ADAM17 Confers Temozolomide Resistance in Human Glioblastoma Cells and miR-145 Regulates Its Expression
Abstract
1. Introduction
2. Results
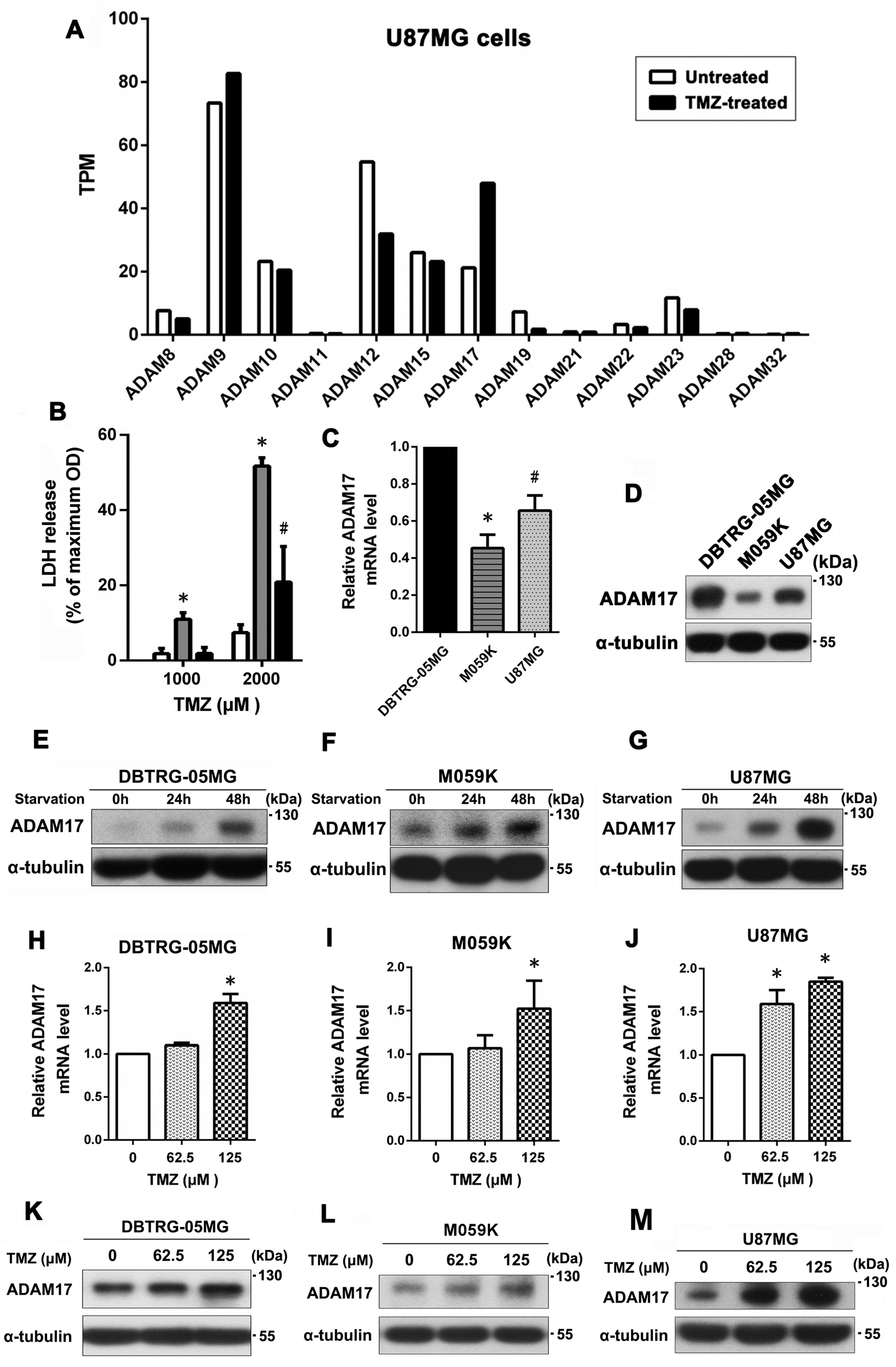
2.1. Serum Starvation and TMZ Treatment Enhanced ADAM17 Expression, Which May Contribute to the Development of TMZ Resistance in GBM Cells
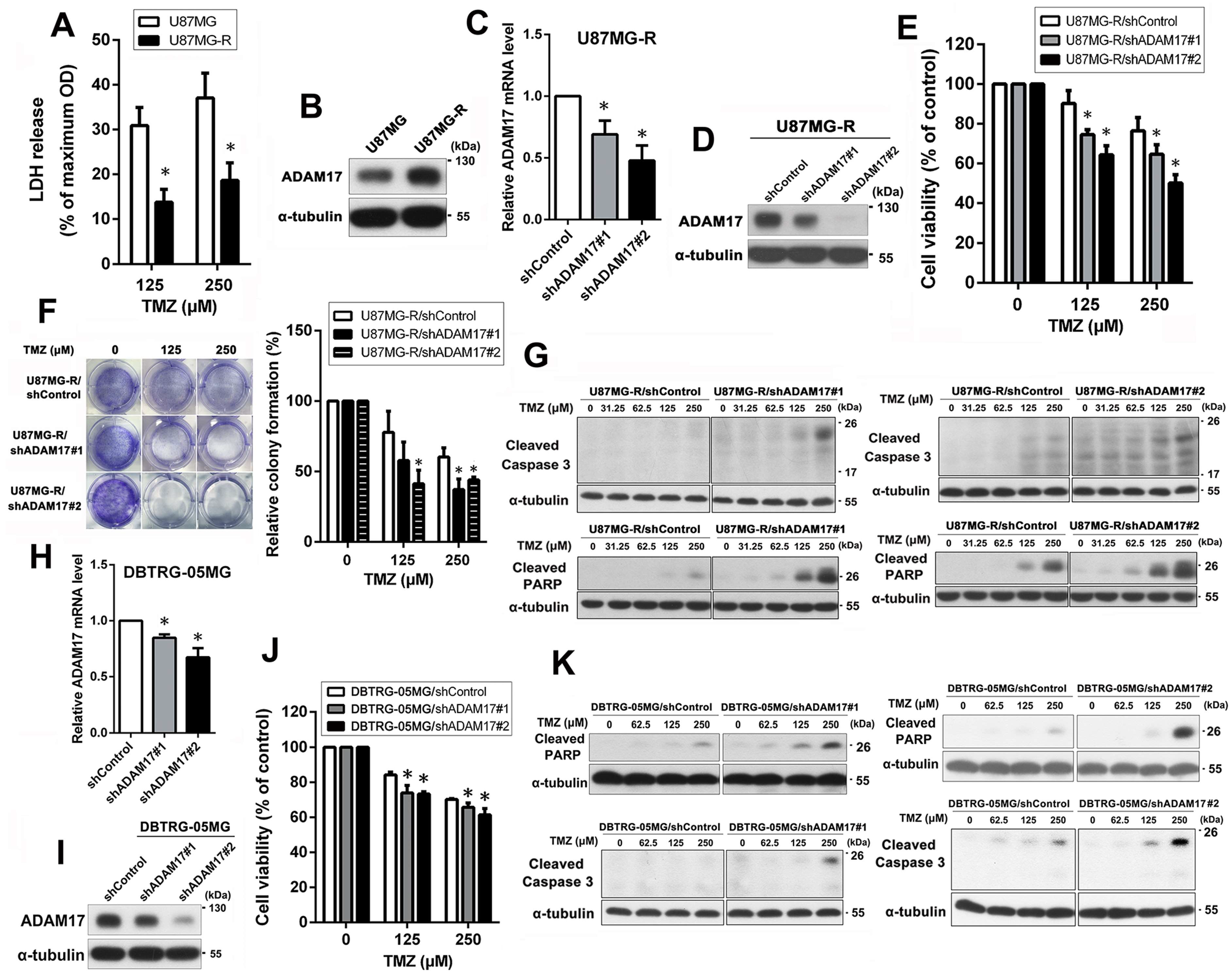
2.2. ADAM17 Knockdown Restored the Chemosensitivity of TMZ-Resistant GBM Cells
2.3. miR-145-5p Regulates ADAM17 Expression by Targeting Its 3′-Untranslated Region (UTR) to Enhance TMZ Sensitivity in GBM Cells
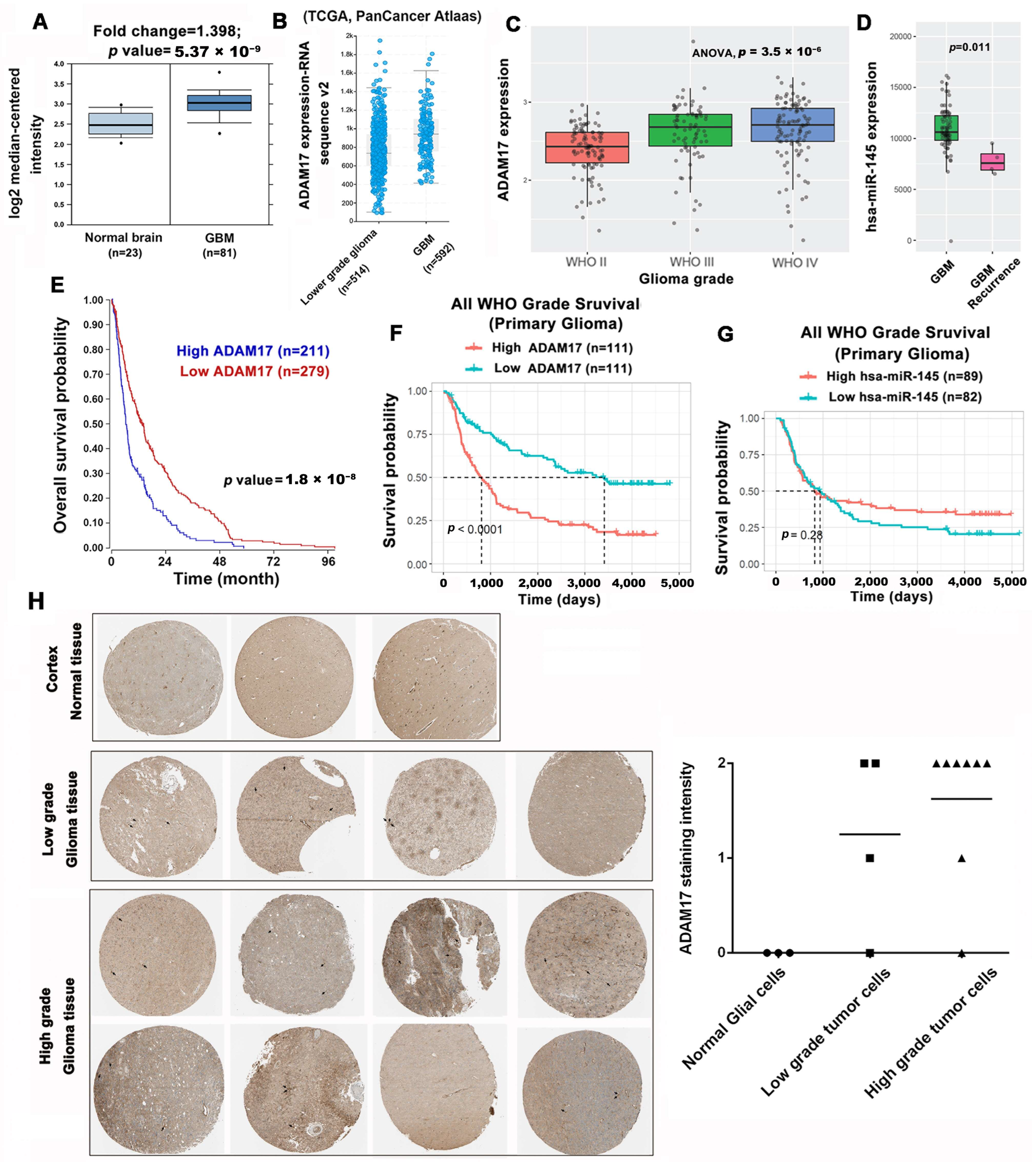
2.4. ADAM17 Expression Is Correlated to GBM Progression and Poor Prognosis
2.5. ADAM17 Knockdown Enhanced TMZ Sensitivity In Vivo
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Reagents, and Chemicals
4.2. Cell Culture and Drug Treatment
4.3. RNA Sequencing
4.4. Lactate Dehydrogenase (LDH) Assay
4.5. Quantitative Real-Time PCR
4.6. Western Blotting
4.7. Establishment of TMZ-Resistant Cells
4.8. Lentiviral Systems for ADAM17 Knockdown
4.9. MTT Assay
4.10. Colony Formation Assay
4.11. Bioinformatic Prediction of miRNAs Targeting ADAM17
4.12. miRNA Expression Analysis
4.13. Cell Transfection with miRNA Mimics
4.14. Public Database Analysis
4.15. Xenograft Mouse Model
4.16. IHC
4.17. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015–2019. Neuro Oncol. 2022, 24 (Suppl. S5), v1–v95. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.R.; O’Neill, B.P. Glioblastoma survival in the United States before and during the temozolomide era. J. Neurooncol. 2012, 107, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Villano, J.L.; Seery, T.E.; Bressler, L.R. Temozolomide in malignant gliomas: Current use and future targets. Cancer Chemother. Pharm. 2009, 64, 647–655. [Google Scholar] [CrossRef]
- Johannessen, T.C.; Bjerkvig, R. Molecular mechanisms of temozolomide resistance in glioblastoma multiforme. Expert Rev. Anticancer Ther. 2012, 12, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Atkins, R.J.; Ng, W.; Stylli, S.S.; Hovens, C.M.; Kaye, A.H. Repair mechanisms help glioblastoma resist treatment. J. Clin. Neurosci. Off. J. Neurosurg. Soc. Australas. 2015, 22, 14–20. [Google Scholar] [CrossRef]
- Stavrovskaya, A.A.; Shushanov, S.S.; Rybalkina, E.Y. Problems of Glioblastoma Multiforme Drug Resistance. Biochem. Biokhim. 2016, 81, 91–100. [Google Scholar] [CrossRef]
- Black, R.A.; Rauch, C.T.; Kozlosky, C.J.; Peschon, J.J.; Slack, J.L.; Wolfson, M.F.; Castner, B.J.; Stocking, K.L.; Reddy, P.; Srinivasan, S.; et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 1997, 385, 729–733. [Google Scholar] [CrossRef]
- Moss, M.L.; Jin, S.L.; Milla, M.E.; Bickett, D.M.; Burkhart, W.; Carter, H.L.; Chen, W.J.; Clay, W.C.; Didsbury, J.R.; Hassler, D.; et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature 1997, 385, 733–736. [Google Scholar] [CrossRef]
- Grotzinger, J.; Lorenzen, I.; Dusterhoft, S. Molecular insights into the multilayered regulation of ADAM17: The role of the extracellular region. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2088–2095. [Google Scholar] [CrossRef]
- Saad, M.I.; Rose-John, S.; Jenkins, B.J. ADAM17: An Emerging Therapeutic Target for Lung Cancer. Cancers 2019, 11, 1218. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Sha, L.; Wang, Y.; Xu, W.; Yu, Y.; Feng, F.; Sun, C.; Xia, L. Diagnostic and prognostic value of a disintegrin and metalloproteinase-17 in patients with gliomas. Oncol. Lett. 2014, 8, 2616–2620. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; He, X.; Zhang, M.; Qu, Y.; Gu, C.; Ren, M.; Wang, H.; Ning, W.; Li, J.; Yu, C.; et al. Reciprocal control of ADAM17/EGFR/Akt signaling and miR-145 drives GBM invasiveness. J. Neurooncol. 2020, 147, 327–337. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Beylerli, O.; Gareev, I.; Sufianov, A.; Ilyasova, T.; Zhang, F. The role of microRNA in the pathogenesis of glial brain tumors. Noncoding RNA Res. 2022, 7, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.P.; Castresana, J.S.; Shahi, M.H. Glioblastoma and MiRNAs. Cancers 2021, 13, 1581. [Google Scholar] [CrossRef] [PubMed]
- Gareev, I.; Beylerli, O.; Liang, Y.; Xiang, H.; Liu, C.; Xu, X.; Yuan, C.; Ahmad, A.; Yang, G. The Role of MicroRNAs in Therapeutic Resistance of Malignant Primary Brain Tumors. Front. Cell Dev. Biol. 2021, 9, 740303. [Google Scholar] [CrossRef]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.G.; Alder, H.; et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef]
- Arndt, G.M.; Dossey, L.; Cullen, L.M.; Lai, A.; Druker, R.; Eisbacher, M.; Zhang, C.; Tran, N.; Fan, H.; Retzlaff, K.; et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer 2009, 9, 374. [Google Scholar] [CrossRef]
- Zaman, M.S.; Chen, Y.; Deng, G.; Shahryari, V.; Suh, S.O.; Saini, S.; Majid, S.; Liu, J.; Khatri, G.; Tanaka, Y.; et al. The functional significance of microRNA-145 in prostate cancer. Br. J. Cancer 2010, 103, 256–264. [Google Scholar] [CrossRef]
- Chiyomaru, T.; Enokida, H.; Tatarano, S.; Kawahara, K.; Uchida, Y.; Nishiyama, K.; Fujimura, L.; Kikkawa, N.; Seki, N.; Nakagawa, M. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br. J. Cancer 2010, 102, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Kano, M.; Seki, N.; Kikkawa, N.; Fujimura, L.; Hoshino, I.; Akutsu, Y.; Chiyomaru, T.; Enokida, H.; Nakagawa, M.; Matsubara, H. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int. J. Cancer 2010, 127, 2804–2814. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chopp, M.; Zheng, X.; Katakowski, M.; Buller, B.; Jiang, F. MiR-145 reduces ADAM17 expression and inhibits in vitro migration and invasion of glioma cells. Oncol. Rep. 2013, 29, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Seals, D.F.; Courtneidge, S.A. The ADAMs family of metalloproteases: Multidomain proteins with multiple functions. Genes Dev. 2003, 17, 7–30. [Google Scholar] [CrossRef]
- Reiss, K.; Saftig, P. The “a disintegrin and metalloprotease” (ADAM) family of sheddases: Physiological and cellular functions. Semin. Cell Dev. Biol. 2009, 20, 126–137. [Google Scholar] [CrossRef]
- Huovila, A.P.; Turner, A.J.; Pelto-Huikko, M.; Karkkainen, I.; Ortiz, R.M. Shedding light on ADAM metalloproteinases. Trends Biochem. Sci. 2005, 30, 413–422. [Google Scholar] [CrossRef]
- Bolik, J.; Krause, F.; Stevanovic, M.; Gandraß, M.; Thomsen, I.; Schacht, S.S.; Rieser, E.; Müller, M.; Schumacher, N.; Fritsch, J.; et al. Inhibition of ADAM17 impairs endothelial cell necroptosis and blocks metastasis. J. Exp. Med. 2022, 219, e20201039. [Google Scholar] [CrossRef]
- Xiang, Y.; Liu, L.; Wang, Y.; Li, B.; Peng, J.; Feng, D. ADAM17 promotes the invasion of hepatocellular carcinoma via upregulation MMP21. Cancer Cell Int. 2020, 20, 516. [Google Scholar] [CrossRef]
- Zheng, X.; Jiang, F.; Katakowski, M.; Lu, Y.; Chopp, M. ADAM17 promotes glioma cell malignant phenotype. Mol. Carcinog. 2012, 51, 150–164. [Google Scholar] [CrossRef]
- Rogmans, C.; Kuhlmann, J.D.; Hugendieck, G.; Link, T.; Arnold, N.; Weimer, J.P.; Flörkemeier, I.; Rambow, A.C.; Lieb, W.; Maass, N.; et al. ADAM17-A Potential Blood-Based Biomarker for Detection of Early-Stage Ovarian Cancer. Cancers 2021, 13, 5563. [Google Scholar] [CrossRef]
- Hedemann, N.; Herz, A.; Schiepanski, J.H.; Dittrich, J.; Sebens, S.; Dempfle, A.; Feuerborn, J.; Rogmans, C.; Tribian, N.; Flörkemeier, I.; et al. ADAM17 Inhibition Increases the Impact of Cisplatin Treatment in Ovarian Cancer Spheroids. Cancers 2021, 13, 2039. [Google Scholar] [CrossRef] [PubMed]
- Moss, M.L.; Minond, D. Recent Advances in ADAM17 Research: A Promising Target for Cancer and Inflammation. Mediat. Inflamm. 2017, 2017, 9673537. [Google Scholar] [CrossRef]
- Rose-John, S. ADAM17, shedding, TACE as therapeutic targets. Pharmacol. Res. 2013, 71, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Arribas, J.; Esselens, C. ADAM17 as a therapeutic target in multiple diseases. Curr. Pharm. Des. 2009, 15, 2319–2335. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Wang, F.; Gao, H.; Hu, W. Dihydroartemisinin suppresses glioma proliferation and invasion via inhibition of the ADAM17 pathway. Neurol. Sci. 2015, 36, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Saha, N.; Xu, K.; Zhu, Z.; Robev, D.; Kalidindi, T.; Xu, Y.; Himanen, J.; de Stanchina, E.; Pillarsetty, N.V.K.; Dimitrov, D.S.; et al. Inhibitory monoclonal antibody targeting ADAM17 expressed on cancer cells. Transl. Oncol. 2022, 15, 101265. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Shen, Z.; Zhou, S. Function of microRNA-145 and mechanisms underlying its role in malignant tumor diagnosis and treatment. Cancer Manag. Res. 2019, 11, 969–979. [Google Scholar] [CrossRef]
- Doberstein, K.; Steinmeyer, N.; Hartmetz, A.K.; Eberhardt, W.; Mittelbronn, M.; Harter, P.N.; Juengel, E.; Blaheta, R.; Pfeilschifter, J.; Gutwein, P. MicroRNA-145 targets the metalloprotease ADAM17 and is suppressed in renal cell carcinoma patients. Neoplasia 2013, 15, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.W.; Zhang, L.J.; Huang, X.H.; Chen, L.Z.; Su, Q.; Zeng, W.T.; Li, W.; Wang, Q. miR-145 suppresses cell invasion in hepatocellular carcinoma cells: miR-145 targets ADAM17. Hepatol. Res. 2014, 44, 551–559. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, C.; Wang, Y.; Wen, S.; Wang, J.; Chen, Z.; He, Q.; Feng, D. MicroRNA-145 inhibits cell proliferation by directly targeting ADAM17 in hepatocellular carcinoma. Oncol. Rep. 2014, 32, 1923–1930. [Google Scholar] [CrossRef]
- Chen, W.; Huang, B.; Wang, E.; Wang, X. MiR-145 inhibits EGF-induced epithelial-to-mesenchymal transition via targeting Smad2 in human glioblastoma. OncoTargets Ther. 2019, 12, 3099–3107. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Wang, B.; Zou, Y.; Zhang, Y.; Hu, X.; Sun, W.; Xiao, H.; Liu, H.; Shi, L. MicroRNA 145 enhances chemosensitivity of glioblastoma stem cells to demethoxycurcumin. Cancer Manag. Res. 2019, 11, 6829–6840. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, Z.; Jiang, B.; Huo, L.; Liu, J.; Lu, J. Synthetic miR-145 Mimic Enhances the Cytotoxic Effect of the Antiangiogenic Drug Sunitinib in Glioblastoma. Cell Biochem. Biophys. 2015, 72, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wang, B.; Gu, X.; Zhang, S.; Li, X.; Zhu, H. miR-145 is a potential biomarker for predicting clinical outcome in glioblastomas. J. Cell. Biochem. 2019, 120, 8016–8020. [Google Scholar] [CrossRef]
- Zhao, H.; Shen, J.; Hodges, T.R.; Song, R.; Fuller, G.N.; Heimberger, A.B. Serum microRNA profiling in patients with glioblastoma: A survival analysis. Mol. Cancer 2017, 16, 59. [Google Scholar] [CrossRef]
- AmeliMojarad, M.; AmeliMojarad, M.; Pourmahdian, A. Circular RNA circ_0051620 sponges miR-338-3p and regulates ADAM17 to promote the gastric cancer progression. Pathol. Res. Pract. 2022, 233, 153887. [Google Scholar] [CrossRef]
- Hong, Y.; Chen, X.; Liang, Z.; Xu, Z.; Li, Y.; Pan, Y. MiR-338-3p inhibits cell migration and invasion in human hypopharyngeal cancer via downregulation of ADAM17. Anticancer Drugs 2020, 31, 925–931. [Google Scholar] [CrossRef]
- Fan, C.; Yuan, Q.; Liu, G.; Zhang, Y.; Yan, M.; Sun, Q.; Zhu, C. Long non-coding RNA MALAT1 regulates oxaliplatin-resistance via miR-324-3p/ADAM17 axis in colorectal cancer cells. Cancer Cell Int. 2020, 20, 473. [Google Scholar] [CrossRef]
- Xu, K.; Liang, X.; Shen, K.; Sun, L.; Cui, D.; Zhao, Y.; Tian, J.; Ni, L.; Liu, J. MiR-222 modulates multidrug resistance in human colorectal carcinoma by down-regulating ADAM-17. Exp Cell Res 2012, 318, 2168–2177. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, M.; Zhao, Y.; Pan, Y.; Kan, M.; Li, J.; He, K.; Zhang, X. HNF-4α inhibits hepatocellular carcinoma cell proliferation through mir-122-adam17 pathway. PLoS ONE 2020, 15, e0230450. [Google Scholar] [CrossRef]
- Yang, B.; Wang, C.; Xie, H.; Wang, Y.; Huang, J.; Rong, Y.; Zhang, H.; Kong, H.; Yang, Y.; Lu, Y. MicroRNA-3163 targets ADAM-17 and enhances the sensitivity of hepatocellular carcinoma cells to molecular targeted agents. Cell Death Dis. 2019, 10, 784. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Huang, W.; Chen, H.; Wei, H.; Luo, A.; Xia, G.; Deng, X.; Zhang, G. MicroRNA-224, negatively regulated by c-jun, inhibits growth and epithelial-to-mesenchymal transition phenotype via targeting ADAM17 in oral squamous cell carcinoma. J. Cell. Mol. Med. 2019, 23, 4913–4920. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Tan, S.; Song, L.; Song, L.; Wang, Y. LncRNA XIST knockdown suppresses the malignancy of human nasopharyngeal carcinoma through XIST/miRNA-148a-3p/ADAM17 pathway in vitro and in vivo. Biomed. Pharmacother. 2020, 121, 109620. [Google Scholar] [CrossRef] [PubMed]
- Fei, Q.; Du, M.Y.; Qian, L.X.; Chen, H.B.; Chen, J.; Zhu, H.M.; Tian, X.K.; Jiang, N.; Gu, J.J.; He, X.; et al. Feedback loop in miR-449b-3p/ADAM17/NF-κB promotes metastasis in nasopharyngeal carcinoma. Cancer Med. 2019, 8, 6049–6063. [Google Scholar] [CrossRef]
- Su, Y.; Wang, Y.; Zhou, H.; Lei, L.; Xu, L. MicroRNA-152 targets ADAM17 to suppress NSCLC progression. FEBS Lett. 2014, 588, 1983–1988. [Google Scholar] [CrossRef]
- Cai, M.; Wang, Z.; Zhang, J.; Zhou, H.; Jin, L.; Bai, R.; Weng, Y. Adam17, a Target of Mir-326, Promotes Emt-Induced Cells Invasion in Lung Adenocarcinoma. Cell Physiol. Biochem. 2015, 36, 1175–1185. [Google Scholar] [CrossRef]
- Lee, I.N.; Yang, J.T.; Huang, C.; Huang, H.C.; Wu, Y.P.; Chen, J.C. Elevated XRCC5 expression level can promote temozolomide resistance and predict poor prognosis in glioblastoma. Oncol. Lett. 2021, 21, 443. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.-T.; Lee, I.-N.; Huang, C.; Huang, H.-C.; Wu, Y.-P.; Chong, Z.-Y.; Chen, J.-C. ADAM17 Confers Temozolomide Resistance in Human Glioblastoma Cells and miR-145 Regulates Its Expression. Int. J. Mol. Sci. 2023, 24, 7703. https://doi.org/10.3390/ijms24097703
Yang J-T, Lee I-N, Huang C, Huang H-C, Wu Y-P, Chong Z-Y, Chen J-C. ADAM17 Confers Temozolomide Resistance in Human Glioblastoma Cells and miR-145 Regulates Its Expression. International Journal of Molecular Sciences. 2023; 24(9):7703. https://doi.org/10.3390/ijms24097703
Chicago/Turabian StyleYang, Jen-Tsung, I-Neng Lee, Cheng Huang, Hsiu-Chen Huang, Yu-Ping Wu, Zhi-Yong Chong, and Jui-Chieh Chen. 2023. "ADAM17 Confers Temozolomide Resistance in Human Glioblastoma Cells and miR-145 Regulates Its Expression" International Journal of Molecular Sciences 24, no. 9: 7703. https://doi.org/10.3390/ijms24097703
APA StyleYang, J.-T., Lee, I.-N., Huang, C., Huang, H.-C., Wu, Y.-P., Chong, Z.-Y., & Chen, J.-C. (2023). ADAM17 Confers Temozolomide Resistance in Human Glioblastoma Cells and miR-145 Regulates Its Expression. International Journal of Molecular Sciences, 24(9), 7703. https://doi.org/10.3390/ijms24097703




