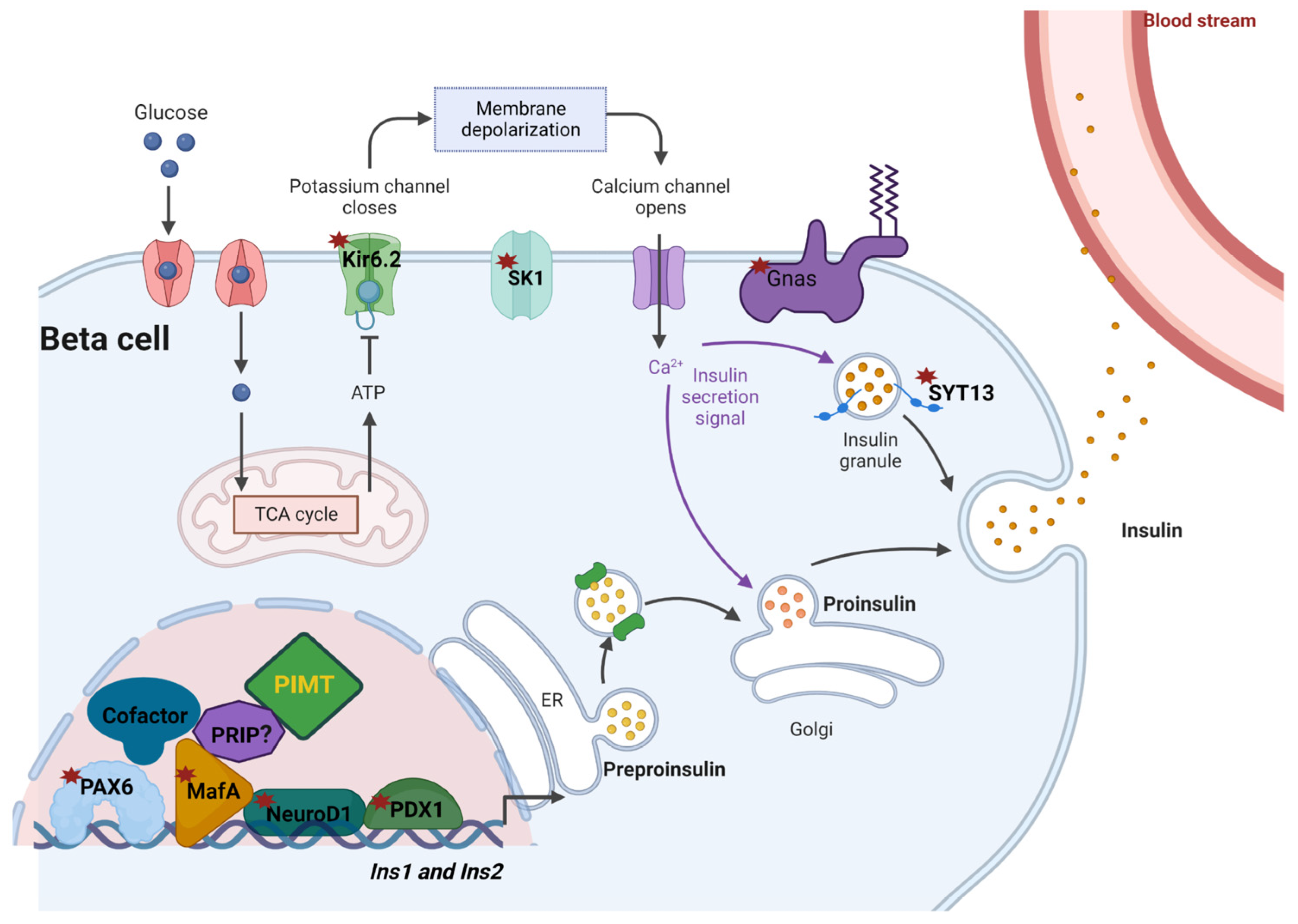
3. Discussion
4. Materials and Methods
4.1. Cells Line and Animals
4.2. High Fat Fed Mouse Model
4.3. Cell Lysis and Western Blotting
4.4. Real-Time PCR
4.5. Cloning of shRNA and Generation of Stable PIMT Knockdown Cells
4.6. RNA Sequencing and Data Analysis
4.7. Islet Isolation
4.8. Lentivirus Generation and Transduction
4.9. GSIS, ELISA, and CRE-Luc Assay
4.10. Immunoprecipitation and ChIP (Chromatin Immunoprecipitation)
4.11. Immunofluorescence
4.12. Nuclear and Cytoplasmic Fractionation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, J.-W.; Yoon, K.-H. Glucolipotoxicity in Pancreatic β-Cells. Diabetes Metab. J. 2011, 35, 444–450. [Google Scholar] [CrossRef]
- Poitout, V.; Amyot, J.; Semache, M.; Zarrouki, B.; Hagman, D.; Fontés, G. Glucolipotoxicity of the pancreatic beta cell. Biochim. Et Biophys. Acta BBA-Mol. Cell Biol. Lipids 2010, 1801, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, J.; Carlsson, L.; Edlund, T.; Edlund, H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 1994, 371, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Sussel, L.; Kalamaras, J.; Hartigan-O’Connor, D.J.; Meneses, J.J.; Pedersen, R.A.; Rubenstein, J.L.R.; German, M.S. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic β cells. Development 1998, 125, 2213–2221. [Google Scholar] [CrossRef] [PubMed]
- Sosa-Pineda, B.; Chowdhury, K.; Torres, M.; Oliver, G.; Gruss, P. The Pax4 gene is essential for differentiation of insulin-producing β cells in the mammalian pancreas. Nature 1997, 386, 399–402. [Google Scholar] [CrossRef] [PubMed]
- Gradwohl, G.; Dierich, A.; LeMeur, M.; Guillemot, F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. USA 2000, 97, 1607–1611. [Google Scholar] [CrossRef] [PubMed]
- Naya, F.J.; Huang, H.-P.; Qiu, Y.; Mutoh, H.; DeMayo, F.J.; Leiter, A.B.; Tsai, M.-J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes Dev. 1997, 11, 2323–2334. [Google Scholar] [CrossRef] [PubMed]
- Staffers, D.A.; Ferrer, J.; Clarke, W.L.; Habener, J.F. Early-onset type-ll diabetes mellitus (MODY4) linked to IPF1. Nat. Genet. 1997, 17, 138–139. [Google Scholar] [CrossRef]
- Malecki, M.T.; Jhala, U.S.; Antonellis, A.; Fields, L.; Doria, A.; Orban, T.; Saad, M.; Warram, J.H.; Montminy, M.; Krolewski, A.S. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat. Genet. 1999, 23, 323–328. [Google Scholar] [CrossRef]
- Yoon, J.; Xu, G.; Deeney, J.T.; Yang, S.-N.; Rhee, J.; Puigserver, P.; Levens, A.R.; Yang, R.; Zhang, C.-Y.; Lowell, B.B.; et al. Suppression of β Cell Energy Metabolism and Insulin Release by PGC-1α. Dev. Cell 2003, 5, 73–83. [Google Scholar] [CrossRef]
- Yeom, S.-Y.; Kim, G.H.; Kim, C.H.; Jung, H.D.; Kim, S.-Y.; Park, J.-Y.; Pak, Y.K.; Rhee, D.-K.; Kuang, S.-Q.; Xu, J.; et al. Regulation of Insulin Secretion and β-Cell Mass by Activating Signal Cointegrator 2. Mol. Cell. Biol. 2006, 26, 4553–4563. [Google Scholar] [CrossRef]
- Misra, P.; Qi, C.; Yu, S.; Shah, S.H.; Cao, W.-Q.; Rao, M.S.; Thimmapaya, B.; Zhu, Y.; Reddy, J.K. Interaction of PIMT with Transcriptional Coactivators CBP, p300, and PBP Differential Role in Transcriptional Regulation. J. Biol. Chem. 2002, 277, 20011–20019. [Google Scholar] [CrossRef] [PubMed]
- Mouaikel, J.; Verheggen, C.; Bertrand, E.; Tazi, J.; Bordonné, R. Hypermethylation of the Cap Structure of Both Yeast snRNAs and snoRNAs Requires a Conserved Methyltransferase that Is Localized to the Nucleolus. Mol. Cell 2002, 9, 891–901. [Google Scholar] [CrossRef]
- Wurth, L.; Gribling-Burrer, A.-S.; Verheggen, C.; Leichter, M.; Takeuchi, A.; Baudrey, S.; Martin, F.; Krol, A.; Bertrand, E.; Allmang, C. Hypermethylated-capped selenoprotein mRNAs in mammals. Nucleic Acids Res. 2014, 42, 8663–8677. [Google Scholar] [CrossRef] [PubMed]
- Mouaikel, J.; Bujnicki, J.M.; Tazi, J.; Bordonné, R. Sequence–structure–function relationships of Tgs1, the yeast snRNA/snoRNA cap hypermethylase. Nucleic Acids Res. 2003, 31, 4899–4909. [Google Scholar] [CrossRef] [PubMed]
- Kapadia, B.; Viswakarma, N.; Parsa, K.V.L.; Kain, V.; Behera, S.; Suraj, S.K.; Babu, P.P.; Kar, A.; Panda, S.; Zhu, Y.-J.; et al. ERK2-Mediated Phosphorylation of Transcriptional Coactivator Binding Protein PIMT/NCoA6IP at Ser298 Augments Hepatic Gluconeogenesis. PLoS ONE 2013, 8, e83787. [Google Scholar] [CrossRef]
- Kain, V.; Kapadia, B.; Viswakarma, N.; Seshadri, S.; Prajapati, B.; Jena, P.K.; Meda, C.L.T.; Subramanian, M.; Suraj, S.K.; Kumar, S.T.; et al. Co-activator binding protein PIMT mediates TNF-α induced insulin resistance in skeletal muscle via the transcriptional down-regulation of MEF2A and GLUT4. Sci. Rep. 2015, 5, 15197. [Google Scholar] [CrossRef] [PubMed]
- Blandino-Rosano, M.; Llacer, P.R.; Lin, A.; Reddy, J.K.; Bernal-Mizrachi, E. Trimethylguanosine synthase 1 is a novel regulator of pancreatic beta-cell mass and function. J. Biol. Chem. 2022, 298, 101592. [Google Scholar] [CrossRef]
- Andersson, S.A.; Olsson, A.H.; Esguerra, J.L.; Heimann, E.; Ladenvall, C.; Edlund, A.; Salehi, A.; Taneera, J.; Degerman, E.; Groop, L.; et al. Reduced insulin secretion correlates with decreased expression of exocytotic genes in pancreatic islets from patients with type 2 diabetes. Mol. Cell. Endocrinol. 2012, 364, 36–45. [Google Scholar] [CrossRef]
- Mitchell, R.K.; Nguyen-Tu, M.-S.; Chabosseau, P.; Callingham, R.M.; Pullen, T.J.; Cheung, R.; Leclerc, I.; Hodson, D.J.; Rutter, G.A. The transcription factor Pax6 is required for pancreatic β cell identity, glucose-regulated ATP synthesis, and Ca2+ dynamics in adult mice. J. Biol. Chem. 2017, 292, 8892–8906. [Google Scholar] [CrossRef]
- Chang-Chen, K.J.; Mullur, R.; Bernal-Mizrachi, E. β-cell failure as a complication of diabetes. Rev. Endocr. Metab. Disord. 2008, 9, 329–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Moriguchi, T.; Kajihara, M.; Esaki, R.; Harada, A.; Shimohata, H.; Oishi, H.; Hamada, M.; Morito, N.; Hasegawa, K.; et al. MafA Is a Key Regulator of Glucose-Stimulated Insulin Secretion. Mol. Cell. Biol. 2005, 25, 4969–4976. [Google Scholar] [CrossRef] [PubMed]
- Juliana, C.A.; Yang, J.; Cannon, C.E.; Good, A.L.; Haemmerle, M.W.; Stoffers, D.A. A PDX1-ATF transcriptional complex governs β cell survival during stress. Mol. Metab. 2018, 17, 39–48. [Google Scholar] [CrossRef]
- Gosmain, Y.; Katz, L.S.; Masson, M.H.; Cheyssac, C.; Poisson, C.; Philippe, J. Pax6 Is Crucial for β-Cell Function, Insulin Biosynthesis, and Glucose-Induced Insulin Secretion. Mol. Endocrinol. 2012, 26, 696–709. [Google Scholar] [CrossRef]
- Delépine, M.; Nicolino, M.; Barrett, T.; Golamaully, M.; Lathrop, G.M.; Julier, C. EIF2AK3, encoding translation initiation factor 2-α kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet. 2000, 25, 406–409. [Google Scholar] [CrossRef]
- Harada, S.; Smith, R.M.; Jarett, L. Mechanisms of nuclear translocation of insulin. Cell Biochem. Biophys. 1999, 31, 307–319. [Google Scholar] [CrossRef]
- Carrat, G.R.; Hu, M.; Nguyen-Tu, M.-S.; Chabosseau, P.; Gaulton, K.J.; van de Bunt, M.; Siddiq, A.; Falchi, M.; Thurner, M.; Canouil, M.; et al. Decreased STARD10 Expression Is Associated with Defective Insulin Secretion in Humans and Mice. Am. J. Hum. Genet. 2017, 100, 238–256. [Google Scholar] [CrossRef]
- Cunningham, C.N.; He, K.; Arunagiri, A.; Paton, A.W.; Paton, J.C.; Arvan, P.; Tsai, B. Chaperone-Driven Degradation of a Misfolded Proinsulin Mutant in Parallel With Restoration of Wild-Type Insulin Secretion. Diabetes 2016, 66, 741–753. [Google Scholar] [CrossRef]
- Christensen, D.P.; Dahllöf, M.; Lundh, M.; Rasmussen, D.N.; Nielsen, M.D.; Billestrup, N.; Grunnet, L.G.; Mandrup-Poulsen, T. Histone Deacetylase (HDAC) Inhibition as a Novel Treatment for Diabetes Mellitus. Mol. Med. 2011, 17, 378–390. [Google Scholar] [CrossRef]
- Zhu, Y.; Qi, C.; Cao, W.-Q.; Yeldandi, A.V.; Rao, M.S.; Reddy, J.K. Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proc. Natl. Acad. Sci. USA 2001, 98, 10380–10385. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Cui, S.; Zong, C.; Gao, W.; Xu, T.; Gao, P.; Chen, J.; Qin, D.; Guan, Q.; Liu, Y.; et al. The Orphan Nuclear Receptor NR4A1 Protects Pancreatic β-Cells from Endoplasmic Reticulum (ER) Stress-mediated Apoptosis. J. Biol. Chem. 2015, 290, 20687–20699. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Alamuru-Yellapragada, N.P.; Kapadia, B.; Parsa, K.V. In-house made nucleofection buffer for efficient and cost effective transfection of RAW 264.7 macrophages. Biochem. Biophys. Res. Commun. 2017, 487, 247–254. [Google Scholar] [CrossRef]
- Jiang, W.; Hua, R.; Wei, M.; Li, C.; Qiu, Z.; Yang, X.; Zhang, C. An optimized method for high-titer lentivirus preparations without ultracentrifugation. Sci. Rep. 2015, 5, 13875. [Google Scholar] [CrossRef]
- Jiménez-Moreno, C.M.; Herrera-Gomez, I.D.G.; Noriega, L.L.; Lorenzo, P.I.; Cobo-Vuilleumier, N.; Fuente-Martin, E.; Mellado-Gil, J.M.; Parnaud, G.; Bosco, D.; Gauthier, B.R.; et al. A Simple High Efficiency Intra-Islet Transduction Protocol Using Lentiviral Vectors. Curr. Gene Ther. 2015, 15, 436–446. [Google Scholar] [CrossRef]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H.; Bannert, K.; Berlin, P.; Reiner, J.; Lemcke, H.; David, R.; Engelmann, R.; Lamprecht, G.; et al. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef]
- Suzuki, K.; Bose, P.; Leong-Quong, R.Y.; Fujita, D.J.; Riabowol, K. REAP: A two minute cell fractionation method. BMC Res. Notes 2010, 3, 294. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, R.; Maity, S.K.; Chakrabarti, P.; Katika, M.R.; Kapettu, S.; Parsa, K.V.L.; Misra, P. PIMT Controls Insulin Synthesis and Secretion through PDX1. Int. J. Mol. Sci. 2023, 24, 8084. https://doi.org/10.3390/ijms24098084
Sharma R, Maity SK, Chakrabarti P, Katika MR, Kapettu S, Parsa KVL, Misra P. PIMT Controls Insulin Synthesis and Secretion through PDX1. International Journal of Molecular Sciences. 2023; 24(9):8084. https://doi.org/10.3390/ijms24098084
Chicago/Turabian StyleSharma, Rahul, Sujay K. Maity, Partha Chakrabarti, Madhumohan R. Katika, Satyamoorthy Kapettu, Kishore V. L. Parsa, and Parimal Misra. 2023. "PIMT Controls Insulin Synthesis and Secretion through PDX1" International Journal of Molecular Sciences 24, no. 9: 8084. https://doi.org/10.3390/ijms24098084
APA StyleSharma, R., Maity, S. K., Chakrabarti, P., Katika, M. R., Kapettu, S., Parsa, K. V. L., & Misra, P. (2023). PIMT Controls Insulin Synthesis and Secretion through PDX1. International Journal of Molecular Sciences, 24(9), 8084. https://doi.org/10.3390/ijms24098084







