DoMYB5 and DobHLH24, Transcription Factors Involved in Regulating Anthocyanin Accumulation in Dendrobium officinale
Abstract
1. Introduction
2. Results
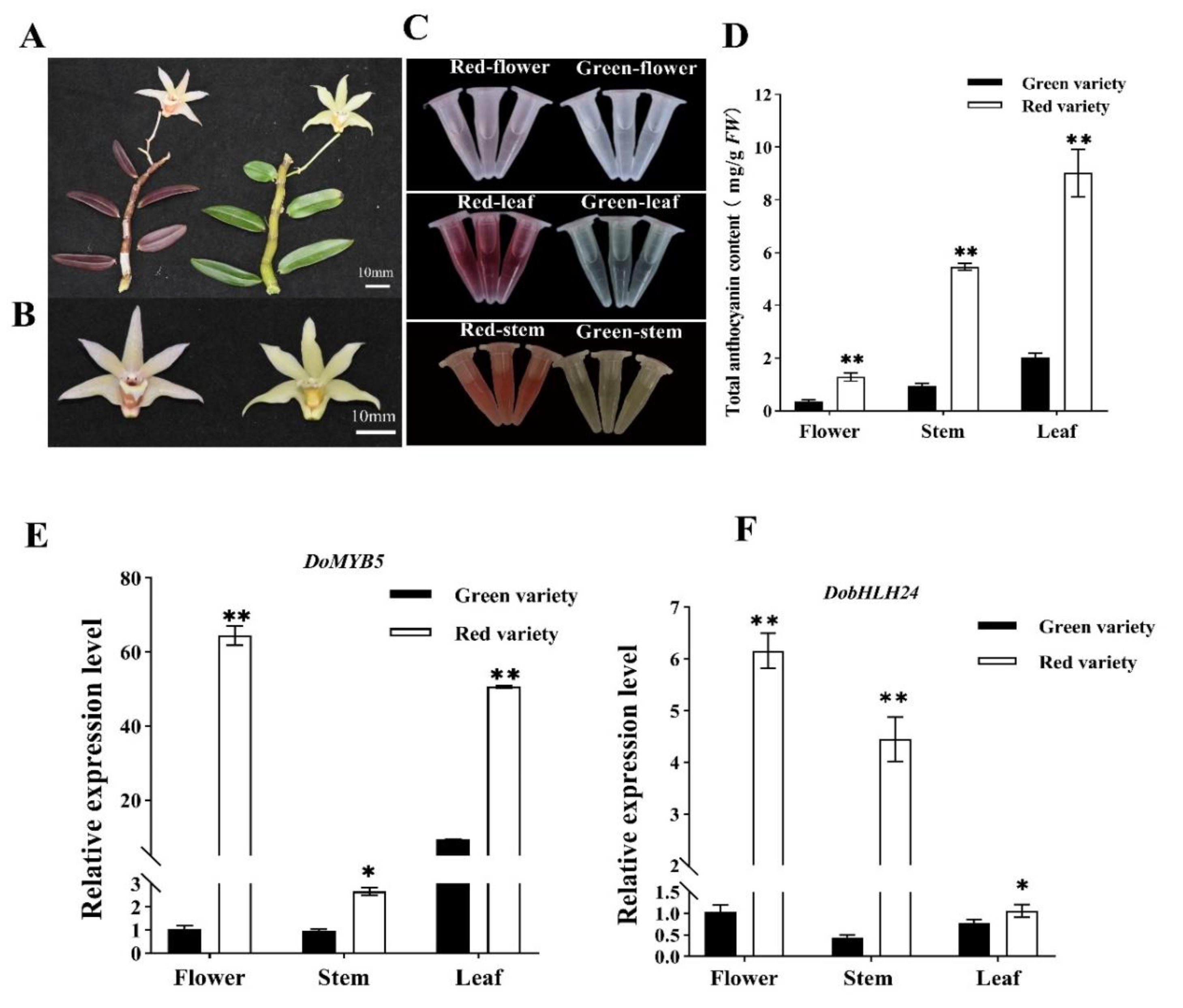
2.1. The Expression Level of DoMYB5 and DobHLH24 Was Correlated with Anthocyanin Biosynthesis
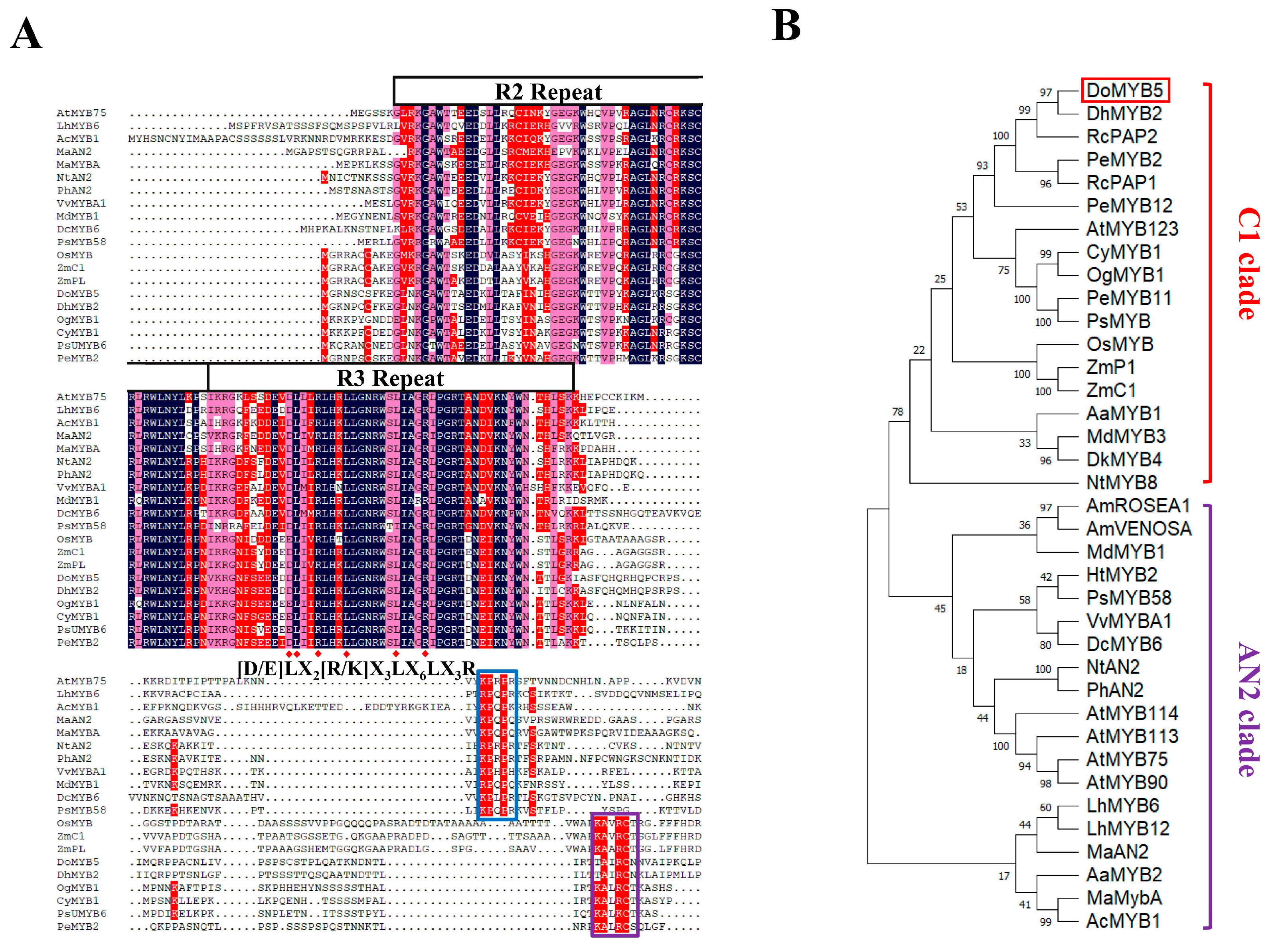
2.2. Cloning and Characterization of DoMYB5 and DobHLH24
2.3. Transient Overexpression of DoMYB5 and DobHLH24 in the Leaf of D. officinale
2.4. Stable Transformation of DoMYB5 and DobHLH24 in Tobacco
2.5. Verification of the Interaction between DobHLH24 and DoMYB5
2.6. Regulation of DoMYB5 and DobHLH24 on the Promoters of DoCHS and DoDFR
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Flavonoid Extraction and Measurement
4.3. DNA, RNA Isolation, and cDNA Synthesis
4.4. Isolation of the Full-Length cDNA of DoMYB5 and DobHLH24 from D. officinale
4.5. Transient Expression of DoMYB5 and DobHLH24 in D. officinale Leaves
4.6. Stable Transformation of DoMYB5 and DobHLH24 in Tobacco
4.7. Real-Time Quantitative PCR
4.8. Yeast One-Hybrid (Y1H) Assay
4.9. Yeast Two-Hybrid (Y2H) Assay
4.10. Dual Luciferase Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jin, Q.; Jiao, C.; Sun, S.; Song, C.; Cai, Y.; Lin, Y.; Fan, H.; Zhu, Y. Metabolic Analysis of Medicinal Dendrobium officinale and Dendrobium huoshanense during Different Growth Years. PLoS ONE 2016, 11, e0146607. [Google Scholar] [CrossRef]
- Zhao, D.; Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015, 6, 261. [Google Scholar] [CrossRef] [PubMed]
- Okitsu, N.; Noda, N.; Chandler, S.; Tanaka, Y. Flower Color and Its Engineering by Genetic Modification. Ornam. Crops Handb. Plant Breed. 2018, 11, 29–62. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Navas, M.J.; Jiménez-Moreno, A.M.; Bueno, J.M.; Sáez-Plaza, P.; Asuero, A.G. Analysis and Antioxidant Capacity of Anthocyanin Pigments. Part IV: Extraction of Anthocyanins. Crit. Rev. Anal. Chem. 2012, 42, 313–342. [Google Scholar] [CrossRef]
- Kong, J.M.; Chia, L.S.; Goh, N.K.; Chia, T.F.; Brouillard, R. Analysis and biological activities of anthocyanins. Phytochemistry 2003, 64, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.M. Modifying Anthocyanin Production in Flowers. Anthocyanins Biosynth. Funct. Appl. 2009, 49–80. [Google Scholar] [CrossRef]
- Mouradov, A.; Spangenberg, G. Flavonoids: A metabolic network mediating plants adaptation to their real estate. Front. Plant Sci. 2014, 5, 620. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef]
- Khoo, H.E.; Ng, H.S.; Yap, W.S.; Goh, H.J.H.; Yim, H.S. Nutrients for Prevention of Macular Degeneration and Eye-Related Diseases. Antioxidants 2019, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A Comprehensive Review of Their Chemical Properties and Health Effects on Cardiovascular and Neurodegenerative Diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Li, Y.; Yang, S.; Gao, R.; Zhou, L.; Bao, T.; Han, T.; Wang, S.; Gao, X.; Wang, L. A functional homologue of Arabidopsis TTG1 from Freesia interacts with bHLH proteins to regulate anthocyanin and proanthocyanidin biosynthesis in both Freesia hybrida and Arabidopsis thaliana. Plant Physiol. Biochem. 2019, 141, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, A. Genomic Characterization and Expression Analysis of Basic Helix-Loop-Helix (bHLH) Family Genes in Traditional Chinese Herb Dendrobium officinale. Plants 2020, 9, 1044. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Ma, H.; Duan, X.; Gao, S.; Zhou, X.; Cheng, Y. The R2R3-MYB gene PsMYB58 positively regulates anthocyanin biosynthesis in tree peony flowers. Plant Physiol. Biochem. 2021, 164, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.S.; Shimoyamada, Y.; Nakayama, M.; Yamagishi, M. Pigment accumulation and transcription of LhMYB12 and anthocyanin biosynthesis genes during flower development in the Asiatic hybrid lily (Lilium spp.). Plant Sci. 2012, 193–194, 136–147. [Google Scholar] [CrossRef]
- Chen, K.; Liu, H.; Lou, Q.; Liu, Y. Ectopic Expression of the Grape Hyacinth (Muscari armeniacum) R2R3-MYB transcription factor gene, MaAN2, induces anthocyanin accumulation in tobacco. Front. Plant Sci. 2017, 8, 965. [Google Scholar] [CrossRef]
- Li, X.; Cao, L.; Jiao, B.; Yang, H.; Ma, C.; Liang, Y. The bHLH transcription factor AcB2 regulates anthocyanin biosynthesis in onion (Allium cepa L.). Hortic. Res. 2022, 9, uhac128. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhou, L.; Han, L.; Zou, H.; Miao, K.; Wang, Y. PsbHLH1, a novel transcription factor involved in regulating anthocyanin biosynthesis in tree peony (Paeonia suffruticosa). Plant Physiol. Biochem. 2020, 154, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Wang, J.; Lu, C.; Li, Y.; Kong, D.; Hong, Y.; Huang, H.; Dai, S. CcMYB6-1 and CcbHLH1, two novel transcription factors synergistically involved in regulating anthocyanin biosynthesis in cornflower. Plant Physiol. Biochem. 2020, 151, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shan, X.; Zhou, L.; Gao, R.; Yang, S.; Wang, S.; Wang, L.; Gao, X. The R2R3-MYB factor FhMYB5 From Freesia hybrida contributes to the regulation of anthocyanin and proanthocyanidin Biosynthesis. Front. Plant Sci. 2018, 9, 1935. [Google Scholar] [CrossRef] [PubMed]
- Chiou, C.Y.; Yeh, K.W. Differential expression of MYB gene (OgMYB1) determines color patterning in floral tissue of Oncidium Gower Ramsey. Plant Mol. Biol. 2008, 66, 379–388. [Google Scholar] [CrossRef]
- Ma, H.; Pooler, M.; Griesbach, R. Anthocyanin regulatory/structural gene expression in Phalaenopsis. J. Am. Soc. Hortic. Sci. 2009, 134, 88–96. [Google Scholar] [CrossRef]
- Hsu, C.C.; Chen, Y.Y.; Tsai, W.C.; Chen, W.H.; Chen, H.H. Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 2015, 168, 175–191. [Google Scholar] [CrossRef]
- Li, C.; Qiu, J.; Ding, L.; Huang, M.; Huang, S.; Yang, G.; Yin, J. Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals. Plant Physiol. Biochem. 2017, 112, 335–345. [Google Scholar] [CrossRef]
- Zhao, A.; Cui, Z.; Li, T.; Pei, H.; Sheng, Y.; Li, X.; Zhao, Y.; Zhou, Y.; Huang, W.; Song, X.; et al. mRNA and miRNA Expression Analysis Reveal the Regulation for Flower Spot Patterning in Phalaenopsis ‘Panda’. Int. J. Mol. Sci. 2019, 20, 4250. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Hung, Y.C.; Tsai, W.C.; Chen, W.H.; Chen, H.H. PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids. J. Exp. Bot. 2018, 69, 4363–4377. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Suzuki, T.; Harada, K.; Kobayashi, Y.; Dohra, H.; Ohno, H. Floral organ- and temperature-dependent regulation of anthocyanin biosynthesis in Cymbidium hybrid flowers. Plant Sci. 2019, 287, 110173. [Google Scholar] [CrossRef] [PubMed]
- Jia, N.; Wang, J.J.; Liu, J.; Jiang, J.; Sun, J.; Yan, P.; Sun, Y.; Wan, P.; Ye, W.; Fan, B. DcTT8, a bHLH transcription factor, regulates anthocyanin biosynthesis in Dendrobium candidum. Plant Physiol. Biochem. 2021, 162, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhang, G.; Teixeira da Silva, J.A.; Zhao, C.; Duan, J. The methyl jasmonate-responsive transcription factor DobHLH4 promotes DoTPS10, which is involved in linalool biosynthesis in Dendrobium officinale during floral development. Plant Sci. 2021, 309, 110952. [Google Scholar] [CrossRef]
- Yu, Z.; Liao, Y.; Teixeira da Silva, J.A.; Yang, Z.; Duan, J. Differential accumulation of anthocyanins in Dendrobium officinale stems with red and green peels. Int. J. Mol. Sci. 2018, 19, 2857. [Google Scholar] [CrossRef]
- Ganesh, A.; Shukla, V.; Mohapatra, A.; George, A.P.; Bhukya, D.P.N.; Das, K.K.; Kola, V.S.R.; Suresh, A.; Ramireddy, E. Root cap to soil interface: A driving force toward plant adaptation and development. Plant Cell Physiol. 2022, 63, 1038–1051. [Google Scholar] [CrossRef]
- Chen, J.; Zhan, J.; Wang, H.; Zhao, Y.; Zhang, D.; Chen, X.; Su, N.; Cui, J. VrMYB90 Functions synergistically with VrbHLHA and VrMYB3 to regulate anthocyanin biosynthesis in mung bean. Plant Cell Physiol. 2022, 64, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rivas, F.J.; Blanco-Portales, R.; Serratosa, M.P.; Ric-Varas, P.; Guerrero-Sanchez, V.; Medina-Puche, L.; Moyano, L.; Mercado, J.A.; Alseekh, S.; Caballero, J.L.; et al. FaMYB123 interacts with FabHLH3 to regulate the late steps of anthocyanin and flavonol biosynthesis during ripening. Plant J. 2023. [Google Scholar] [CrossRef]
- Khan, I.A.; Rahman, M.U.; Sakhi, S.; Nawaz, G.; Khan, A.A.; Ahmad, T.; Adnan, M.; Khan, S.M. PpMYB39 activates PpDFR to modulate anthocyanin biosynthesis during peach fruit maturation. Horticulturae 2022, 8, 332. [Google Scholar] [CrossRef]
- Wang, L.; Albert, N.W.; Zhang, H.; Arathoon, S.; Boase, M.R.; Ngo, H.; Schwinn, K.E.; Davies, K.M.; Lewis, D.H. Temporal and spatial regulation of anthocyanin biosynthesis provide diverse flower colour intensities and patterning in Cymbidium orchid. Planta 2014, 240, 983–1002. [Google Scholar] [CrossRef]
- Zhou, Z.; Ying, Z.; Wu, Z.; Yang, Y.; Fu, S.; Xu, W.; Yao, L.; Zeng, A.; Huang, J.; Lan, S.; et al. Anthocyanin genes involved in the flower coloration mechanisms of Cymbidium kanran. Front. Plant Sci. 2021, 12, 737815. [Google Scholar] [CrossRef]
- Lotkowska, M.E.; Tohge, T.; Fernie, A.R.; Xue, G.P.; Balazadeh, S.; Mueller-Roeber, B. The Arabidopsis Transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 2015, 169, 1862–1880. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Yang, Q.Q.; Feng, K.; Yu, X.; Xiong, A.S. DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol. J. 2020, 18, 1585–1597. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; He, Y.; Ge, H.; Liu, Y.; Chen, H. Functional characterization of SmMYB86, a negative regulator of anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Sci. 2021, 302, 110696. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wen, J.; Xia, Y.; Zhang, L.; Du, H. Evolution and functional diversification of R2R3-MYB transcription factors in plants. Hortic. Res. 2022, 9, uhac058. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Tang, Y.; Pang, B.; Li, X.; Yang, Y.; Deng, J.; Feng, C.; Li, L.; Ren, G.; Wang, Y.; et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic. Res. 2020, 7, 78. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; Romero De La Fuente, I.; van Houwelingen, A.; de Vos, R.C.H.; Jonker, H.H.; Xu, W.; Routaboul, J.M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria x ananassa) fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef]
- Li, P.; Chen, B.; Zhang, G.; Chen, L.; Dong, Q.; Wen, J.; Mysore, K.S.; Zhao, J. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 2016, 210, 905–921. [Google Scholar] [CrossRef]
- Li, Y.; Shan, X.; Gao, R.; Han, T.; Zhang, J.; Wang, Y.; Kimani, S.; Wang, L.; Gao, X. MYB repressors and MBW activation complex collaborate to fine-tune flower coloration in Freesia hybrida. Commun. Biol. 2020, 3, 396. [Google Scholar] [CrossRef]
- Yu, C.; Huang, J.; Wu, Q.; Zhang, C.; Li, X.L.; Xu, X.; Feng, S.; Zhan, X.; Chen, Z.; Wang, H.; et al. Role of female-predominant MYB39-bHLH13 complex in sexually dimorphic accumulation of taxol in Taxus media. Hortic. Res. 2022, 9, uhac062. [Google Scholar] [CrossRef]
- Li, B.J.; Zheng, B.Q.; Wang, J.Y.; Tsai, W.C.; Lu, H.C.; Zou, L.H.; Wan, X.; Zhang, D.Y.; Qiao, H.J.; Liu, Z.J.; et al. New insight into the molecular mechanism of colour differentiation among floral segments in orchids. Commun. Biol. 2020, 3, 89. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Qiu, J.; Yang, G.; Huang, S.; Yin, J. Isolation and characterization of a R2R3-MYB transcription factor gene related to anthocyanin biosynthesis in the spathes of Anthurium andraeanum (Hort.). Plant Cell Rep. 2016, 35, 2151–2165. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, M.; Toda, S.; Tasaki, K. The novel allele of the LhMYB12 gene is involved in splatter-type spot formation on the flower tepals of Asiatic hybrid lilies (Lilium spp.). New Phytol. 2014, 201, 1009–1020. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Espley, R.V.; Allan, H. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Cone, K.C.; Cocciolone, S.M.; Burr, F.A.; Burr, B. Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 1993, 5, 1795–1805. [Google Scholar] [CrossRef]
- Chen, K.; Du, L.; Liu, H.; Liu, Y. A novel R2R3-MYB from grape hyacinth, MaMybA, which is different from MaAN2, confers intense and magenta anthocyanin pigmentation in tobacco. BMC Plant Biol. 2019, 19, 390. [Google Scholar] [CrossRef]
- Schwinn, K.E.; Ngo, H.; Kenel, F.; Brummell, D.A.; Albert, N.W.; McCallum, J.A.; Pither-Joyce, M.; Crowhurst, R.N.; Eady, C.; Davies, K.M. The onion (Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin Biosynthesis. Front. Plant Sci. 2016, 7, 1865. [Google Scholar] [CrossRef]
- Yamagishi, M.; Shimoyamada, Y.; Nakatsuka, T.; Masuda, K. Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower Tepals, tepal spots and leaves of asiatic hybrid lily. Plant Cell Physiol. 2010, 51, 463–474. [Google Scholar] [CrossRef]
- Yin, X.; Zhang, Y.; Zhang, L.; Wang, B.; Zhao, Y.; Irfan, M.; Chen, L.; Feng, Y. Regulation of MYB transcription factors of anthocyanin synthesis in lily flowers. Front. Plant Sci. 2021, 12, 761668. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Qiu, J.; Yang, G.; Huang, S.; Yin, J. Ectopic expression of the Anthurium andraeanum (Hort.) R2R3-MYB genes AaMYB4 and AaMYB5 enhance the flower color in transgenic tobacco. Plant Cell Tissue Organ Cult. PCTOC 2019, 139, 105–117. [Google Scholar] [CrossRef]
- Li, C.; Yu, W.; Xu, J.; Lu, X.; Liu, Y. Anthocyanin Biosynthesis Induced by MYB Transcription Factors in Plants. Int. J. Mol. Sci. 2022, 23, 11701. [Google Scholar] [CrossRef]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Khaldun, A.B.; Lv, H.; Du, L.; Zhang, C.; Wang, Y. Isolation and functional characterization of a R2R3-MYB regulator of the anthocyanin biosynthetic pathway from Epimedium sagittatum. Plant Cell Rep. 2016, 35, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020, 225, 2048–2063. [Google Scholar] [CrossRef]
- Ji, X.; Ren, J.; Zhang, Y.; Lang, S.; Wang, D.; Song, X. Integrated analysis of the metabolome and transcriptome on anthocyanin biosynthesis in four developmental stages of Cerasus humilis peel coloration. Int. J. Mol. Sci. 2021, 22, 11880. [Google Scholar] [CrossRef]
- Yamagishi, M.; Ihara, H.; Arakawa, K.; Toda, S.; Suzuki, K. The origin of the LhMYB12 gene, which regulates anthocyanin pigmentation of tepals, in Oriental and Asiatic hybrid lilies (Lilium spp.). Sci. Hortic. 2014, 174, 119–125. [Google Scholar] [CrossRef]
- Dooner, H.K.; Robbins, T.P.; Jorgensen, R.A. Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 1991, 25, 173–199. [Google Scholar] [CrossRef]
- Feng, C.Y.; Li, S.S.; Yin, D.D.; Zhang, H.J.; Tian, D.K.; Wu, Q.; Wang, L.J.; Su, S.; Wang, L.S. Rapid determination of flavonoids in plumules of sacred lotus cultivars and assessment of their antioxidant activities. Ind. Crop. Prod. 2016, 87, 96–104. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuka, T.; Haruta, K.S.; Pitaksutheepong, C.; Abe, Y.; Kakizaki, Y.; Yamamoto, K.; Shimada, N.; Yamamura, S.; Nishihara, M. Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Physiol. 2008, 49, 1818–1829. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.R.; Allan, A.C.; Chen, K.S.; Ferguson, I.B. Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol. 2010, 153, 1280–1292. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, K.; Hou, Y.; Wu, M.; Pan, Q.; Xie, Y.; Zhang, Y.; Sun, F.; Zhang, Z.; Wu, J. DoMYB5 and DobHLH24, Transcription Factors Involved in Regulating Anthocyanin Accumulation in Dendrobium officinale. Int. J. Mol. Sci. 2023, 24, 7552. https://doi.org/10.3390/ijms24087552
Yang K, Hou Y, Wu M, Pan Q, Xie Y, Zhang Y, Sun F, Zhang Z, Wu J. DoMYB5 and DobHLH24, Transcription Factors Involved in Regulating Anthocyanin Accumulation in Dendrobium officinale. International Journal of Molecular Sciences. 2023; 24(8):7552. https://doi.org/10.3390/ijms24087552
Chicago/Turabian StyleYang, Kun, Yibin Hou, Mei Wu, Qiuyu Pan, Yilong Xie, Yusen Zhang, Fenghang Sun, Zhizhong Zhang, and Jinghua Wu. 2023. "DoMYB5 and DobHLH24, Transcription Factors Involved in Regulating Anthocyanin Accumulation in Dendrobium officinale" International Journal of Molecular Sciences 24, no. 8: 7552. https://doi.org/10.3390/ijms24087552
APA StyleYang, K., Hou, Y., Wu, M., Pan, Q., Xie, Y., Zhang, Y., Sun, F., Zhang, Z., & Wu, J. (2023). DoMYB5 and DobHLH24, Transcription Factors Involved in Regulating Anthocyanin Accumulation in Dendrobium officinale. International Journal of Molecular Sciences, 24(8), 7552. https://doi.org/10.3390/ijms24087552





