The miR166d/TaCPK7-D Signaling Module Is a Critical Mediator of Wheat (Triticum aestivum L.) Tolerance to K+ Deficiency
Abstract
1. Introduction
2. Results
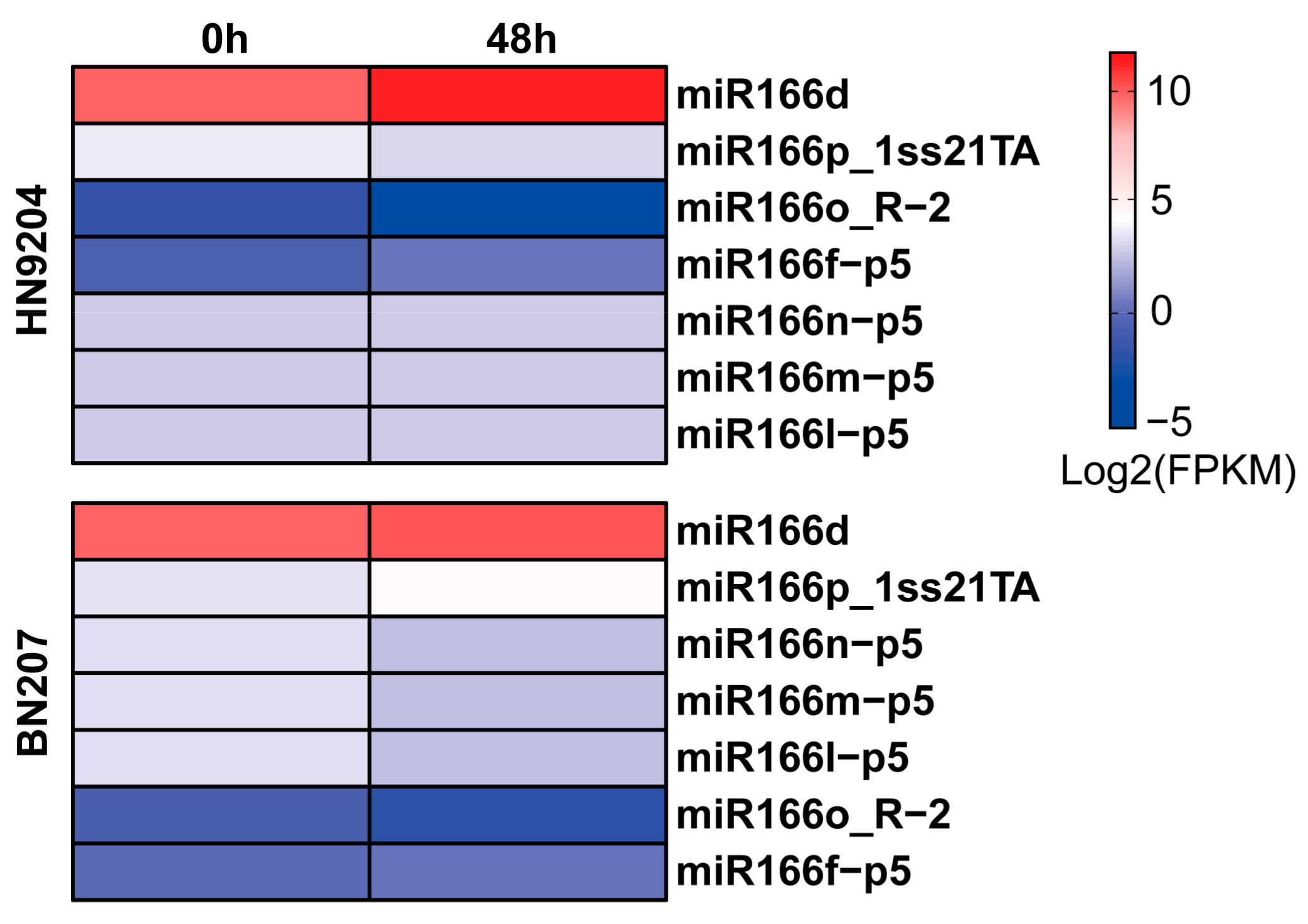
2.1. Expression of miR166d in Wheat Exposed to LK Stress
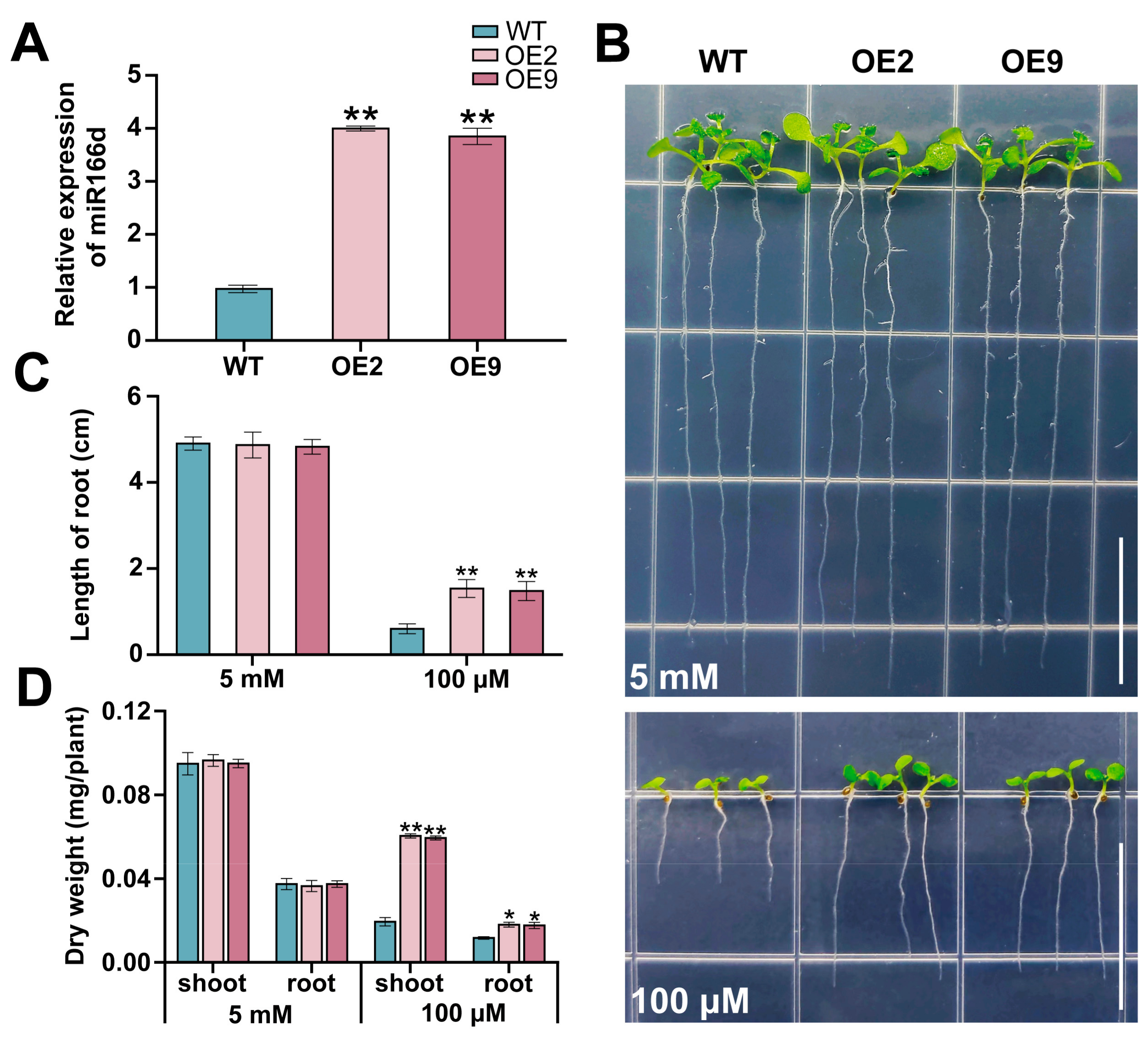
2.2. miR166d Positively Regulates Plant Tolerance to LK Stress
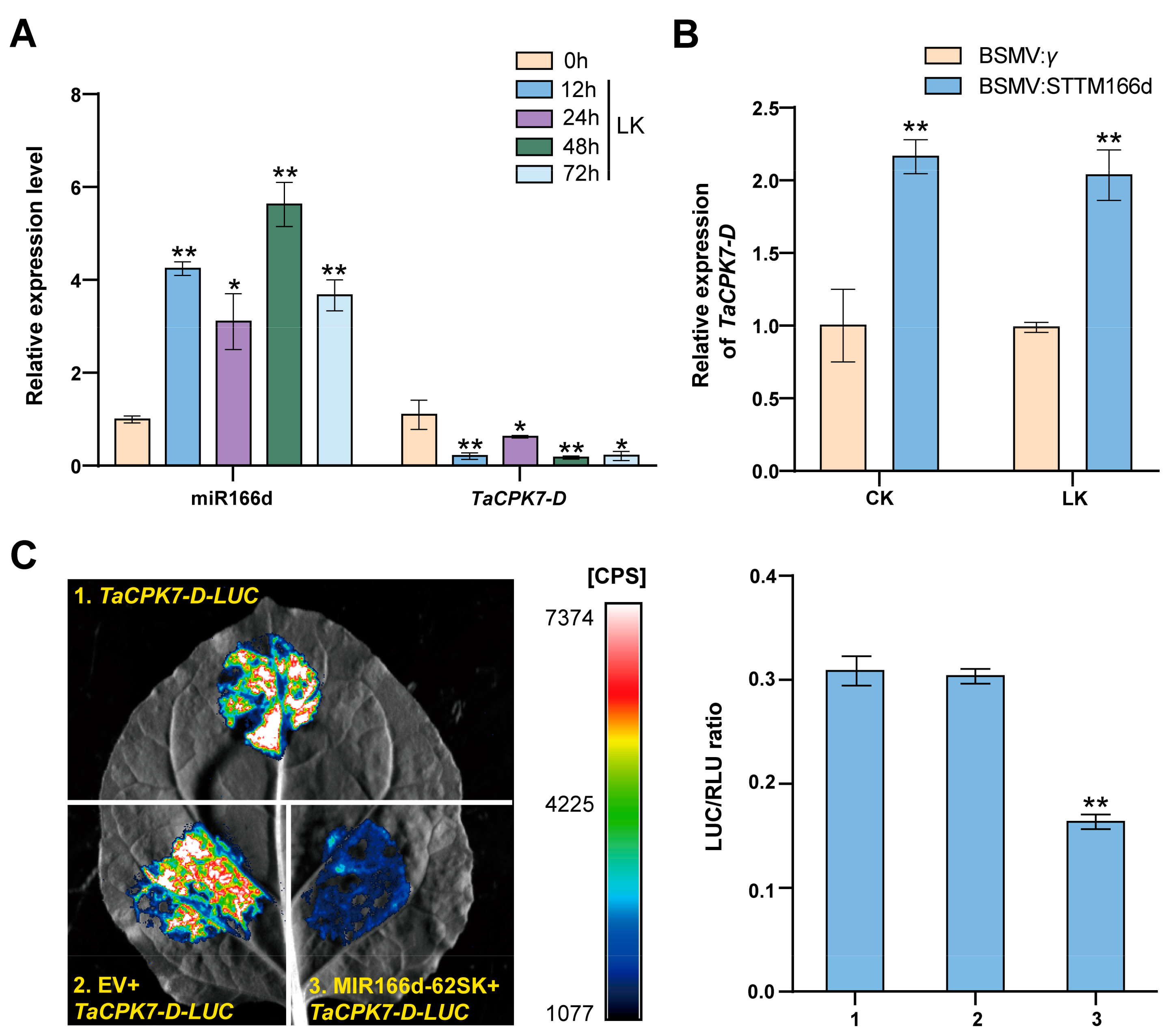
2.3. Identification of the Downstream Genes Targeted by miR166d
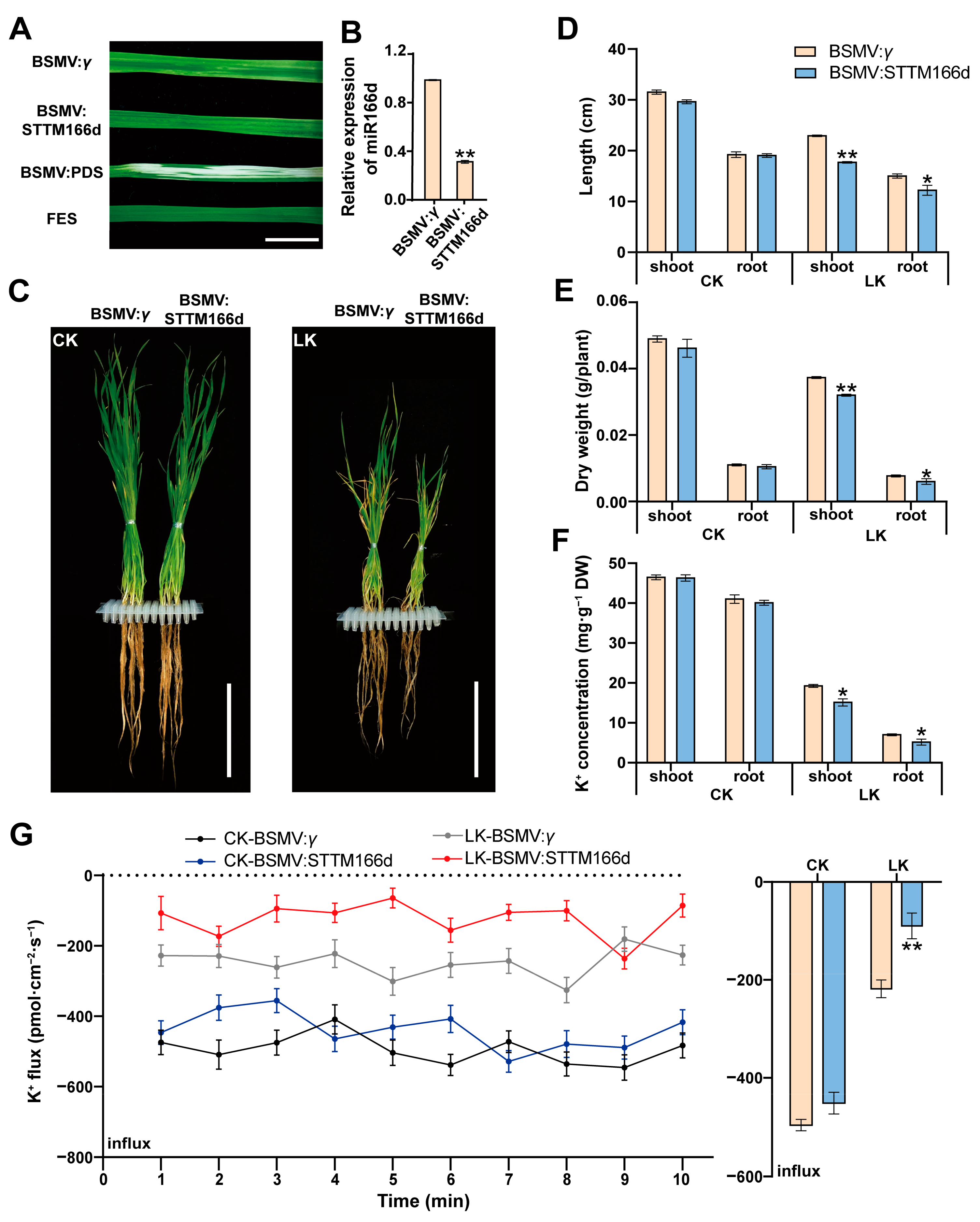
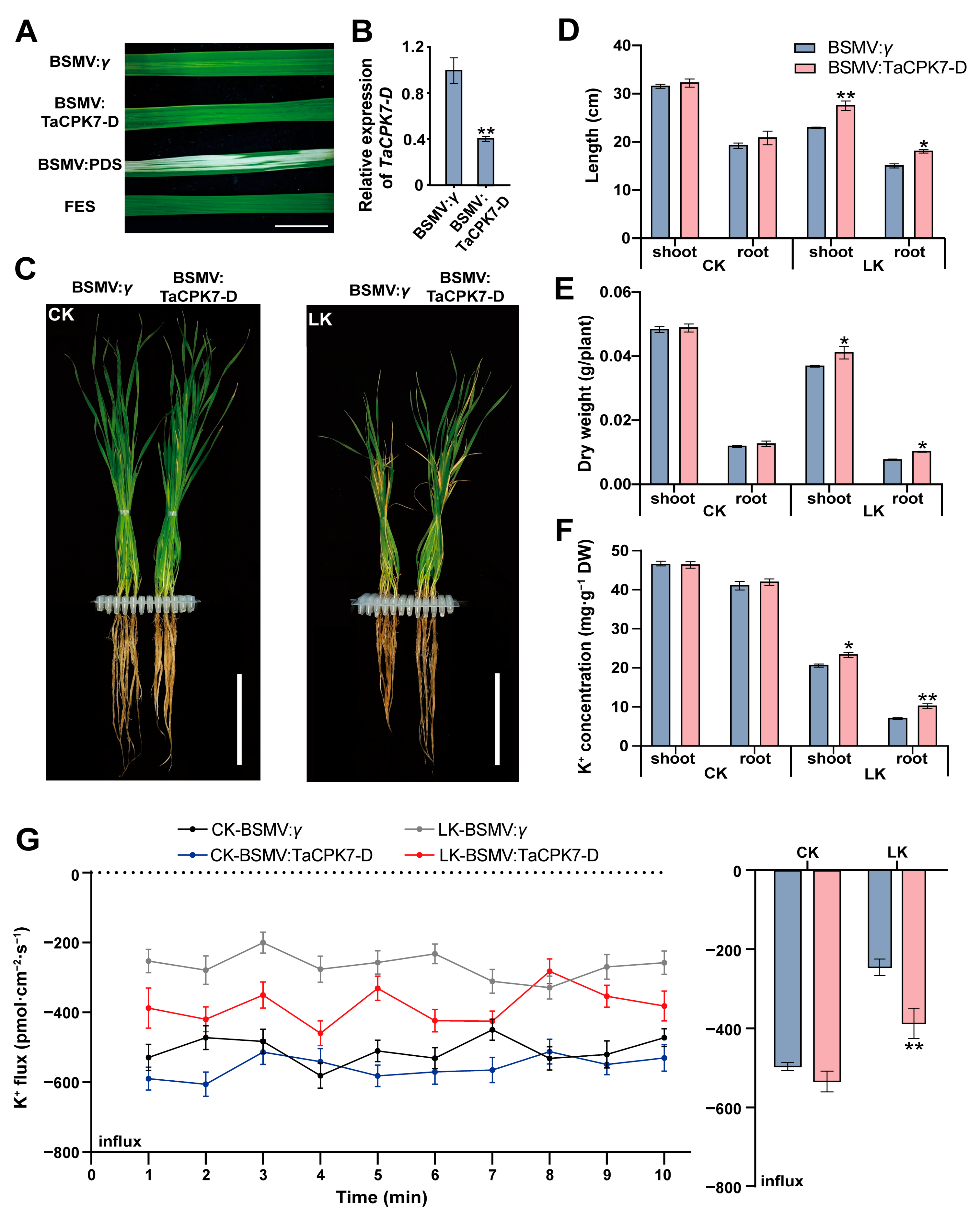
2.4. TaCPK7-D Negatively Regulates Wheat Tolerance to LK Stress
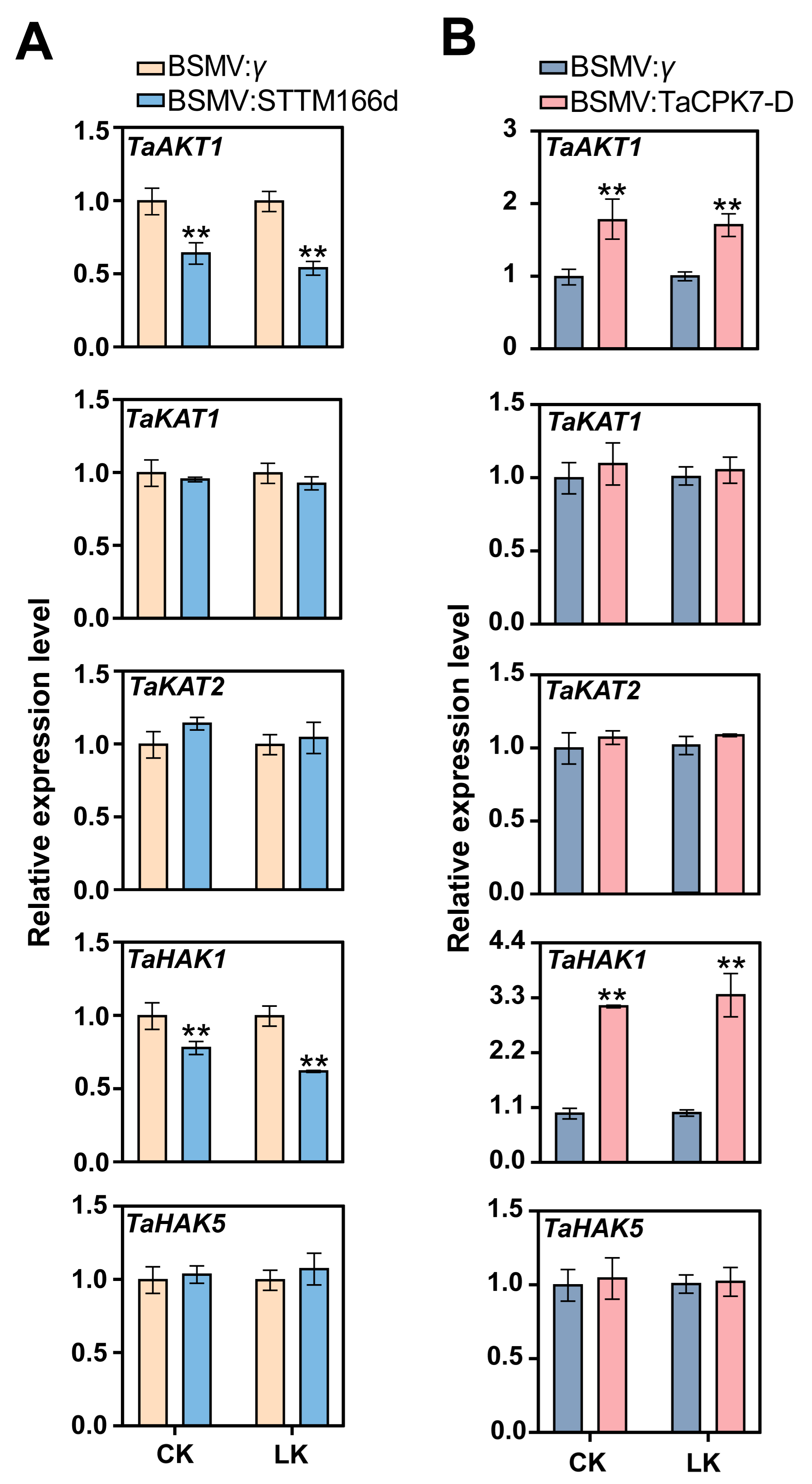
2.5. The miR166d/TaCPK7-D Module Affects TaAKT1 and TaHAK1 Expression in Response to LK Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Vector Construction, Plant Transformation, and Analysis of Arabidopsis Growth
4.3. VIGS Assay of mir166d and TaCPK7-D
4.4. Gene Expression Analysis
4.5. Dual-Luciferase Assay
4.6. Measurement of the Net K+ Flux in Wheat Plants
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clarkson, D.T. The mineral nutrition of higher plants. Annu. Rev. Plant Physiol. 1980, 31, 239–298. [Google Scholar] [CrossRef]
- Leigh, R.A.; Wyn Jones, R.G. A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol. 1984, 97, 1–13. [Google Scholar] [CrossRef]
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Stein, N.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, 6403. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef]
- Gao, S.; Guo, C.; Zhang, Y.; Zhang, F.; Du, X.; Gu, J.; Xiao, K. Wheat microRNA member TamiR444a is nitrogen deprivation-responsive and involves plant adaptation to the nitrogen-starvation stress. Plant Mol. Biol. Rep. 2016, 34, 931–946. [Google Scholar] [CrossRef]
- Bai, Q.; Wang, X.; Chen, X.; Shi, G.; Liu, Z.; Guo, C.; Xiao, K. Wheat miRNA TaemiR408 acts as an essential mediator in plant tolerance to Pi deprivation and salt stress via modulating stress-associated physiological processes. Front. Plant Sci. 2018, 9, 499. [Google Scholar] [CrossRef]
- Jiao, X.; Wang, H.; Yan, J.; Kong, X.; Liu, Y.; Chu, J.; Chen, X.; Fang, R.; Yan, Y. Promotion of BR biosynthesis by miR444 is required for ammonium-triggered inhibition of root growth. Plant Physiol. 2020, 182, 1454–1466. [Google Scholar] [CrossRef]
- Cheng, X.; He, Q.; Tang, S.; Wang, H.; Zhang, X.; Lv, M.; Liu, H.; Gao, Q.; Zhou, Y.; Wang, Q.; et al. The miR172/IDS1 signaling module confers salt tolerance through maintaining ROS homeostasis in cereal crops. New Phytol. 2021, 230, 1017–1033. [Google Scholar] [CrossRef]
- Liu, X.; Liu, S.; Chen, X.; Prasanna, B.M.; Ni, Z.; Li, X.; He, Y.; Fan, Z.; Zhou, T. Maize miR167-ARF3/30-polyamine oxidase 1 module-regulated H2O2 production confers resistance to maize chlorotic mottle virus. Plant Physiol. 2022, 189, 1065–1082. [Google Scholar] [CrossRef]
- Pachamuthu, K.; Hari Sundar, V.; Narjala, A.; Singh, R.R.; Das, S.; Avik Pal, H.C.Y.; Shivaprasad, P.V. Nitrate-dependent regulation of miR444-OsMADS27 signalling cascade controls root development in rice. J. Exp. Bot. 2022, 73, 3511–3530. [Google Scholar] [CrossRef]
- Feng, T.; Zhang, Z.; Gao, P.; Feng, Z.; Zuo, S.; Ouyang, S. Suppression of rice osa-miR444.2 improves the resistance to sheath blight in rice mediating through the phytohormone pathway. Int. J. Mol. Sci. 2023, 24, 3653. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, H.; Hamera, S.; Chen, X.; Fang, R. MiR444a has multiple functions in the rice nitrate-signaling pathway. Plant J. 2014, 78, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Thornburg, T.E.; Liu, J.; Li, Q.; Xue, H.; Wang, G.; Li, L.; Fontana, J.E.; Davis, K.E.; Liu, W.; Zhang, B.; et al. Potassium deficiency significantly affected plant growth and development as well as microRNA-mediated mechanism in wheat (Triticum aestivum L.). Front. Plant Sci. 2020, 11, 1219. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, K.; Liu, G.; Li, S.; Zhao, S.; Liu, X.; Yang, X.; Xiao, K. Global identification and characterization of miRNA family members responsive to potassium deprivation in wheat (Triticum aestivum L.). Sci. Rep. 2020, 10, 15812. [Google Scholar] [CrossRef] [PubMed]
- Floyd, S.K.; Bowman, J.L. Gene regulation: Ancient microRNA target sequences in plants. Nature 2004, 428, 485–486. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Hu, F.; Wang, R.; Zhou, X.; Sze, S.H.; Liou, L.W.; Barefoot, A.; Dickman, M.; Zhang, X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 2011, 145, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Honda, M.; Zhu, H.; Zhang, Z.; Guo, X.; Li, T.; Li, Z.; Peng, X.; Nakajima, K.; Duan, L.; et al. Spatiotemporal sequestration of miR165/166 by Arabidopsis Argonaute10 promotes shoot apical meristem maintenance. Cell Rep. 2015, 10, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singh, S.; Panigrahi, K.C.; Reski, R.; Sarkar, A.K. Balanced activity of microRNA166/165 and its target transcripts from the class III homeodomain-leucine zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 945–953. [Google Scholar] [CrossRef]
- Yan, J.; Gu, Y.; Jia, X.; Kang, W.; Pan, S.; Tang, X.; Chen, X.; Tang, G. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 2012, 24, 415–427. [Google Scholar] [CrossRef]
- Clepet, C.; Devani, R.S.; Boumlik, R.; Hao, Y.; Morin, H.; Marcel, F.; Verdenaud, M.; Mania, B.; Brisou, G.; Citerne, S.; et al. The miR166-SlHB15A regulatory module controls ovule development and parthenocarpic fruit set under adverse temperatures in tomato. Mol. Plant 2021, 14, 1185–1198. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Ma, J.; Zhang, Y.; Yang, S.; Feng, X.; Yan, J. The miR166 mediated regulatory module controls plant height by regulating gibberellic acid biosynthesis and catabolism in soybean. J. Integr. Plant Biol. 2022, 64, 995–1006. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Gong, S.; Wang, Y.; Wang, F.; Bao, H.; Sun, J.; Cai, C.; Yi, K.; Chen, Z.; Zhu, C. MicroRNA166 modulates cadmium tolerance and accumulation in rice. Plant Physiol. 2018, 177, 1691–1703. [Google Scholar] [CrossRef]
- Iwamoto, M.; Tagiri, A. MicroRNA-targeted transcription factor gene RDD1 promotes nutrient ion uptake and accumulation in rice. Plant J. 2016, 85, 466–477. [Google Scholar] [CrossRef]
- Valmonte, G.R.; Arthur, K.; Higgins, C.M.; MacDiarmid, R.M. Calcium-dependent protein kinases in plants: Evolution, expression and function. Plant Cell Physiol. 2014, 55, 551–569. [Google Scholar] [CrossRef]
- Ding, Y.; Yang, H.; Wu, S.; Fu, D.; Li, M.; Gong, Z.; Yang, S. CPK28-NLP7 module integrates cold-induced Ca2+ signal and transcriptional reprogramming in Arabidopsis. Sci. Adv. 2022, 8, eabn7901. [Google Scholar] [CrossRef] [PubMed]
- Geiger, D.; Scherzer, S.; Mumm, P.; Marten, I.; Ache, P.; Matschi, S.; Liese, A.; Wellmann, C.; Al-Rasheid, K.A.; Grill, E.; et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA 2010, 107, 8023–8028. [Google Scholar] [CrossRef]
- Gutermuth, T.; Lassig, R.; Portes, M.T.; Maierhofer, T.; Romeis, T.; Borst, J.W.; Hedrich, R.; Feijó, J.A.; Konrad, K.R. Pollen tube growth regulation by free anions depends on the interaction between the anion channel SLAH3 and calcium-dependent protein kinases CPK2 and CPK20. Plant Cell 2013, 25, 4525–4543. [Google Scholar] [CrossRef]
- Huimin, R.; Hussain, J.; Li, W.; Yao, F.; Guo, J.; Kong, Y.; Liu, S.; Qi, G. The expression of constitutively active CPK3 impairs potassium uptake and transport in Arabidopsis under low K+ stress. Cell Calcium. 2021, 98, 102447. [Google Scholar] [CrossRef]
- Ronzier, E.; Corratgé-Faillie, C.; Sanchez, F.; Brière, C.; Xiong, T.C. Ca2+-dependent protein kinase 6 enhances KAT2 shaker channel activity in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 1596. [Google Scholar] [CrossRef]
- Ronzier, E.; Corratgé-Faillie, C.; Sanchez, F.; Prado, K.; Brière, C.; Leonhardt, N.; Thibaud, J.B.; Xiong, T.C. CPK13, a noncanonical Ca2+-dependent protein kinase, specifically inhibits KAT2 and KAT1 shaker K+ channels and reduces stomatal opening. Plant Physiol. 2014, 166, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Borkiewicz, L.; Polkowska-Kowalczyk, L.; Cieśla, J.; Sowiński, P.; Jończyk, M.; Rymaszewski, W.; Szymańska, K.P.; Jaźwiec, R.; Muszyńska, G.; Szczegielniak, J. Expression of maize calcium-dependent protein kinase (ZmCPK11) improves salt tolerance in transgenic Arabidopsis plants by regulating sodium and potassium homeostasis and stabilizing photosystem II. Physiol. Plant 2020, 168, 38–57. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S. Whole-Transcriptome Profile Reveals the ceRNA Network Response to Low Potassium of Seedlings Stage in Wheat. Master’s Thesis, Hebei Agricultural University, Baoding, China, 2021. [Google Scholar]
- Römheld, V.; Kirkby, E.A. Research on potassium in agriculture: Needs and prospects. Plant Soil. 2010, 335, 155–180. [Google Scholar] [CrossRef]
- Zeng, J.; Ye, Z.; He, X.; Zhang, G. Identification of microRNAs and their targets responding to low-potassium stress in two barley genotypes differing in low-K tolerance. J. Plant Physiol. 2019, 234–235, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef]
- Zhou, G.; Kubo, M.; Zhong, R.; Demura, T.; Ye, Z. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 2007, 48, 391–404. [Google Scholar] [CrossRef]
- Prasad, A.; Sharma, N.; Chirom, O.; Prasad, M. The sly-miR166-SlyHB module acts as a susceptibility factor during ToLCNDV infection. Theor. Appl. Genet. 2022, 135, 233–242. [Google Scholar] [CrossRef]
- Itoh, J.; Hibara, K.; Sato, Y.; Nagato, Y. Developmental role and auxin responsiveness of Class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 2008, 147, 1960–1975. [Google Scholar] [CrossRef]
- Singh, A.; Roy, S.; Singh, S.; Das, S.S.; Gautam, V.; Yadav, S.; Kumar, A.; Singh, A.; Samantha, S.; Sarkar, A.K. Phytohormonal crosstalk modulates the expression of miR166/165s, target Class III HD-ZIPs, and KANADI genes during root growth in Arabidopsis thaliana. Sci. Rep. 2017, 7, 3408. [Google Scholar] [CrossRef]
- Li, A.; Zhu, Y.; Tan, X.; Wang, X.; Wei, B.; Guo, H.; Zhang, Z.; Chen, X.; Zhao, G.; Kong, X.; et al. Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol. Biol. 2008, 66, 429–443. [Google Scholar] [CrossRef]
- Wei, X.; Shen, F.; Hong, Y.; Rong, W.; Du, L.; Liu, X.; Xu, H.; Ma, L.; Zhang, Z. The wheat calcium-dependent protein kinase TaCPK7-D positively regulates host resistance to sharp eyespot disease. Mol. Plant Pathol. 2016, 17, 1252–1254. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, P.H.; Vaidyanathan, R.; Gassmann, W.; Schroeder, J.I. Enhancement of Na+ uptake currents, time-dependent inward-rectifying K+ channel currents, and K+ channel transcripts by K+ starvation in wheat root cells. Plant Physiol. 2000, 122, 1387–1397. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Zhao, Y.; Yu, Y.; Sun, R.; Wang, W.; Zhang, S.; Yang, X. Proteomic analysis of roots response to potassium deficiency and the effect of TaHAK1-4A on K+ uptake in wheat. Int. J. Mol. Sci. 2022, 23, 13504. [Google Scholar] [CrossRef] [PubMed]
- Wu, H. Gene Cloning and Functional Verification of Wheat Potassium Transporter Gene TaHAK5. Master’s Thesis, Henan Agricultural University, Henan, China, 2019. [Google Scholar]
- Zhao, L.; Shen, L.; Zhang, W.; Zhang, W.; Wang, Y.; Wu, W. Ca2+-dependent protein kinase11 and 24 modulate the activity of the inward rectifying K+ channels in Arabidopsis pollen tubes. Plant Cell 2013, 25, 649–661. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, R.; Liu, H.; Liu, X.; Xu, K.; Xiao, K.; Zhang, S.; Yang, X.; Xue, C. Multi-Omics analyses reveal the molecular mechanisms underlying the adaptation of wheat (Triticum aestivum L.) to potassium deprivation. Front. Plant Sci. 2020, 11, 588994. [Google Scholar] [CrossRef]
- Guo, Y.; Kong, F.; Xu, Y.; Zhao, Y.; Liang, X.; Wang, Y.; An, D.; Li, S. QTL mapping for seedling traits in wheat grown under varying concentrations of N, P and K nutrients. Theor. Appl. Genet. 2012, 124, 851–865. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Holzberg, S.; Brosio, P.; Gross, C.; Pogue, G.P. Barley stripe mosaic virus-induced gene silencing in a monocot Plant. Plant J. 2002, 30, 315–327. [Google Scholar] [CrossRef]
- Jian, C.; Han, R.; Chi, Q.; Wang, S.; Ma, M.; Liu, X.; Zhao, H. Virus-based microRNA silencing and overexpressing in common wheat (Triticum aestivum L.). Front. Plant Sci. 2017, 8, 500. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, Q.; Axtell, M.J. Quantitating plant microRNA-mediated target repression using a dual-luciferase transient expression system. Methods Mol. Bio. 2015, 1284, 287–303. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, H.; Zhang, Y.; Wang, Y.; Li, M.; Zhang, J.; Duan, L.; Zhang, M.; Li, Z. Increased abscisic acid levels in transgenic maize overexpressing AtLOS5 mediated root ion fluxes and leaf water status under salt stress. J. Exp. Bot. 2016, 67, 1339–1355. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, X.; Chen, M.; Xu, K.; Sun, R.; Zhao, S.; Wu, N.; Zhang, S.; Yang, X.; Xiao, K.; Zhao, Y. The miR166d/TaCPK7-D Signaling Module Is a Critical Mediator of Wheat (Triticum aestivum L.) Tolerance to K+ Deficiency. Int. J. Mol. Sci. 2023, 24, 7926. https://doi.org/10.3390/ijms24097926
Lei X, Chen M, Xu K, Sun R, Zhao S, Wu N, Zhang S, Yang X, Xiao K, Zhao Y. The miR166d/TaCPK7-D Signaling Module Is a Critical Mediator of Wheat (Triticum aestivum L.) Tolerance to K+ Deficiency. International Journal of Molecular Sciences. 2023; 24(9):7926. https://doi.org/10.3390/ijms24097926
Chicago/Turabian StyleLei, Xiaotong, Miaomiao Chen, Ke Xu, Ruoxi Sun, Sihang Zhao, Ningjing Wu, Shuhua Zhang, Xueju Yang, Kai Xiao, and Yong Zhao. 2023. "The miR166d/TaCPK7-D Signaling Module Is a Critical Mediator of Wheat (Triticum aestivum L.) Tolerance to K+ Deficiency" International Journal of Molecular Sciences 24, no. 9: 7926. https://doi.org/10.3390/ijms24097926
APA StyleLei, X., Chen, M., Xu, K., Sun, R., Zhao, S., Wu, N., Zhang, S., Yang, X., Xiao, K., & Zhao, Y. (2023). The miR166d/TaCPK7-D Signaling Module Is a Critical Mediator of Wheat (Triticum aestivum L.) Tolerance to K+ Deficiency. International Journal of Molecular Sciences, 24(9), 7926. https://doi.org/10.3390/ijms24097926



