Proteomic Exploration of Paraoxonase 1 Function in Health and Disease
Abstract
1. Introduction
2. PON1, Lipid Oxidation, and CVD
3. Mechanistic Bases of PON1 Involvement in CVD
3.1. PON1 Controls NO Synthesis
3.2. PON1 Is Not a Redox Protein
3.3. PON1 Interacts with Redox-Related Proteins
3.4. HHcy Diet Exacerbates Pro-Oxidative and Pro-Atherogenic Changes in Mouse Proteome
4. PON1, Lipid Oxidation, and Alzheimer’s Disease
4.1. PON1 and Cognition
4.2. Mechanistic Bases of PON1 Involvement in AD
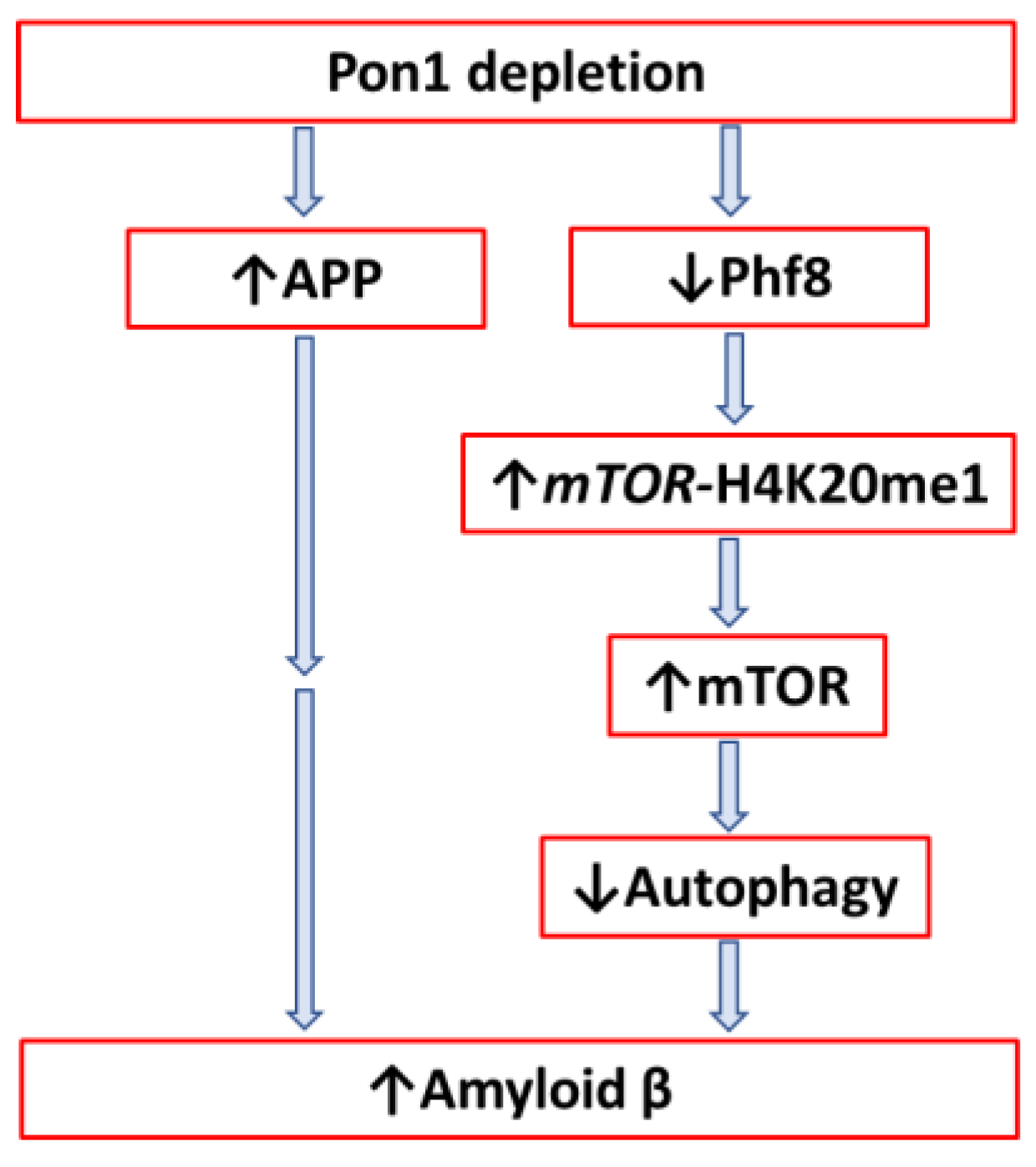
4.2.1. Pon1 Depletion Induces Pro-Neurodegenerative Changes in Mouse Brain Proteome
4.2.2. Pon1 Depletion Induces Accumulation of Amyloid β in Mouse Brain
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mackness, B.; Beltran-Debon, R.; Aragones, G.; Joven, J.; Camps, J.; Mackness, M. Human tissue distribution of paraoxonases 1 and 2 mRNA. IUBMB Life 2010, 62, 480–482. [Google Scholar] [CrossRef]
- Leduc, V.; Legault, V.; Dea, D.; Poirier, J. Normalization of gene expression using SYBR green qPCR: A case for paraoxonase 1 and 2 in Alzheimer’s disease brains. J. Neurosci. Methods 2011, 200, 14–19. [Google Scholar] [CrossRef]
- Witucki, L.; Jakubowski, H. Depletion of Paraoxonase 1 (Pon1) Dysregulates mTOR, Autophagy, and Accelerates Amyloid Beta Accumulation in Mice. Cells 2023, 12, 746. [Google Scholar] [CrossRef] [PubMed]
- Marsillach, J.; Mackness, B.; Mackness, M.; Riu, F.; Beltran, R.; Joven, J.; Camps, J. Immunohistochemical analysis of paraoxonases-1, 2, and 3 expression in normal mouse tissues. Free. Radic. Biol. Med. 2008, 45, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Moren, X.; Lhomme, M.; Bulla, A.; Sanchez, J.-C.; Kontush, A.; James, R.W. Proteomic and lipidomic analyses of paraoxonase defined high density lipoprotein particles: Association of paraoxonase with the anti-coagulant, protein S. Proteom. Clin. Appl. 2016, 10, 230–238. [Google Scholar] [CrossRef]
- Humbert, R.; Adler, D.A.; Disteche, C.M.; Hassett, C.; Omiecinski, C.J.; Furlong, C.E. The molecular basis of the human serum paraoxonase activity polymorphism. Nat. Genet. 1993, 3, 73–76. [Google Scholar] [CrossRef]
- Jakubowski, H.; Ambrosius, W.T.; Pratt, J. Genetic determinants of homocysteine thiolactonase activity in humans: Implications for atherosclerosis. FEBS Lett. 2001, 491, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Loscalzo, J. Paraoxonase and coronary heart disease risk: Language misleads, linkage misinforms, function clarifies. Circ. Cardiovasc. Genet. 2008, 1, 79–80. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, Z.; Xu, Y. HDL and Oxidation. Adv. Exp. Med. Biol. 2022, 1377, 63–77. [Google Scholar]
- Marsillach, J.; Adorni, M.P.; Zimetti, F.; Papotti, B.; Zuliani, G.; Cervellati, C. HDL Proteome and Alzheimer’s Disease: Evidence of a Link. Antioxidants 2020, 9, 1224. [Google Scholar] [CrossRef]
- Jakubowski, H. Calcium-dependent Human Serum Homocysteine Thiolactone Hydrolase. A protective mechanism against protein N-homocysteinylation. J. Biol. Chem. 2000, 275, 3957–3962. [Google Scholar] [CrossRef] [PubMed]
- Borowczyk, K.; Shih, D.M.; Jakubowski, H. Metabolism and Neurotoxicity of Homocysteine Thiolactone in Mice: Evidence for a Protective Role of Paraoxonase 1. J. Alzheimer’s Dis. 2012, 30, 225–231. [Google Scholar] [CrossRef]
- Jakubowski, H.; Goldman, E. Synthesis of homocysteine thiolactone by methionyl-tRNA synthetase in cultured mammalian cells. FEBS Lett. 1993, 317, 237–240. [Google Scholar] [CrossRef]
- Jakubowski, H. Quality control in tRNA charging. Wiley Interdiscip. Rev. RNA 2012, 3, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H. Homocysteine Modification in Protein Structure/Function and Human Disease. Physiol. Rev. 2019, 99, 555–604. [Google Scholar] [CrossRef]
- Borowczyk, K.; Piechocka, J.; Głowacki, R.; Dhar, I.; Midtun, O.; Tell, G.S.; Ueland, P.M.; Nygård, O.; Jakubowski, H. Urinary excretion of homocysteine thiolactone and the risk of acute myocardial infarction in coronary artery disease patients: The WENBIT trial. J. Intern. Med. 2019, 285, 232–244. [Google Scholar] [CrossRef]
- Sauls, D.L.; Lockhart, E.; Warren, M.E.; Lenkowski, A.; Wilhelm, S.E.; Hoffman, M. Modification of Fibrinogen by Homocysteine Thiolactone Increases Resistance to Fibrinolysis: A Potential Mechanism of the Thrombotic Tendency in Hyperhomocysteinemia. Biochemistry 2006, 45, 2480–2487. [Google Scholar] [CrossRef] [PubMed]
- Sikora, M.; Skrzydlewski, P.; Perła-Kaján, J.; Jakubowski, H. Homocysteine thiolactone contributes to the prognostic value of fibrin clot structure/function in coronary artery disease. PLoS ONE 2022, 17, e0275956. [Google Scholar] [CrossRef]
- Lacinski, M.; Skorupski, W.; Cieslinski, A.; Sokolowska, J.; Trzeciak, W.H.; Jakubowski, H. Determinants of homocysteine-thiolactonase activity of the paraoxonase-1 (PON1) protein in humans. Cell Mol. Biol. 2004, 50, 885–893. [Google Scholar]
- Domagala, T.; Łacinski, M.; Trzeciak, W.H.; Mackness, B.; Mackness, M.I.; Jakubowski, H. The correlation of homocysteine-thiolactonase activity of the paraoxonase (PON1) protein with coronary heart disease status. Cell Mol. Biol. 2006, 52, 4–10. [Google Scholar]
- Perla-Kajan, J.; Jakubowski, H. Paraoxonase 1 protects against protein N-homocysteinylation in humans. FASEB J. 2010, 24, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Zimny, J.; Sikora, M.; Guranowski, A.; Jakubowski, H. Protective mechanisms against homocysteine toxicity: The role of bleomycin hydrolase. J. Biol. Chem. 2006, 281, 22485–22492. [Google Scholar] [CrossRef] [PubMed]
- Borowczyk, K.; Tisonczyk, J.; Jakubowski, H. Metabolism and neurotoxicity of homocysteine thiolactone in mice: Protective role of bleomycin hydrolase. Amino Acids 2012, 43, 1339–1348. [Google Scholar] [CrossRef]
- Perła-Kaján, J.; Borowczyk, K.; Głowacki, R.; Nygård, O.; Jakubowski, H. Paraoxonase 1 Q192R genotype and activity affect homocysteine thiolactone levels in humans. FASEB J. 2018, 32, 6019–6024. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.G.; Cole, T.B.; Jarvik, G.P.; Furlong, C.E. Functional genomic of the paraoxonase (PON1) polymorphisms: Effects on pesticide sensitivity, cardiovascular disease, and drug metabolism. Annu. Rev. Med. 2003, 54, 371–392. [Google Scholar] [CrossRef]
- Bhattacharyya, T.; Nicholls, S.J.; Topol, E.J.; Zhang, R.; Yang, X.; Schmitt, D.; Fu, X.; Shao, M.; Brennan, D.M.; Ellis, S.G.; et al. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA 2008, 299, 1265–1276. [Google Scholar] [CrossRef]
- Tang, W.W.; Hartiala, J.; Fan, Y.; Wu, Y.; Stewart, A.F.; Erdmann, J.; Kathiresan, S.; The CARDIoGRAM Consortium; Roberts, R.; McPherson, R.; et al. Clinical and Genetic Association of Serum Paraoxonase and Arylesterase Activities with Cardiovascular Risk. Arter. Thromb. Vasc. Biol. 2012, 32, 2803–2812. [Google Scholar] [CrossRef]
- Gan, K.N.; Smolen, A.; Eckerson, H.W.; La Du, B.N. Purification of human serum paraoxonase/arylesterase. Evidence for one esterase catalyzing both activities. Drug Metab. Dispos. 1991, 19, 100–106. [Google Scholar]
- Ben-David, M.; Elias, M.; Filippi, J.-J.; Duñach, E.; Silman, I.; Sussman, J.L.; Tawfik, D.S. Catalytic Versatility and Backups in Enzyme Active Sites: The Case of Serum Paraoxonase 1. J. Mol. Biol. 2012, 418, 181–196. [Google Scholar] [CrossRef]
- Khersonsky, O.; Tawfik, D.S. Structure−Reactivity Studies of Serum Paraoxonase PON1 Suggest that Its Native Activity Is Lactonase. Biochemistry 2005, 44, 6371–6382. [Google Scholar] [CrossRef]
- Draganov, D.I.; Teiber, J.F.; Speelman, A.; Osawa, Y.; Sunahara, R.; La Du, B.N. Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. J. Lipid Res. 2005, 46, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, C.J.; Lamichhane, S.; Connolly, J.A.; Soehnlen, S.M.; Khalaf, F.K.; Malhotra, D.; Haller, S.T.; Isailovic, D.; Kennedy, D.J. A PON for All Seasons: Comparing Paraoxonase Enzyme Substrates, Activity and Action including the Role of PON3 in Health and Disease. Antioxidants 2022, 11, 590. [Google Scholar] [CrossRef]
- Eryanni-Levin, S.; Khatib, S.; Levy-Rosenzvig, R.; Tamir, S.; Szuchman-Sapir, A. 5,6-δ-DHTL, a stable metabolite of arachidonic acid, is a potential substrate for paraoxonase 1. Biochim. Et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2015, 1851, 1118–1122. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.A.S.; Khatib, S.; Szuchman-Sapir, A. Fishing for lipid lactones using selective reaction and characteristic fragmentation pattern. J. Chromatogr. B 2022, 1197, 123201. [Google Scholar] [CrossRef]
- Costa, L.G.; Giordano, G.; Cole, T.B.; Marsillach, J.; Furlong, C.E. Paraoxonase 1 (PON1) as a genetic determinant of susceptibility to organophosphate toxicity. Toxicology 2013, 307, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Shih, D.M.; Gu, L.; Xia, Y.-R.; Navab, M.; Li, W.-F.; Hama, S.; Castellani, L.W.; Furlong, C.E.; Costa, L.G.; Fogelman, A.M.; et al. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature 1998, 394, 284–287. [Google Scholar] [CrossRef]
- Shih, D.M.; Xia, Y.-R.; Wang, X.-P.; Miller, E.; Castellani, L.W.; Subbanagounder, G.; Cheroutre, H.; Faull, K.F.; Berliner, J.A.; Witztum, J.L.; et al. Combined Serum Paraoxonase Knockout/Apolipoprotein E Knockout Mice Exhibit Increased Lipoprotein Oxidation and Atherosclerosis. J. Biol. Chem. 2000, 275, 17527–17535. [Google Scholar] [CrossRef]
- Hong, C.G.; Florida, E.; Li, H.; Parel, P.M.; Mehta, N.N.; Sorokin, A.V. Oxidized low-density lipoprotein associates with cardiovascular disease by a vicious cycle of atherosclerosis and inflammation: A systematic review and meta-analysis. Front. Cardiovasc. Med. 2022, 9, 1023651. [Google Scholar] [CrossRef] [PubMed]
- Mackness, B.; Quarck, R.; Verreth, W.; Mackness, M.; Holvoet, P. Human Paraoxonase-1 Overexpression Inhibits Atherosclerosis in a Mouse Model of Metabolic Syndrome. Arter. Thromb. Vasc. Biol. 2006, 26, 1545–1550. [Google Scholar] [CrossRef]
- She, Z.-G.; Zheng, W.; Wei, Y.-S.; Chen, H.-Z.; Wang, A.-B.; Li, H.-L.; Liu, G.; Zhang, R.; Liu, J.-J.; Stallcup, W.B.; et al. Human Paraoxonase Gene Cluster Transgenic Overexpression Represses Atherogenesis and Promotes Atherosclerotic Plaque Stability in ApoE-Null Mice. Circ. Res. 2009, 104, 1160–1168. [Google Scholar] [CrossRef]
- Kresanov, P.; Vasankari, T.; Ahotupa, M.; Kaikkonen, J.; Hutri-Kähönen, N.; Juonala, M.; Kähönen, M.; Lehtimäki, T.; Viikari, J.; Raitakari, O.T. Paraoxonase-1 and oxidized lipoprotein lipids. The Cardiovascular Risk in Young Finns Study. Atherosclerosis 2015, 241, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Ahotupa, M.; Marniemi, J.; Lehtimäki, T.; Talvinen, K.; Raitakari, O.T.; Vasankari, T.; Viikari, J.; Luoma, J.; Ylä-Herttuala, S. Baseline Diene Conjugation in LDL Lipids as a Direct Measure of In Vivo LDL Oxidation. Clin. Biochem. 1998, 31, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Esterbauer, H.; Striegl, G.; Puhl, H.; Rotheneder, M. Continuous monitoring of in vitro oxidation of human low density lipoprotein. Free. Radic. Res. Commun. 1989, 6, 67–75. [Google Scholar] [CrossRef]
- Breton, C.V.; Yin, F.; Wang, X.; Avol, E.; Gilliland, F.D.; Araujo, J.A. HDL anti-oxidant function associates with LDL level in young adults. Atherosclerosis 2014, 232, 165–170. [Google Scholar] [CrossRef]
- Kuchta, A.; Strzelecki, A.; Ćwiklińska, A.; Totoń, M.; Gruchała, M.; Zdrojewski, Z.; Kortas-Stempak, B.; Gliwińska, A.; Dąbkowski, K.; Jankowski, M. PON-1 Activity and Plasma 8-Isoprostane Concentration in Patients with Angiographically Proven Coronary Artery Disease. Oxidative Med. Cell Longev. 2015, 2016, 5136937. [Google Scholar] [CrossRef]
- Kunutsor, S.K.; Bakker, S.J.; James, R.W.; Dullaart, R.P. Serum paraoxonase-1 activity and risk of incident cardiovascular disease: The PREVEND study and meta-analysis of prospective population studies. Atherosclerosis 2016, 245, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Zuin, M.; Trentini, A.; Marsillach, J.; D’amuri, A.; Bosi, C.; Roncon, L.; Passaro, A.; Zuliani, G.; Mackness, M.; Cervellati, C. Paraoxonase-1 (PON-1) Arylesterase Activity Levels in Patients with Coronary Artery Disease: A Meta-Analysis. Dis. Markers 2022, 2022, 4264314. [Google Scholar] [CrossRef]
- Giusti, B.; Saracini, C.; Bolli, P.; Magi, A.; Sestini, I.; Sticchi, E.; Pratesi, G.; Pulli, R.; Pratesi, C.; Abbate, R. Genetic analysis of 56 polymorphisms in 17 genes involved in methionine metabolism in patients with abdominal aortic aneurysm. J. Med. Genet. 2008, 45, 721–730. [Google Scholar] [CrossRef]
- McCormick, M.L.; Gavrila, D.; Weintraub, N.L. Role of Oxidative Stress in the Pathogenesis of Abdominal Aortic Aneurysms. Arter. Thromb. Vasc. Biol. 2007, 27, 461–469. [Google Scholar] [CrossRef]
- Shih, D.M.; Lusis, A.J. The roles of PON1 and PON2 in cardiovascular disease and innate immunity. Curr. Opin. Infect. Dis. 2009, 20, 288–292. [Google Scholar] [CrossRef]
- Grzegorzewska, A.E.; Adamska, P.; Iwanczyk-Skalska, E.; Ostromecka, K.; Niepolski, L.; Marcinkowski, W.; Mostowska, A.; Warchol, W.; Zaba, C.; Jagodzinski, P.P. Paraoxonase 1 concerning dyslipidaemia, cardiovascular diseases, and mortality in haemodialysis patients. Sci. Rep. 2021, 11, 6773. [Google Scholar] [CrossRef]
- Grzegorzewska, A.E.; Ostromecka, K.; Świderska, M.K.; Adamska, P.; Mostowska, A.; Jagodziński, P.P. Paraoxonase 1 gene variants concerning cardiovascular mortality in conventional cigarette smokers and non-smokers treated with hemodialysis. Sci. Rep. 2021, 11, 19467. [Google Scholar] [CrossRef] [PubMed]
- Yuhanna, I.S.; Zhu, Y.; Cox, B.E.; Hahner, L.D.; Osborne-Lawrence, S.; Lu, P.; Marcel, Y.L.; Anderson, R.G.; Mendelsohn, M.E.; Hobbs, H.H.; et al. High-density lipoprotein binding to scavenger receptor-BI activates endothelial nitric oxide synthase. Nat. Med. 2001, 7, 853–857. [Google Scholar] [CrossRef] [PubMed]
- Trigatti, B.L.; Fuller, M. HDL signaling and protection against coronary artery atherosclerosis in mice. J. Biomed. Res. 2016, 30, 94–100. [Google Scholar] [PubMed]
- Seetharam, D.; Mineo, C.; Gormley, A.K.; Gibson, L.L.; Vongpatanasin, W.; Chambliss, K.L.; Hahner, L.D.; Cummings, M.L.; Kitchens, R.L.; Marcel, Y.L.; et al. High-Density Lipoprotein Promotes Endothelial Cell Migration and Reendothelialization via Scavenger Receptor-B Type I. Circ. Res. 2006, 98, 63–72. [Google Scholar] [CrossRef]
- Besler, C.; Heinrich, K.; Rohrer, L.; Doerries, C.; Riwanto, M.; Shih, D.M.; Chroni, A.; Yonekawa, K.; Stein, S.; Schaefer, N.; et al. Mechanisms underlying adverse effects of HDL on eNOS-activating pathways in patients with coronary artery disease. J. Clin. Investig. 2011, 121, 2693–2708. [Google Scholar] [CrossRef]
- Mackness, M.I.; Durrington, P.N.; Mackness, B. The role of paraoxonase 1 activity in cardiovascular disease: Potential for therapeutic intervention. Am. J. Cardiovasc. Drugs 2004, 4, 211–217. [Google Scholar] [CrossRef]
- Mackness, M.I.; Arrol, S.; Durrington, P.N. Paraoxonase prevents accumulation of lipoperoxides in low-density lipoprotein. FEBS Lett. 1991, 286, 152–154. [Google Scholar] [CrossRef]
- Watson, A.D.; Berliner, J.A.; Hama, S.Y.; La Du, B.N.; Faull, K.F.; Fogelman, A.M.; Navab, M. Protective effect of high density lipoprotein associated paraoxonase. Inhibition of the biological activity of minimally oxidized low density lipoprotein. J. Clin. Investig. 1995, 96, 2882–2891. [Google Scholar] [CrossRef]
- Aviram, M.; Rosenblat, M.; Bisgaier, C.L.; Newton, R.S.; Primo-Parmo, S.L.; La Du, B.N. Paraoxonase inhibits high-density lipoprotein oxidation and preserves its functions. A possible peroxidative role for paraoxonase. J. Clin. Investig. 1998, 101, 1581–1590. [Google Scholar] [CrossRef]
- Aviram, M.; Billecke, S.; Sorenson, R.; Bisgaier, C.; Newton, R.; Rosenblat, M.; Erogul, J.; Hsu, C.; Dunlop, C.; La Du, B. Paraoxonase active site required for protection against LDL oxidation involves its free sulfhydryl group and is different from that required for its arylesterase/paraoxonase activities: Selective action of human paraoxonase allozymes Q and R. Arter. Thromb. Vasc. Biol. 1998, 18, 1617–1624. [Google Scholar] [CrossRef]
- Bayrak, A.; Bayrak, T.; Bodur, E.; Kılınç, K.; Demirpençe, E. The effect of HDL-bound and free PON1 on copper-induced LDL oxidation. Chem. Interact. 2016, 257, 141–146. [Google Scholar] [CrossRef]
- Brophy, V.H.; Jampsa, R.L.; Clendenning, J.B.; McKinstry, L.A.; Jarvik, G.P.; Furlong, C.E. Effects of 5′ Regulatory-Region Polymorphisms on Paraoxonase-Gene (PON1) Expression. Am. J. Hum. Genet. 2001, 68, 1428–1436. [Google Scholar] [CrossRef] [PubMed]
- Sikora, M.; Bretes, E.; Perła-Kaján, J.; Lewandowska, I.; Marczak, L.; Jakubowski, H. Genetic Attenuation of Paraoxonase 1 Activity Induces Proatherogenic Changes in Plasma Proteomes of Mice and Humans. Antioxidants 2020, 9, 1198. [Google Scholar] [CrossRef]
- Marathe, G.K.; Zimmerman, G.A.; McIntyre, T.M. Platelet-activating Factor Acetylhydrolase, and Not Paraoxonase-1, Is the Oxidized Phospholipid Hydrolase of High Density Lipoprotein Particles. J. Biol. Chem. 2003, 278, 3937–3947. [Google Scholar] [CrossRef]
- Teiber, J.F.; Draganov, D.I.; La Du, B.N. Purified human serum PON1 does not protect LDL against oxidation in the in vitro assays initiated with copper or AAPH. J. Lipid Res. 2004, 45, 2260–2268. [Google Scholar] [CrossRef]
- Connelly, P.W.; Draganov, A.; Maguire, G.F. Paraoxonase-1 does not reduce or modify oxidation of phospholipids by peroxynitrite. Free. Radic. Biol. Med. 2005, 38, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Carlson, C.S.; Heagerty, P.J.; Hatsukami, T.S.; Richter, R.J.; Ranchalis, J.; Lewis, J.; Bacus, T.J.; McKinstry, L.A.; Schellenberg, G.D.; Rieder, M.; et al. TagSNP analyses of the PON gene cluster: Effects on PON1 activity, LDL oxidative susceptibility, and vascular disease. J. Lipid Res. 2006, 47, 1014–1024. [Google Scholar] [CrossRef]
- Tanzi, R.E. The genetics of Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006296. [Google Scholar] [CrossRef]
- Sikora, M.; Jakubowski, H. Changes in redox plasma proteome of Pon1−/− mice are exacerbated by a hyperhomocysteinemic diet. Free Radic. Biol. Med. 2021, 169, 169–180. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, J.C. Alzheimer disease as a vascular disorder: Nosological evidence. Stroke 2002, 33, 1152–1162. [Google Scholar] [CrossRef]
- Stampfer, M.J. Cardiovascular disease and Alzheimer’s disease: Common links. J. Intern. Med. 2006, 260, 211–223. [Google Scholar] [CrossRef]
- Menini, T.; Gugliucci, A. Paraoxonase 1 in neurological disorders. Redox Rep. 2013, 19, 49–58. [Google Scholar] [CrossRef]
- Erlich, P.M.; Lunetta, K.L.; Cupples, L.A.; Abraham, C.R.; Green, R.C.; Baldwin, C.T.; Farrer, L.A. Serum paraoxonase activity is associated with variants in the PON gene cluster and risk of Alzheimer disease. Neurobiol. Aging 2012, 33, 1015.e7–1015.e23. [Google Scholar] [CrossRef] [PubMed]
- Bednarska-Makaruk, M.E.; Krzywkowski, T.; Graban, A.; Lipczyńska-Łojkowska, W.; Bochyńska, A.; Rodo, M.; Wehr, H.; Ryglewicz, D.K. Original article Paraoxonase 1 (PON1) gene -108C>T and p.Q192R polymorphisms and arylesterase activity of the enzyme in patients with dementia. Folia Neuropathol. 2013, 2, 111–119. [Google Scholar] [CrossRef]
- Dantoine, T.F.; Debord, J.; Merle, L.; Lacroix-Ramiandrisoa, H.; Bourzeix, L.; Charmes, J.P. Paraoxonase 1 activity: A new vascular marker of dementia? Ann. N. Y. Acad. Sci. 2002, 977, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Paragh, G.; Balla, P.; Katona, E.; Seres, I.; Egerhazi, A.; Degrell, I. Serum paraoxonase activity changes in patients with Alzheimer’s disease and vascular dementia. Eur. Arch. Psychiatry Clin. Neurosci. 2002, 252, 63–67. [Google Scholar] [CrossRef]
- Bednarz-Misa, I.; Berdowska, I.; Zboch, M.; Misiak, B.; Zieliński, B.; Płaczkowska, S.; Fleszar, M.; Wiśniewski, J.; Gamian, A.; Krzystek-Korpacka, M. Paraoxonase 1 decline and lipid peroxidation rise reflect a degree of brain atrophy and vascular impairment in dementia. Adv. Clin. Exp. Med. 2020, 29, 71–78. [Google Scholar] [CrossRef]
- Perla-Kajan, J.; Wloczkowska, O.; Ziola-Frankowska, A.; Frankowski, M.; Smith, A.D.; de Jager, C.A.; Refsum, H.; Jakubowski, H. Paraoxonase 1, B Vitamins Supplementation, and Mild Cognitive Impairment. J. Alzheimer’s Dis. 2021, 81, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- Narasimhulu, C.A.; Mitra, C.; Bhardwaj, D.; Burge, K.Y.; Parthasarathy, S. Alzheimer’s Disease Markers in Aged ApoE-PON1 Deficient Mice. J. Alzheimer’s Dis. 2019, 67, 1353–1365. [Google Scholar] [CrossRef]
- Salazar, J.G.; Marsillach, J.; Reverte, I.; Mackness, B.; Mackness, M.; Joven, J.; Camps, J.; Colomina, M.T. Paraoxonase-1 and -3 Protein Expression in the Brain of the Tg2576 Mouse Model of Alzheimer’s Disease. Antioxidants 2021, 10, 339. [Google Scholar] [CrossRef]
- Li, H.; Wetten, S.; Li, L.; Jean, P.L.S.; Upmanyu, R.; Surh, L.; Hosford, D.; Barnes, M.R.; Briley, J.D.; Borrie, M.; et al. Candidate Single-Nucleotide Polymorphisms From a Genomewide Association Study of Alzheimer Disease. Arch. Neurol. 2008, 65, 45–53. [Google Scholar] [CrossRef]
- Leduc, V.; Théroux, L.; Dea, D.; Robitaille, Y.; Poirier, J. Involvement of paraoxonase 1 genetic variants in Alzheimer’s disease neuropathology. Eur. J. Neurosci. 2009, 30, 1823–1830. [Google Scholar] [CrossRef]
- Erlich, P.M.; Lunetta, K.L.; Cupples, L.A.; Huyck, M.; Green, R.C.; Baldwin, C.T.; Farrer, L.A.; Auerbach, S.; Akomolafe, A.; Griffith, P.; et al. Polymorphisms in the PON gene cluster are associated with Alzheimer disease. Hum. Mol. Genet. 2006, 15, 77–85. [Google Scholar] [CrossRef]
- Chapuis, J.; Boscher, M.; Bensemain, F.; Cottel, D.; Amouyel, P.; Lambert, J.C. Association study of the paraoxonase 1 gene with the risk of developing Alzheimer’s disease. Neurobiol. Aging 2009, 30, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Luo, D.; Yang, M.; Wang, Y.; Xiong, L.; Gao, L.; Liu, Y.; Liu, H. A Meta-Analysis on the Relationship of the PON Genes and Alzheimer Disease. J. Geriatr. Psychiatry Neurol. 2017, 30, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Bacchetti, T.; Vignini, A.; Giulietti, A.; Nanetti, L.; Provinciali, L.; Luzzi, S.; Mazzanti, L.; Ferretti, G. Higher Levels of Oxidized Low Density Lipoproteins in Alzheimer’s Disease Patients: Roles for Platelet Activating Factor Acetyl Hydrolase and Paraoxonase-1. J. Alzheimer’s Dis. 2015, 46, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Cagnin, A.; Leon, A.; Vianello, D.; Colavito, D.; Favaretto, S.; Zarantonello, G.; Stecca, A.; Ermani, M.; Zambon, A. LDL density and oxidation are modulated by PON1 promoter genotype in patients with Alzheimer’s disease. J. Alzheimer’s Dis. 2013, 34, 377–385. [Google Scholar] [CrossRef]
- Gauthier, S.; Reisberg, B.; Zaudig, M.; Petersen, R.C.; Ritchie, K.; Broich, K.; Belleville, S.; Brodaty, H.; Bennett, D.; Chertkow, H.; et al. International Psychogeriatric Association Expert Conference on mild cognitive, i., Mild cognitive impairment. Lancet 2006, 367, 1262–1270. [Google Scholar] [CrossRef]
- Graham, J.E.; Rockwood, K.; Beattie, B.L.; Eastwood, R.; Gauthier, S.; Tuokko, H.; McDowell, I. Prevalence and severity of cognitive impairment with and without dementia in an elderly population. Lancet 1997, 349, 1793–1796. [Google Scholar] [CrossRef]
- Petersen, R.C.; Roberts, R.O.; Knopman, D.S.; Boeve, B.F.; Geda, Y.E.; Ivnik, R.J.; Smith, G.E.; Jack, C.R., Jr. Mild cognitive impairment: Ten years later. Arch. Neurol. 2009, 66, 1447–1455. [Google Scholar] [CrossRef]
- Wehr, H.; Bednarska-Makaruk, M.; Graban, A.; Lipczynska-Lojkowska, W.; Rodo, M.; Bochynska, A.; Ryglewicz, D. Paraoxonase activity and dementia. J. Neurol. Sci. 2009, 283, 107–108. [Google Scholar] [CrossRef]
- Cervellati, C.; Trentini, A.; Romani, A.; Bellini, T.; Bosi, C.; Ortolani, B.; Zurlo, A.; Passaro, A.; Seripa, D.; Zuliani, G. Serum paraoxonase and arylesterase activities of paraoxonase-1 (PON-1), mild cognitive impairment, and 2-year conversion to dementia: A pilot study. J. Neurochem. 2015, 135, 395–401. [Google Scholar] [CrossRef]
- Jakubowski, H.; Zioła-Frankowska, A.; Frankowski, M.; Perła-Kaján, J.; Refsum, H.; de Jager, C.A.; Smith, A.D. B Vitamins Prevent Iron-Associated Brain Atrophy and Domain-Specific Effects of Iron, Copper, Aluminum, and Silicon on Cognition in Mild Cognitive Impairment. J. Alzheimer’s Dis. 2021, 84, 1039–1055. [Google Scholar] [CrossRef]
- Wloczkowska, O.; Perla-Kajan, J.; Smith, A.D.; de Jager, C.; Refsum, H.; Jakubowski, H. Anti-N-homocysteine-protein autoantibodies are associated with impaired cognition. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2021, 7, e12159. [Google Scholar] [CrossRef]
- Hayden, K.M.; Norton, M.C.; Darcey, D.; Ostbye, T.; Zandi, P.P.; Breitner, J.C.S.; Welsh-Bohmer, K.A. Occupational exposure to pesticides increases the risk of incident AD: The Cache County Study. Neurology 2010, 74, 1524–1530. [Google Scholar] [CrossRef] [PubMed]
- Jones, N. Alzheimer disease: Risk of dementia and Alzheimer disease increases with occupational pesticide exposure. Nat. Rev. Neurol. 2010, 6, 353. [Google Scholar] [CrossRef] [PubMed]
- Suszyńska-Zajczyk, J.; Łuczak, M.; Marczak, L.; Jakubowski, H. Inactivation of the Paraoxonase 1 Gene Affects the Expression of Mouse Brain Proteins Involved in Neurodegeneration. J. Alzheimer’s Dis. 2014, 42, 247–260. [Google Scholar] [CrossRef]
- Blatter, M.-C.; James, R.W.; Messmer, S.; Barja, F.; Pometta, D. Identification of a distinct human high-density lipoprotein subspecies defined by a lipoprotein-associated protein, K-45. Identity of K-45 with paraoxonase. JBIC J. Biol. Inorg. Chem. 1993, 211, 871–879. [Google Scholar] [CrossRef]
- Oakley, H.; Cole, S.L.; Logan, S.; Maus, E.; Shao, P.; Craft, J.; Guillozet-Bongaarts, A.; Ohno, M.; Disterhoft, J.; Van Eldik, L.; et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J. Neurosci. 2006, 26, 10129–10140. [Google Scholar] [CrossRef] [PubMed]
- Khayati, K.; Antikainen, H.; Bonder, E.M.; Weber, G.F.; Kruger, W.D.; Jakubowski, H.; Dobrowolski, R. The amino acid metabolite homocysteine activates mTORC1 to inhibit autophagy and form abnormal proteins in human neurons and mice. FASEB J. 2017, 31, 598–609. [Google Scholar] [CrossRef]
- Yates, S.C.; Zafar, A.; Hubbard, P.; Nagy, S.; Durant, S.; Bicknell, R.; Wilcock, G.; Christie, S.; Esiri, M.M.; Smith, A.D.; et al. Dysfunction of the mTOR pathway is a risk factor for Alzheimer’s disease. Acta Neuropathol. Commun. 2013, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, S.; Zhou, Y.; Han, Y.; Li, S.; Xu, Q.; Xu, L.; Zhu, Z.; Deng, Y.; Yu, L.; et al. Phf8 histone demethylase deficiency causes cognitive impairments through the mTOR pathway. Nat. Commun. 2018, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Witucki, Ł.; Jakubowski, H. Homocysteine Metabolites Inhibit Autophagy and Elevate Amyloid Beta by Impairing Phf8/H4K20me1-dependent Epigenetic Regulation of mTOR in Cystathionine β-Synthase-Deficient Mice. bioRxiv. [CrossRef]
- Jakubowski, H.; Perła-Kaján, J.; Finnell, R.H.; Cabrera, R.M.; Wang, H.; Gupta, S.; Kruger, W.D.; Kraus, J.P.; Shih, D.M. Genetic or nutritional disorders in homocysteine or folate metabolism increase protein N-homocysteinylation in mice. FASEB J. 2009, 23, 1721–1727. [Google Scholar] [CrossRef]
- Smith, A.D.; Refsum, H. Homocysteine—From disease biomarker to disease prevention. J. Intern. Med. 2021, 290, 826–854. [Google Scholar] [CrossRef] [PubMed]
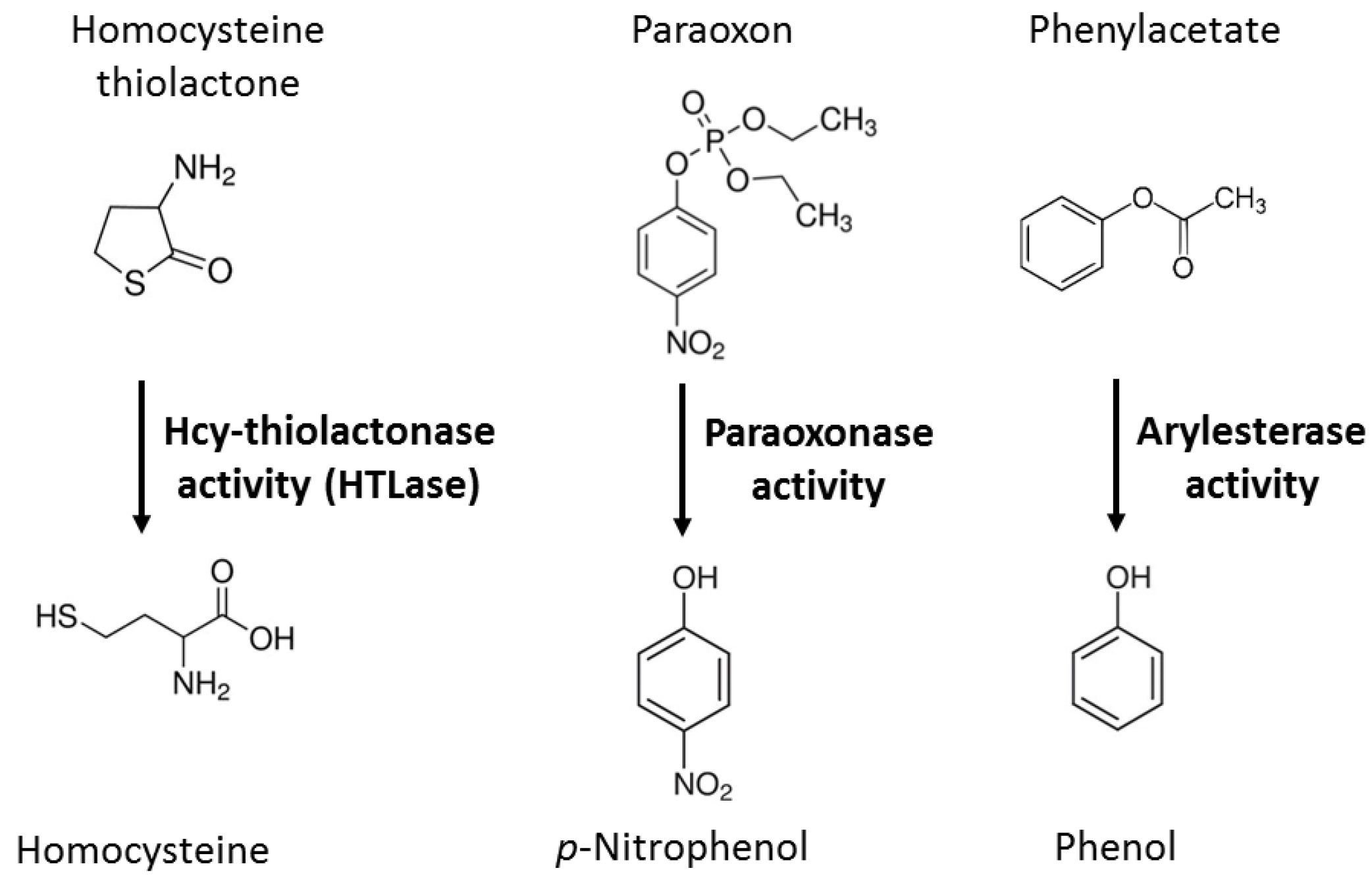
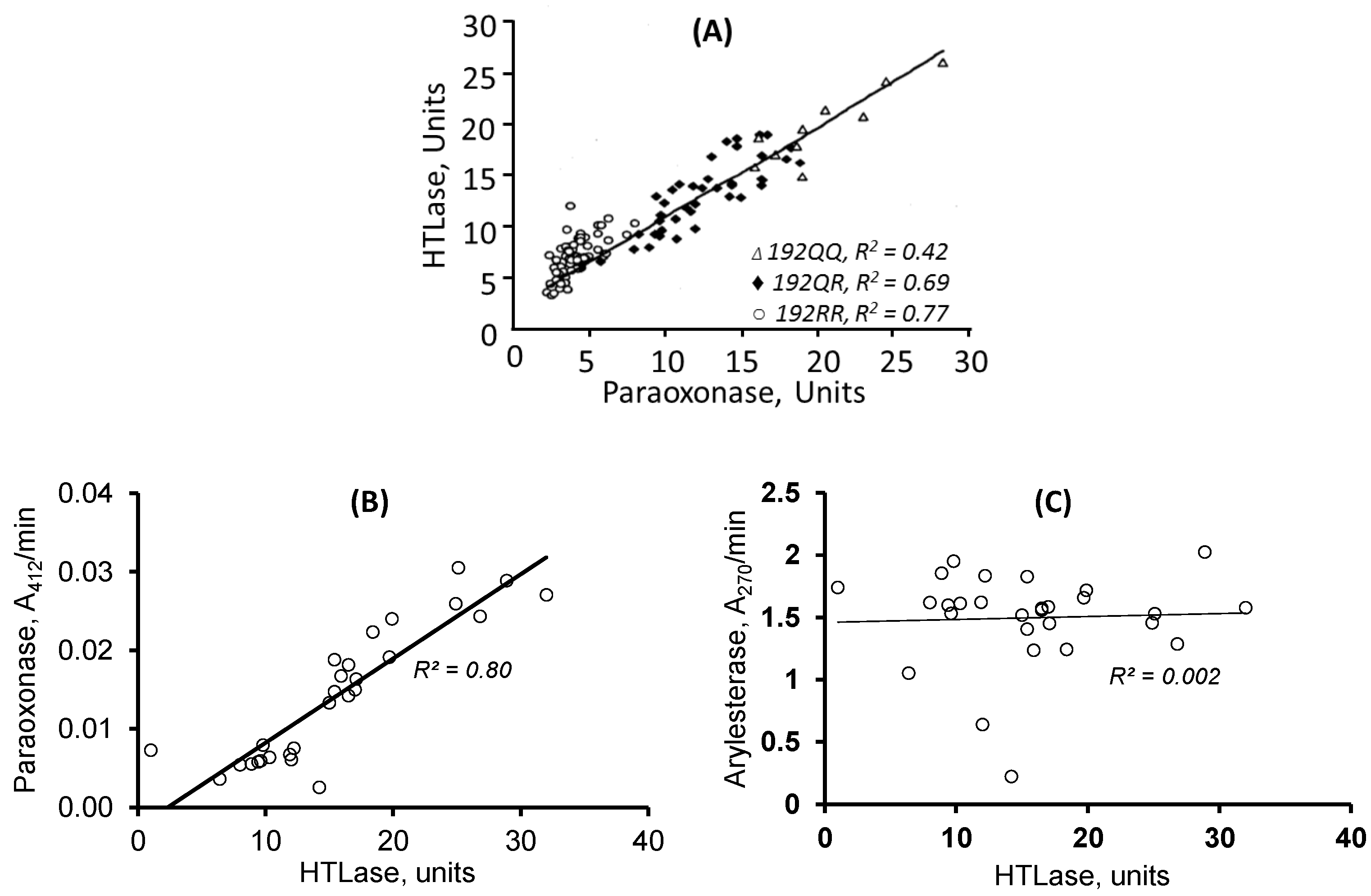

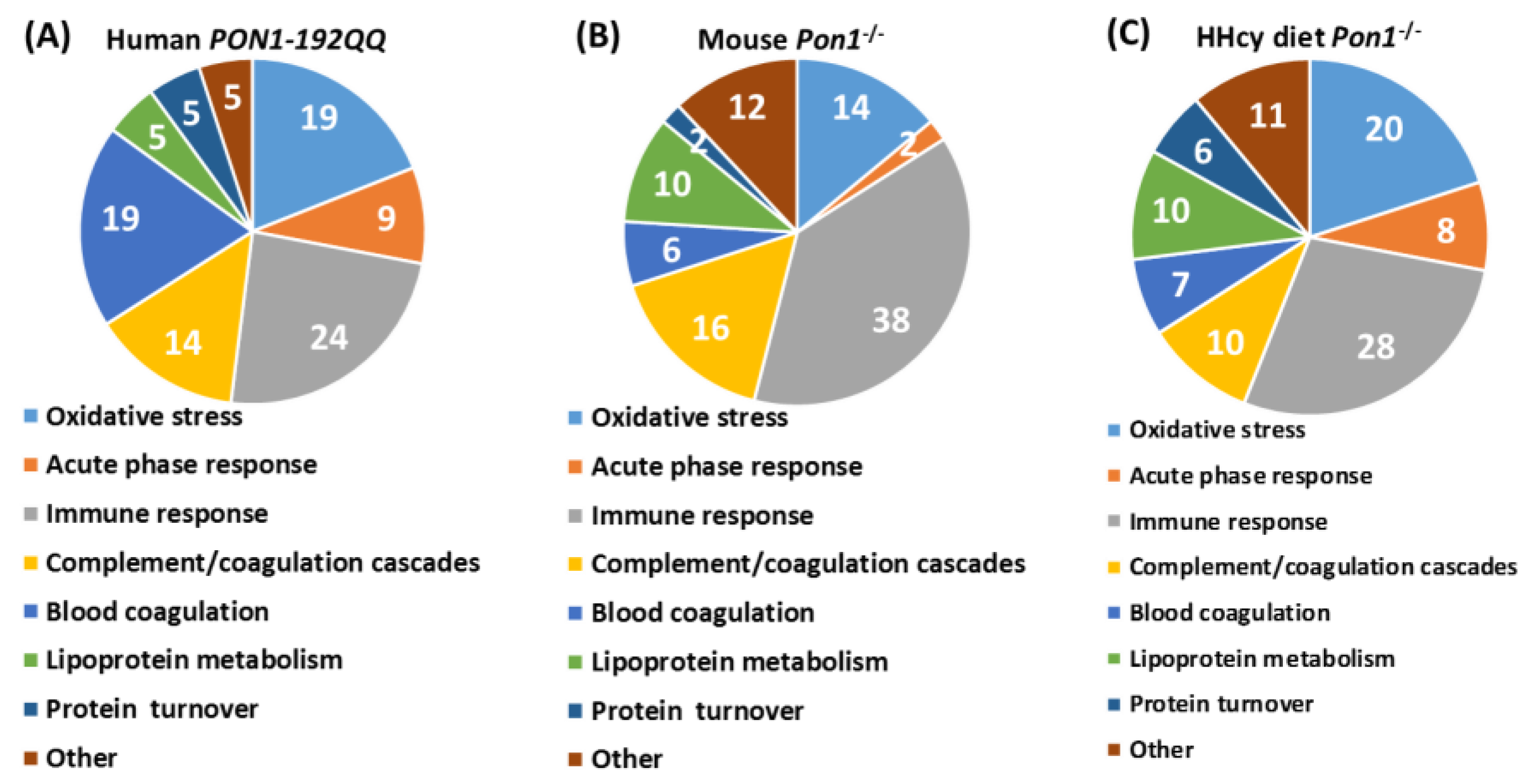

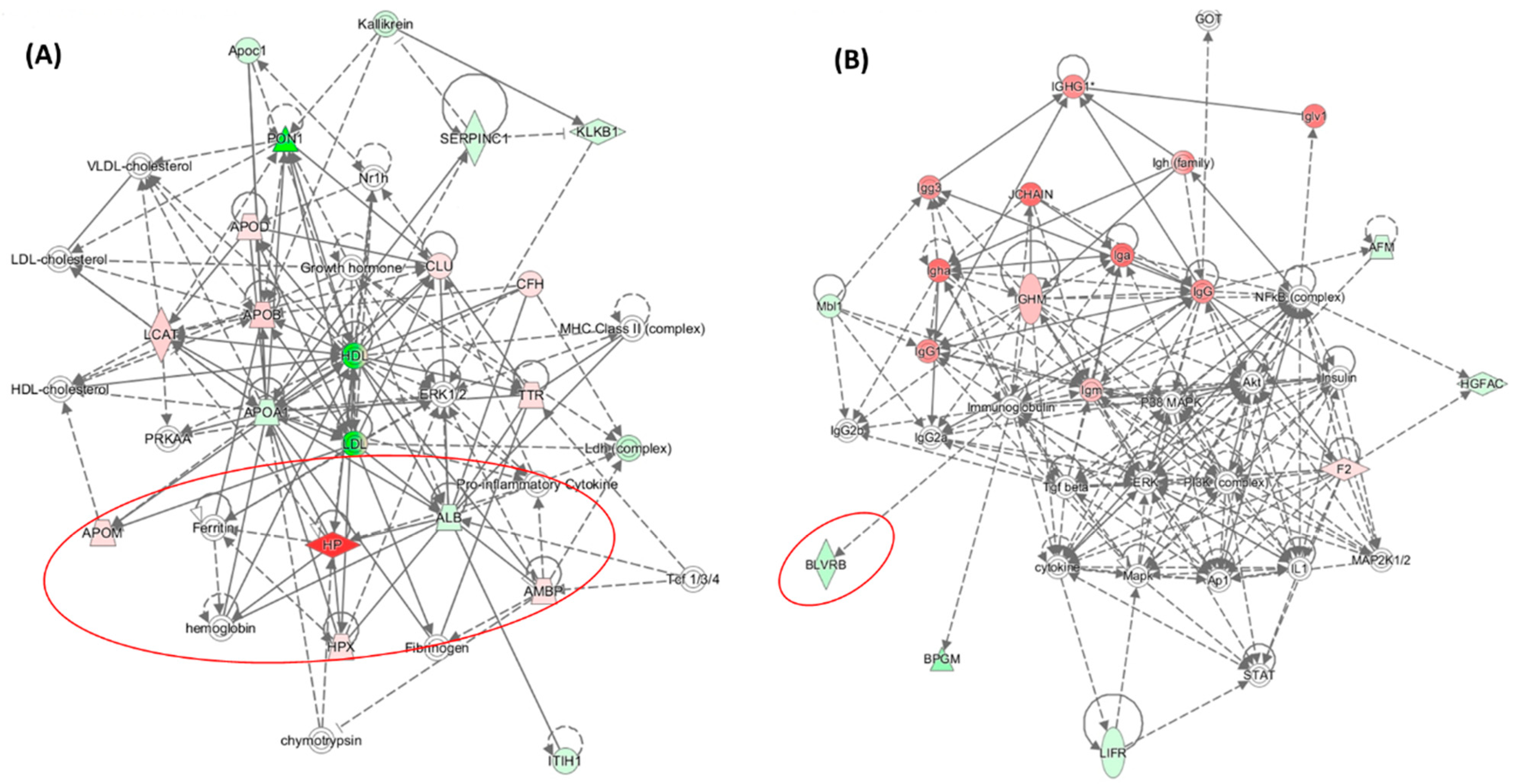
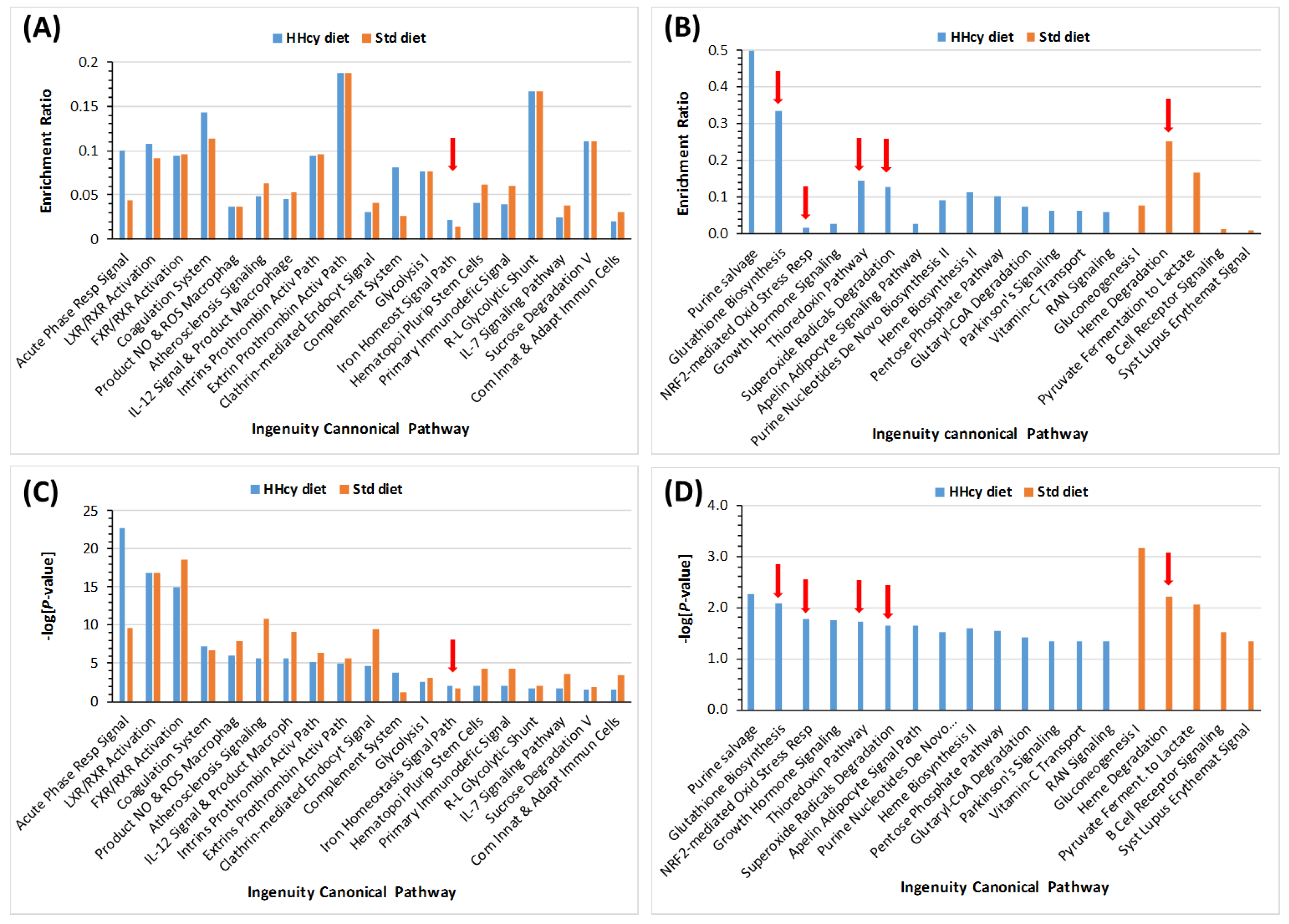
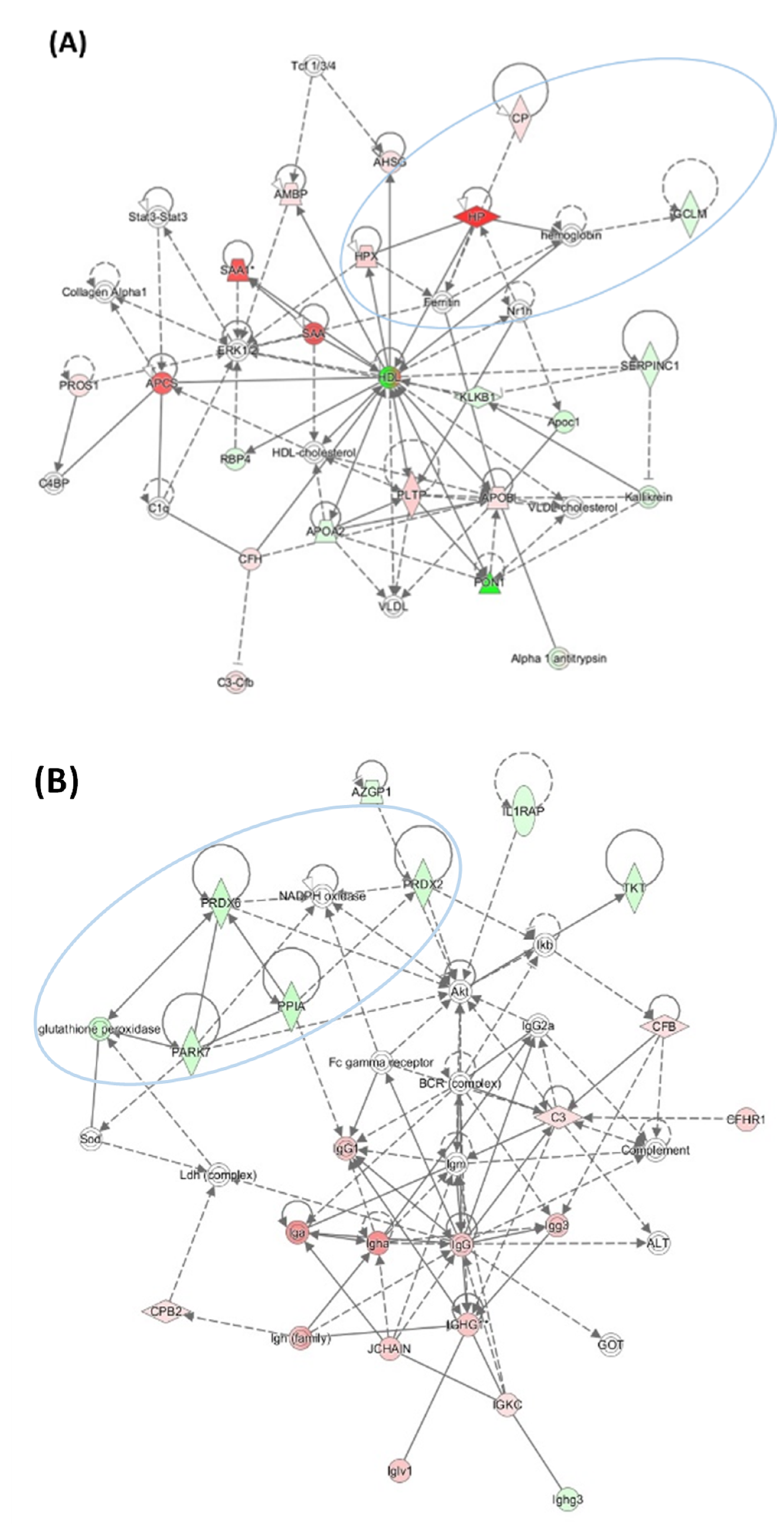
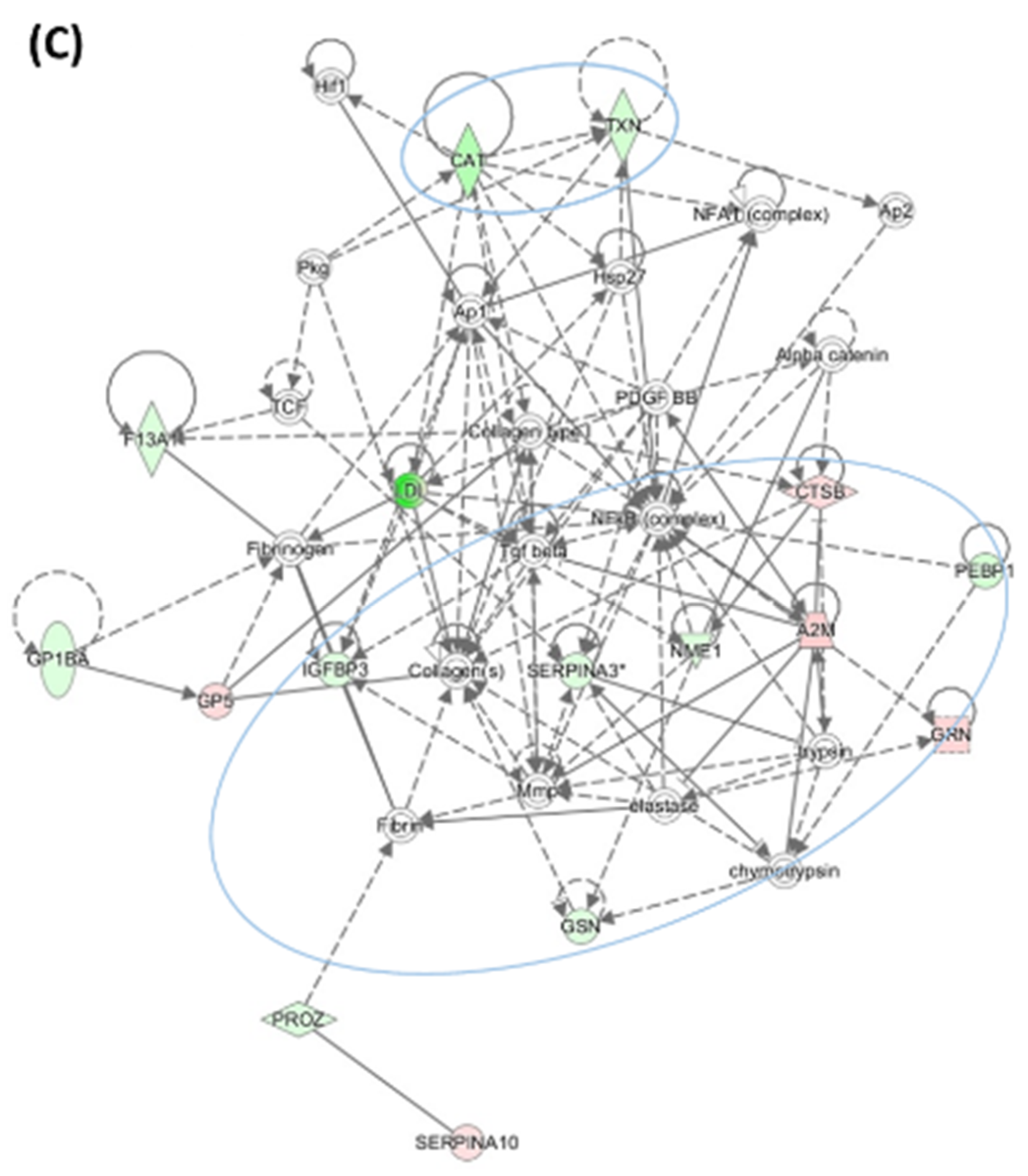
| Unique to Mice (n = 41) | Unique to Humans (n = 12) | Proteins Affected Both in Mice and Humans (n = 9) # |
|---|---|---|
| Oxidative stress † (n = 4): ↓Alb $, ↓Blvrb, ↑α-1-microglubulin (Ambp), ↑Hemopexin (Hpx) | Oxidative stress † (n = 1): ↑Glutathione peroxidase 3 (GPX3) | Oxidative stress † (n = 3): ↑APOD↑, ↑APOM ‡↑, ↑haptoglobin (HP)↓ |
| Immune response (n = 18): ↑Igh (n = 9), ↑Igj, ↑Igk (n = 6), ↑Igl (n = 2) | Immune response (n = 4): ↑CFP, ↓N/A, ↑PGLYRP2, ↑V2-6 (IGL) | Immune response (n = 1): ↓IGHG3↑ |
| Acute phase response (n = 5): ↑Ahsg, ↑Orm1, ↑Orm2, ↑Saa1, ↑Saa2 | Acute phase response (n = 1): ↑Ttr | Acute phase response (n = 1): ↑Ambp |
| Complement/coagulation (n = 7): ↑Al182371, ↑Cfh, ↑Clu $, ↑F2 (prothrombin), ↓Klkb1, ↓Mbl1; ↓Serpinc1 (antithrombin III) | Complement/coagulation (n = 2): ↑C9, ↑V2-17 (IGL) | |
| Blood coagulation (n = 2): ↑Hrg ‡, ↓Itih1 | Blood coagulation (n = 3): ↓PLG, ↓SERPINA10, ↓VTN | Blood coagulation (n = 1): ↓F13B↓ |
| Lipoprotein/lipid metabolism (n = 5): ↓ApoA2, ↓ApoC2, ↓Azgp1, ↓Pgp, ↑Pltp | Lipoprotein metabolism (n = 4): ↓ApoA1, ↑ApoB, ↓ApoC1, ↓Pon1 | |
| Protein turnover (n = 1): ↑Mug1 | Protein turnover (n = 1): ↑FETUB↓ | |
| Other (n = 6): ↓Afm, ↓Aldoa, ↓Bpgm, ↓Ica, ↓Ldha, ↓Lifr | Other (n = 1): ↓RBP4 |
| Unique to HHcy Diet Mice (n = 66) | Unique to Control Diet Mice (n = 27) | Proteins Affected Both in HHcy and Control Diet Mice (n = 23) # |
|---|---|---|
| Oxidative stress (n = 15): ↓Alad, ↑Cp, ↓Gclm, ↓Cat, ↑Ctsb, ↓Gsn, ↑Grn, ↓Prdx2 #, ↓Prdx6, ↓Txn, ↓Igfbp3, ↓Park7 #, ↓Pebp1 #, ↓Ppia, ↓Serpina3k | Oxidative stress (n = 1): ↓Blvrb | Oxidative stress (n = 3): ↓Alb $, ↑Hp, ↑Hpx |
| Immune response (n = 15): ↓Il1rap, Igh (n = 10↑, 1↓), ↑Igk (n = 3), | Immune response (n = 10): ↑Clu $, ↑Igh (n = 3↑, 1↓), ↑Igk (n = 3), ↑Igl, ↑Igm | Immune response (n = 9): ↑Igh (n = 4), ↑Igj, ↑Igk (n = 3), ↑Igl |
| Acute phase response (n = 5): ↑Ahsg, ↑Orm1, ↑Orm2, ↑Saa1, ↑Saa2 | Acute phase response (n = 1): ↑Ttr | Acute phase response (n = 1): ↑Ambp |
| Complement/coagulation (n = 6): ↑A2m $, ↑Apcs, ↓F13a1, ↑C3, ↑Cfb, ↑Cfhr1 | Complement/coagulation (n = 4): ↑AI182371, ↑F2, ↓F13b, ↓Mbl1 | Complement/coagulation (n = 3): ↑Cfh, ↓Klkb1, ↓Serpinc1 |
| Blood coagulation (n = 6): ↑Serpina10, ↓Gp1ba, ↑Gp5, ↑Itih3, ↑Pros1 $, ↓Proz | Blood coagulation (n = 3): ↓Hgfac, ↑Hrg ‡, ↓Itih1 | |
| Lipoprotein/lipid metabolism (n = 5): ↓ApoA2, ↓ApoC2, ↓Azgp1, ↓Pgp, ↑Pltp | Lipoprotein metabolism (n = 4): ↓Afm, ↑ApoD, ↑ApoM, ↑Lcat | Lipoprotein metabolism (n = 4): ↓ApoA1, ↑ApoB, ↓ApoC1, ↓Pon1 |
| Protein turnover (n = 5): ↓Apeh, ↓Mug2, ↓Serpina3m, ↓Uba1, ↓Uba52 | Protein turnover (n = 1): ↑Fetub | Protein turnover (n = 1): ↓Mug1; |
| Other proteins (n = 8): ↓Atic, ↓Nme1, ↓Pnp (purine metabo-lism), ↓Tpi, ↓Tkt (glucose metabolism), ↓Ran # (nucleoplasmic transport), ↓Rbp4 (retinol transport), ↓Spp2 (bone remodeling) | Other proteins (n = 3): ↓Aldoa, ↓Ldha (glucose metabolism), ↓Lifr (tissue regeneration) | Other proteins (n = 2): ↓Bpgm (glucose metabolism), ↓Ica (carbonic anhydrase inhibitor) |
| Variable (n = 82–112) | Global Cognition | Episodic Memory | Attention/Processing Speed | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MMSE_2 1 | TICSm_2 2 | HVLT-TR_2 3 | HVLT-DR_2 4 | Trail_Making_A _2 5 | SDMT_2 6 | SDMT_2 7 | ||||||||
| β | p | β | p | β | p | β | p | β | p | β | p | |||
| Arylesterase activity_1 | −0.24 | 0.034 | −0.24 | 0.027 | −0.19 | 0.046 | −0.32 | 0.012 | 0.24 | 0.015 | −0.18 | 0.008 | ||
| Paraoxonase activity_1 | NS # | NS # | NS # | NS # | −0.33 | 0.028 | ||||||||
| PON1-Q192R | NS | NS | NS | NS | 0.049 | 0.29 | 0.047 | |||||||
| Brain atrophy rate | −0.27 | 0.029 | −0.27 | 0.011 | NS | NS | 0.007 | −0.24 | 0.001 | −0.23 | 0.002 | |||
| MMSE_1 | 0.26 | 0.017 | ||||||||||||
| TICS-m_1 | 0.25 | 0.017 | ||||||||||||
| HVLT-TR_1 | 0.45 | 0.000 | ||||||||||||
| HVLT-DR_1 | 0.46 | 0.000 | ||||||||||||
| Trail Making A_1 | 0.32 | 0.001 | ||||||||||||
| SDMT_1 | 0.78 | 0.000 | 0.66 | 0.000 | ||||||||||
| * Log-transformed data were used in analyses. _1—baseline _2—end of study | p = 0.000, R2 = 0.43 | p = 0.000, R2 = 0.51 | p = 0.000, R2 = 0.54 | p = 0.001, R2 = 0.37 | p = 0.000, R2 = 0.57 | p = 0.000, R2 = 0.78 | p = 0.000, R2 = 0.78 | |||||||
| 1−7 Adjusted for sex, age. Additional adjustment for: 1, 4 Anti-N-Hcy, tHcy_1; 1 BDNF V66M genotype; 3 Creatinine, TCN 776CG genotype; 4 APOE genotype; 5 Fe_1, FA_1, TG_1, COMT V158M and DHFR 19bpins genotypes. # Models with or w/o arylesterase. MMSE—Mini-Mental State Examination; TICS-m—Telephone Inventory for Cognitive Status modified; HVLT-TR_1—Hopkins Verbal Learning Test-revised Total Recall; HVLT-DR—Hopkins Verbal Learning Test-revised, Delayed Recall; SDMT—Symbol Digits Modalities Test. | ||||||||||||||
| Variable (n = 82–112) | Global Cognition | Episodic Memory | Attention/Processing Speed | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MMSE_2 1 | TICSm_2 2 | HVLT-TR_2 3 | HVLT-DR_2 4 | Trail_Making_A _2 5 | SDMT_2 6 | SDMT_2 7 | ||||||||
| β | p | β | p | β | p | β | p | β | p | β | p | |||
| Arylesterase activity_1 | NS | NS | NS | NS | NS | NS | ||||||||
| Paraoxonase activity_1 | NS | |||||||||||||
| PON1-Q192R | NS | NS | ||||||||||||
| Brain atrophy rate | NS | NS | NS | NS | NS | NS | NS | |||||||
| MMSE_1 | 0.52 | 0.000 | ||||||||||||
| TICS-m_1 | 0.38 | 0.002 | ||||||||||||
| HVLT-TR_1 | 0.63 | 0.000 | ||||||||||||
| HVLT-DR_1 | 0.47 | 0.001 | ||||||||||||
| Trail Making_1 | 0.62 | 0.000 | ||||||||||||
| SDMT_1 | 0.65 | 0.000 | 0.66 | 0.000 | ||||||||||
| * Log-transformed data were used in analyses. _1—baseline _2—end of study | p = 0.019, R2 = 0.20 | p = 0.001, R2 = 0.28 | P = 0.000, R2 = 0.38 | p = 0.001, R2 = 0.20 | p = 0.005, R2 = 0.30 | P = 0.000, R2 = 0.61 | p = 0.000, R2 = 0.60 | |||||||
| 1−7 Adjusted for sex, age. Additional adjustment for: 1, 4 Anti-N-Hcy, tHcy_1; 1 BDNF V66M genotype; 3 Creatinine, TCN 776CG genotype; 4 APOE genotype; 5 Fe_1, FA_1, TG_1, COMT V158M and DHFR 19bpins genotypes. Neuropsychological test acronyms defined as in Table 3. | ||||||||||||||
| Protein Name | Change in Pon1−/− vs. Pon1+/+ Brain * | Change in 1% Met Diet vs. Std. Diet Brain * | Change in AD Brain (Other Neuropathy or Animal Model) ** | |
|---|---|---|---|---|
| Std. Diet | 1%-Met Diet | Pon1+/+ | ||
| Brain-specific | ||||
| Ncald | – | ↑ | ↓ | ↓, (↓ in Gls−/− mouse) |
| Nrgn | ↓ | ↑ | ↓ | ↓ |
| Stmn1 | – | ↑ | ↓ | ↓, (↑ in MS, TLE, SMA, schizophrenia), (↑ in HD4 mouse model) |
| Antioxidant defense | ||||
| Sod1 | ↓ | – | – | (↑ in ALS) |
| Prdx2 | – | ↑ | ↓ | ↑ |
| DJ-1 (Park7) | ↓ | ↑ | ↓ | ↑ |
| Energy metabolism | ||||
| Ak1 | – | ↑ | ↓ | ↑ |
| Cell cycle | ||||
| GDI1 | – | ↑ | ↓ | (↑ in rat ischemic brain) |
| Ran | – | ↑ | ↓ | ↑ |
| Cytoskeleton assembly | ||||
| Tbcb | ↓ | ↑ | ↑ | (↑ in GAN) |
| CapZa2 | ↑ | – | ↑ | ↑ CapZb2 # |
| Other proteins | ||||
| Hdhd2 | – | ↑ | – | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jakubowski, H. Proteomic Exploration of Paraoxonase 1 Function in Health and Disease. Int. J. Mol. Sci. 2023, 24, 7764. https://doi.org/10.3390/ijms24097764
Jakubowski H. Proteomic Exploration of Paraoxonase 1 Function in Health and Disease. International Journal of Molecular Sciences. 2023; 24(9):7764. https://doi.org/10.3390/ijms24097764
Chicago/Turabian StyleJakubowski, Hieronim. 2023. "Proteomic Exploration of Paraoxonase 1 Function in Health and Disease" International Journal of Molecular Sciences 24, no. 9: 7764. https://doi.org/10.3390/ijms24097764
APA StyleJakubowski, H. (2023). Proteomic Exploration of Paraoxonase 1 Function in Health and Disease. International Journal of Molecular Sciences, 24(9), 7764. https://doi.org/10.3390/ijms24097764








