Signals and Their Perception for Remodelling, Adjustment and Repair of the Plant Cell Wall
Abstract
1. Introduction
2. Structure and Polysaccharides of the Plant Cell Wall
2.1. Cellulose
2.2. Hemicelluloses
2.3. Pectin
2.4. Proteins Associated with the Plant Cell Wall
3. Polysaccharide Degradation
3.1. Degradation of Polysaccharides by Lytic Polysaccharide Monooxygenases (LPMOs)
3.2. Berberine Bridge-Enzyme-like (BBE-like) Proteins Oxidize OGs and Cello-Oligomers to Prevent Hyper-Immunity
4. Perception of Cell Wall Breakdown Products
4.1. OG Perception by WAKs
4.2. Perception of Cellulose Breakdown Products and Mixed Linkage Glucan-Derived Oligosaccharides
5. MAL Domain RLKs
5.1. MALL-RLKs
5.2. MALL-LRR-RLKs
5.3. A Lesson from Symbiosis
5.4. MAL Domain-Containing RLKs
6. Mechanosensors in Monitoring CWI
6.1. Ideas from Fungi
6.2. Mechanosensing in Plants
6.2.1. MCA Proteins Are Mechanical Stress Sensors
6.2.2. OSCA Channels
6.2.3. The MALL-RLK BUPS1 Participates in Mechanoperception
6.2.4. PIEZO Ion Channels Operate at Vacuolar Membranes
6.2.5. MscS
7. The FASCILIN-LIKE ARABINOGALACTAN PROTEIN4 (FLA4)-FEI Pathway
8. eATP-Induced Signaling Occurs during Massive Cell Wall Alterations
9. Extracellular Pyridine Nucleotides Inform Neighboring Cells
10. Crosstalk and Downstream Signaling
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hrmova, M.; Stratilová, B.; Stratilová, E. Broad specific xyloglucan: Xyloglucosyl transferases are formidable players in the re-modelling of plant cell wall structures. Int. J. Mol. Sci. 2022, 23, 1656. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Chane, A.; Jung, M.; Lee, Y. Recent advances in understanding the roles of pectin as an active participant in plant signaling networks. Plants 2021, 10, 1712. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J. Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 2008, 11, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Kozlova, L.V.; Nazipova, A.R.; Gorshkov, O.V.; Petrova, A.A.; Gorshkova, T.A. Elongating maize root: Zone-specific combinations of polysaccharides from type I and type II primary cell walls. Sci. Rep. 2020, 10, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Gigli-Bisceglia, N.; Engelsdorf, T.; Hamann, T. Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cell. Mol. Life Sci. 2020, 77, 2049–2077. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.-H.; Scholz, S.S.; Fliegmann, J.; Krüger, T.; Gandhi, A.; Furch, A.C.; Kniemeyer, O.; Brakhage, A.A.; Oelmüller, R. CORK1, A LRR-malectin receptor kinase, is required for cellooligomer-induced responses in Arabidopsis thaliana. Cells 2022, 11, 2960. [Google Scholar] [CrossRef]
- Martín, M.; Fernández-Calvo, P.; Jiménez-Sandoval, P.; López, G.; Garrido-Arandía, M.; Rebaque, D.; Del Hierro, I.; Berlanga, D.J.; Torres, M.Á.; Kumar, V.; et al. Arabidopsis immune responses triggered by cellulose and mixed-linked glucan oligosaccharides require a group of Leucine-Rich Repeat-Malectin Receptor Kinases. Plant J. 2023, 113, 833–850. [Google Scholar] [CrossRef]
- Zarattini, M.; Corso, M.; Kadowaki, M.A.; Monclaro, A.; Magri, S.; Milanese, I.; Jolivet, S.; de Godoy, M.O.; Hermans, C.; Fagard, M. LPMO-oxidized cellulose oligosaccharides evoke immunity in Arabidopsis conferring resistance towards necrotrophic fungus B. cinerea. Commun. Biol. 2021, 4, 727. [Google Scholar] [CrossRef]
- Souza, C.d.A.; Li, S.; Lin, A.Z.; Boutrot, F.; Grossmann, G.; Zipfel, C.; Somerville, S.C. Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiol. 2017, 173, 2383–2398. [Google Scholar] [CrossRef]
- Johnson, J.M.; Thürich, J.; Petutschnig, E.K.; Altschmied, L.; Meichsner, D.; Sherameti, I.; Dindas, J.; Mrozinska, A.; Paetz, C.; Scholz, S.S. A poly (A) ribonuclease controls the cellotriose-based interaction between Piriformospora indica and its host Arabidopsis. Plant Physiol. 2018, 176, 2496–2514. [Google Scholar] [CrossRef]
- Locci, F.; Benedetti, M.; Pontiggia, D.; Citterico, M.; Caprari, C.; Mattei, B.; Cervone, F.; De Lorenzo, G. An Arabidopsis berberine bridge enzyme-like protein specifically oxidizes cellulose oligomers and plays a role in immunity. Plant J. 2019, 98, 540–554. [Google Scholar] [CrossRef] [PubMed]
- Aziz, A.; Gauthier, A.; Bézier, A.; Poinssot, B.; Joubert, J.-M.; Pugin, A.; Heyraud, A.; Baillieul, F. Elicitor and resistance-inducing activities of β-1, 4 cellodextrins in grapevine, comparison with β-1, 3 glucans and α-1, 4 oligogalacturonides. J. Exp. Bot. 2007, 58, 1463–1472. [Google Scholar] [CrossRef] [PubMed]
- Melida, H.; Sopeña-Torres, S.; Bacete, L.; Garrido-Arandia, M.; Jordá, L.; Lopez, G.; Muñoz-Barrios, A.; Pacios, L.F.; Molina, A. Non-branched β-1, 3-glucan oligosaccharides trigger immune responses in Arabidopsis. Plant J. 2018, 93, 34–49. [Google Scholar] [CrossRef] [PubMed]
- Wanke, A.; Rovenich, H.; Schwanke, F.; Velte, S.; Becker, S.; Hehemann, J.H.; Wawra, S.; Zuccaro, A. Plant species-specific recognition of long and short β-1, 3-linked glucans is mediated by different receptor systems. Plant J. 2020, 102, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Galletti, R.; Denoux, C.; Gambetta, S.; Dewdney, J.; Ausubel, F.M.; De Lorenzo, G.; Ferrari, S. The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol. 2008, 148, 1695–1706. [Google Scholar] [CrossRef] [PubMed]
- Gamir, J.; Minchev, Z.; Berrio, E.; García, J.M.; De Lorenzo, G.; Pozo, M.J. Roots drive oligogalacturonide-induced systemic immunity in tomato. Plant Cell Environ. 2021, 44, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Melida, H.; Bacete, L.; Ruprecht, C.; Rebaque, D.; Del Hierro, I.; Lopez, G.; Brunner, F.; Pfrengle, F.; Molina, A. Arabinoxylan-oligosaccharides act as damage associated molecular patterns in plants regulating disease resistance. Front. Plant Sci. 2020, 11, 1210. [Google Scholar] [CrossRef] [PubMed]
- Rebaque, D.; Del Hierro, I.; López, G.; Bacete, L.; Vilaplana, F.; Dallabernardina, P.; Pfrengle, F.; Jordá, L.; Sánchez-Vallet, A.; Pérez, R. Cell wall-derived mixed-linked β-1, 3/1, 4-glucans trigger immune responses and disease resistance in plants. Plant J. 2021, 106, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Liu, R.; Pang, J.; Ren, B.; Zhou, H.; Wang, G.; Wang, E.; Liu, J. Poaceae-specific cell wall-derived oligosaccharides activate plant immunity via OsCERK1 during Magnaporthe oryzae infection in rice. Nat. Commun. 2021, 12, 2178. [Google Scholar] [CrossRef]
- Barghahn, S.; Arnal, G.; Jain, N.; Petutschnig, E.; Brumer, H.; Lipka, V. Mixed linkage β-1, 3/1, 4-glucan oligosaccharides induce defense responses in Hordeum vulgare and Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 682439. [Google Scholar] [CrossRef]
- Claverie, J.; Balacey, S.; Lemaître-Guillier, C.; Brulé, D.; Chiltz, A.; Granet, L.; Noirot, E.; Daire, X.; Darblade, B.; Héloir, M.-C. The cell wall-derived xyloglucan is a new DAMP triggering plant immunity in Vitis vinifera and Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 1725. [Google Scholar] [CrossRef] [PubMed]
- Zang, H.; Xie, S.; Zhu, B.; Yang, X.; Gu, C.; Hu, B.; Gao, T.; Chen, Y.; Gao, X. Mannan oligosaccharides trigger multiple defence responses in rice and tobacco as a novel danger-associated molecular pattern. Mol. Plant Pathol. 2019, 20, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Polko, J.K.; Kieber, J.J. The regulation of cellulose biosynthesis in plants. Plant Cell 2019, 31, 282–296. [Google Scholar] [CrossRef]
- Rongpipi, S.; Ye, D.; Gomez, E.D.; Gomez, E.W. Progress and opportunities in the characterization of cellulose–an important regulator of cell wall growth and mechanics. Front. Plant Sci. 2019, 9, 1894. [Google Scholar] [CrossRef]
- Daras, G.; Templalexis, D.; Avgeri, F.; Tsitsekian, D.; Karamanou, K.; Rigas, S. Updating Insights into the Catalytic Domain Properties of Plant Cellulose synthase (CesA) and Cellulose synthase-like (Csl) Proteins. Molecules 2021, 26, 4335. [Google Scholar] [CrossRef]
- Guerriero, G.; Fugelstad, J.; Bulone, V. What do we really know about cellulose biosynthesis in higher plants? J. Integr. Plant Biol. 2010, 52, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Atanassov, I.; Turner, S. Functional analysis of cellulose synthase (CESA) protein class specificity. Plant Physiol. 2017, 173, 970–983. [Google Scholar] [CrossRef]
- Griffiths, J.S.; North, H.M. Sticking to cellulose: Exploiting Arabidopsis seed coat mucilage to understand cellulose biosynthesis and cell wall polysaccharide interactions. New Phytol. 2017, 214, 959–966. [Google Scholar] [CrossRef]
- Wilson, T.H.; Kumar, M.; Turner, S.R. The molecular basis of plant cellulose synthase complex organisation and assembly. Biochem. Soc. Trans. 2021, 49, 379–391. [Google Scholar] [CrossRef]
- Jones, D.M.; Murray, C.M.; Ketelaar, K.J.; Thomas, J.J.; Villalobos, J.A.; Wallace, I.S. The emerging role of protein phosphorylation as a critical regulatory mechanism controlling cellulose biosynthesis. Front. Plant Sci. 2016, 7, 684. [Google Scholar] [CrossRef]
- Taylor, N.G.; Laurie, S.; Turner, S.R. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 2000, 12, 2529–2539. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.G.; Scheible, W.-R.; Cutler, S.; Somerville, C.R.; Turner, S.R. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 1999, 11, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ehrhardt, D.W.; Somerville, C.R. Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc. Natl. Acad. Sci. USA 2010, 107, 17188–17193. [Google Scholar] [CrossRef]
- Li, S.; Lei, L.; Yingling, Y.G.; Gu, Y. Microtubules and cellulose biosynthesis: The emergence of new players. Curr. Opin. Plant Biol. 2015, 28, 76–82. [Google Scholar] [CrossRef]
- Endler, A.; Kesten, C.; Schneider, R.; Zhang, Y.; Ivakov, A.; Froehlich, A.; Funke, N.; Persson, S. A mechanism for sustained cellulose synthesis during salt stress. Cell 2015, 162, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Speicher, T.L.; Li, P.Z.; Wallace, I.S. Phosphoregulation of the plant cellulose synthase complex and cellulose synthase-like proteins. Plants 2018, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Li, S.; Gu, Y. Cellulose synthase interactive protein 1 (CSI1) mediates the intimate relationship between cellulose microfibrils and cortical microtubules. Plant Signal. Behav. 2012, 7, 714–718. [Google Scholar] [CrossRef]
- Lei, L.; Singh, A.; Bashline, L.; Li, S.; Yingling, Y.G.; Gu, Y. CELLULOSE SYNTHASE INTERACTIVE1 is required for fast recycling of cellulose synthase complexes to the plasma membrane in Arabidopsis. Plant Cell 2015, 27, 2926–2940. [Google Scholar] [CrossRef]
- Zhu, Y.; McFarlane, H.E. Regulation of cellulose synthesis via exocytosis and endocytosis. Curr. Opin. Plant Biol. 2022, 69, 102273. [Google Scholar] [CrossRef]
- Ellis, C.; Karafyllidis, I.; Wasternack, C.; Turner, J.G. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 2002, 14, 1557–1566. [Google Scholar] [CrossRef]
- Bacete, L.; Melida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Blanco, C.; Feng, D.X.; Hu, J.; Sánchez-Vallet, A.; Deslandes, L.; Llorente, F.; Berrocal-Lobo, M.; Keller, H.; Barlet, X.; Sánchez-Rodríguez, C. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 2007, 19, 890–903. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, V.; Agorio, A.; Coego, A.; García-Andrade, J.; Hernández, M.J.; Balaguer, B.; Ouwerkerk, P.B.; Zarra, I.; Vera, P. MYB46 modulates disease susceptibility to Botrytis cinerea in Arabidopsis. Plant Physiol. 2011, 155, 1920–1935. [Google Scholar] [CrossRef] [PubMed]
- Escudero, V.; Jordá, L.; Sopeña-Torres, S.; Melida, H.; Miedes, E.; Muñoz-Barrios, A.; Swami, S.; Alexander, D.; McKee, L.S.; Sánchez-Vallet, A. Alteration of cell wall xylan acetylation triggers defense responses that counterbalance the immune deficiencies of plants impaired in the β-subunit of the heterotrimeric G-protein. Plant J. 2017, 92, 386–399. [Google Scholar] [CrossRef]
- Chen, Z.; Hong, X.; Zhang, H.; Wang, Y.; Li, X.; Zhu, J.K.; Gong, Z. Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 2005, 43, 273–283. [Google Scholar] [CrossRef]
- Sánchez-Vallet, A.; López, G.; Ramos, B.; Delgado-Cerezo, M.; Riviere, M.-P.; Llorente, F.; Fernández, P.V.; Miedes, E.; Estevez, J.M.; Grant, M. Disruption of abscisic acid signaling constitutively activates Arabidopsis resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant Physiol. 2012, 160, 2109–2124. [Google Scholar] [CrossRef]
- Douchkov, D.; Lueck, S.; Hensel, G.; Kumlehn, J.; Rajaraman, J.; Johrde, A.; Doblin, M.S.; Beahan, C.T.; Kopischke, M.; Fuchs, R. The barley (Hordeum vulgare) cellulose synthase-like D2 gene (HvCslD2) mediates penetration resistance to host-adapted and nonhost isolates of the powdery mildew fungus. New Phytol. 2016, 212, 421–433. [Google Scholar] [CrossRef]
- Caño-Delgado, A.; Penfield, S.; Smith, C.; Catley, M.; Bevan, M. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 2003, 34, 351–362. [Google Scholar] [CrossRef]
- Manfield, I.W.; Orfila, C.; McCartney, L.; Harholt, J.; Bernal, A.J.; Scheller, H.V.; Gilmartin, P.M.; Mikkelsen, J.D.; Paul Knox, J.; Willats, W.G. Novel cell wall architecture of isoxaben-habituated Arabidopsis suspension-cultured cells: Global transcript profiling and cellular analysis. Plant J. 2004, 40, 260–275. [Google Scholar] [CrossRef]
- Hamann, T.; Bennett, M.; Mansfield, J.; Somerville, C. Identification of cell-wall stress as a hexose-dependent and osmosensitive regulator of plant responses. Plant J. 2009, 57, 1015–1026. [Google Scholar] [CrossRef]
- Mélida, H.; Largo-Gosens, A.; Novo-Uzal, E.; Santiago, R.; Pomar, F.; García, P.; García-Angulo, P.; Acebes, J.L.; Álvarez, J.; Encina, A. Ectopic lignification in primary cellulose-deficient cell walls of maize cell suspension cultures. J. Integr. Plant Biol. 2015, 57, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Largo-Gosens, A.; Encina, A.; de Castro, M.; Mélida, H.; Acebes, J.L.; García-Angulo, P.; Álvarez, J.M. Early habituation of maize (Zea mays) suspension-cultured cells to 2, 6-dichlorobenzonitrile is associated with the enhancement of antioxidant status. Physiol. Plant. 2016, 157, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Tateno, M.; Brabham, C.; DeBolt, S. Cellulose biosynthesis inhibitors–a multifunctional toolbox. J. Exp. Bot. 2016, 67, 533–542. [Google Scholar] [CrossRef]
- Julian, J.D.; Zabotina, O.A. Xyloglucan biosynthesis: From genes to proteins and their functions. Front. Plant Sci. 2022, 13, 920494. [Google Scholar] [CrossRef] [PubMed]
- Hv, S.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar]
- Pauly, M.; Gille, S.; Liu, L.; Mansoori, N.; de Souza, A.; Schultink, A.; Xiong, G. Hemicellulose biosynthesis. Planta 2013, 238, 627–642. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Xin, X.; Gu, Y. Cellulose and hemicellulose synthesis and their regulation in plant cells. In Extracellular Sugar-Based Biopolymers Matrices; Springer: Berlin/Heidelberg, Germany, 2019; pp. 303–353. [Google Scholar]
- Zhang, W.; Qin, W.; Li, H.; Wu, A.-M. Biosynthesis and transport of nucleotide sugars for plant hemicellulose. Front. Plant Sci. 2021, 12, 723128. [Google Scholar] [CrossRef]
- Yu, L.; Yoshimi, Y.; Cresswell, R.; Wightman, R.; Lyczakowski, J.J.; Wilson, L.F.; Ishida, K.; Stott, K.; Yu, X.; Charalambous, S.; et al. Eudicot primary cell wall glucomannan is related in synthesis, structure and function to xyloglucan. bioRxiv 2022. [Google Scholar] [CrossRef]
- Ishida, K.; Yokoyama, R. Reconsidering the function of the xyloglucan endotransglucosylase/hydrolase family. J. Plant Res. 2022, 135, 145–156. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, X.; Yao, W.; Gao, Y.; Zhao, K.; Guo, Q.; Zhou, B.; Jiang, T. Genome-wide identification and expression analysis of the xyloglucan endotransglucosylase/hydrolase gene family in poplar. BMC Genom. 2021, 22, 804. [Google Scholar] [CrossRef]
- Kim, S.-J.; Brandizzi, F. Advances in cell wall matrix research with a focus on mixed-linkage glucan. Plant Cell Physiol. 2021, 62, 1839–1846. [Google Scholar] [CrossRef]
- Bulone, V.; Schwerdt, J.G.; Fincher, G.B. Co-evolution of enzymes involved in plant cell wall metabolism in the grasses. Front. Plant Sci. 2019, 10, 1009. [Google Scholar] [CrossRef] [PubMed]
- Tryfona, T.; Bourdon, M.; Delgado Marques, R.; Busse-Wicher, M.; Vilaplana, F.; Stott, K.; Dupree, P. Grass xylan structural variation suggests functional specialisation and distinctive interaction with cellulose and lignin. Plant J. 2023, 113, 100–1020. [Google Scholar] [CrossRef]
- Kozlova, L.; Ageeva, M.; Ibragimova, N.; Gorshkova, T. Arrangement of mixed-linkage glucan and glucuronoarabinoxylan in the cell walls of growing maize roots. Ann. Bot. 2014, 114, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.M.; Zeef, L.A.; Ellis, J.; Goodacre, R.; Turner, S.R. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 2005, 17, 2281–2295. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.A.; Dubos, C.; Surman, C.; Willment, J.; Cullis, I.F.; Mansfield, S.D.; Campbell, M.M. Comparison of lignin deposition in three ectopic lignification mutants. New Phytol. 2005, 168, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Sampedro, J.; Pardo, B.; Gianzo, C.; Guitián, E.; Revilla, G.; Zarra, I. Lack of α-xylosidase activity in Arabidopsis alters xyloglucan composition and results in growth defects. Plant Physiol. 2010, 154, 1105–1115. [Google Scholar]
- Delgado-Cerezo, M.; Sánchez-Rodríguez, C.; Escudero, V.; Miedes, E.; Fernández, P.V.; Jordá, L.; Hernández-Blanco, C.; Sánchez-Vallet, A.; Bednarek, P.; Schulze-Lefert, P. Arabidopsis heterotrimeric G-protein regulates cell wall defense and resistance to necrotrophic fungi. Mol. Plant 2012, 5, 98–114. [Google Scholar] [CrossRef]
- Chowdhury, J.; Lück, S.; Rajaraman, J.; Douchkov, D.; Shirley, N.J.; Schwerdt, J.G.; Schweizer, P.; Fincher, G.B.; Burton, R.A.; Little, A. Altered expression of genes implicated in xylan biosynthesis affects penetration resistance against powdery mildew. Front. Plant Sci. 2017, 8, 445. [Google Scholar] [CrossRef]
- Llorente, F.; Alonso-Blanco, C.; Sánchez-Rodriguez, C.; Jorda, L.; Molina, A. ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 2005, 43, 165–180. [Google Scholar] [CrossRef]
- Sánchez-Rodríguez, C.; Estévez, J.M.; Llorente, F.; Hernández-Blanco, C.; Jordá, L.; Pagán, I.; Berrocal, M.; Marco, Y.; Somerville, S.; Molina, A. The ERECTA receptor-like kinase regulates cell wall–mediated resistance to pathogens in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2009, 22, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Liu, Z.; Xie, H.; Zhu, J.; Zhang, J.; Kraus, J.; Blaschnig, T.; Nehls, R.; Wang, H. Increased drought tolerance through the suppression of ESKMO1 gene and overexpression of CBF-related genes in Arabidopsis. PLoS ONE 2014, 9, e106509. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Mandaokar, A.; Chen, J.; Last, R.L.; Browse, J. Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. Plant J. 2007, 49, 786–799. [Google Scholar] [CrossRef] [PubMed]
- Lugan, R.; NIOGRET, M.F.; Kervazo, L.; Larher, F.R.; Kopka, J.; Bouchereau, A. Metabolome and water status phenotyping of Arabidopsis under abiotic stress cues reveals new insight into ESK1 function. Plant Cell Environ. 2009, 32, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Atmodjo, M.A.; Hao, Z.; Mohnen, D. Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 2013, 64, 747–779. [Google Scholar] [CrossRef]
- Wolf, S.; Mouille, G.; Pelloux, J. Homogalacturonan methyl-esterification and plant development. Mol. Plant 2009, 2, 851–860. [Google Scholar] [CrossRef]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- San Clemente, H.; Kolkas, H.; Canut, H.; Jamet, E. Plant Cell Wall Proteomes: The Core of Conserved Protein Families and the Case of Non-Canonical Proteins. Int. J. Mol. Sci. 2022, 23, 4273. [Google Scholar] [CrossRef]
- Ishida, K.; Noutoshi, Y. The function of the plant cell wall in plant–microbe interactions. Plant Physiol. Biochem. 2022, 192, 273–284. [Google Scholar] [CrossRef]
- Narváez-Barragán, D.A.; Tovar-Herrera, O.E.; Guevara-García, A.; Serrano, M.; Martinez-Anaya, C. Mechanisms of plant cell wall surveillance in response to pathogens, cell wall-derived ligands and the effect of expansins to infection resistance or susceptibility. Front. Plant Sci. 2022, 13, 969343. [Google Scholar] [CrossRef]
- De Lorenzo, G.; Cervone, F. Plant immunity by damage-associated molecular patterns (DAMPs). Essays Biochem. 2022, 66, 459–469. [Google Scholar] [PubMed]
- Wolf, S. Cell wall signaling in plant development and defense. Annu. Rev. Plant Biol. 2022, 73, 323–353. [Google Scholar] [CrossRef] [PubMed]
- Mitsumasu, K.; Seto, Y.; Yoshida, S. Apoplastic interactions between plants and plant root intruders. Front. Plant Sci. 2015, 6, 617. [Google Scholar] [CrossRef] [PubMed]
- Cascallares, M.; Setzes, N.; Marchetti, F.; López, G.A.; Distéfano, A.M.; Cainzos, M.; Zabaleta, E.; Pagnussat, G.C. A complex journey: Cell wall remodeling, interactions, and integrity during pollen tube growth. Front. Plant Sci. 2020, 11, 599247. [Google Scholar] [CrossRef]
- Swaminathan, S.; Lionetti, V.; Zabotina, O. Plant Cell Wall Integrity Perturbations and Priming for Defense. Plants 2022, 11, 3539. [Google Scholar] [CrossRef]
- Lorrai, R.; Ferrari, S. Host cell wall damage during pathogen infection: Mechanisms of perception and role in plant-pathogen interactions. Plants 2021, 10, 399. [Google Scholar] [CrossRef]
- Van Vu, B.; Itoh, K.; Nguyen, Q.B.; Tosa, Y.; Nakayashiki, H. Cellulases belonging to glycoside hydrolase families 6 and 7 contribute to the virulence of Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2012, 25, 1135–1141. [Google Scholar] [CrossRef]
- Chen, Q.; Rehman, S.; Smant, G.; Jones, J.T. Functional analysis of pathogenicity proteins of the potato cyst nematode Globodera rostochiensis using RNAi. Mol. Plant-Microbe Interact. 2005, 18, 621–625. [Google Scholar] [CrossRef]
- Hwang, I.S.; Oh, E.-J.; Lee, H.B.; Oh, C.-S. Functional characterization of two cellulase genes in the Gram-positive pathogenic bacterium Clavibacter michiganensis for wilting in tomato. Mol. Plant-Microbe Interact. 2019, 32, 491–501. [Google Scholar] [CrossRef]
- Villares, A.; Moreau, C.; Bennati-Granier, C.; Garajova, S.; Foucat, L.; Falourd, X.; Saake, B.; Berrin, J.-G.; Cathala, B. Lytic polysaccharide monooxygenases disrupt the cellulose fibers structure. Sci. Rep. 2017, 7, 40262. [Google Scholar] [CrossRef]
- Puchart, V.; Šuchová, K.; Biely, P. Xylanases of glycoside hydrolase family 30–An overview. Biotechnol. Adv. 2021, 47, 107704. [Google Scholar] [CrossRef] [PubMed]
- Manabe, Y.; Nafisi, M.; Verhertbruggen, Y.; Orfila, C.; Gille, S.; Rautengarten, C.; Cherk, C.; Marcus, S.E.; Somerville, S.; Pauly, M. Loss-of-function mutation of reduced wall acetylation 2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 2011, 155, 1068–1078. [Google Scholar] [CrossRef] [PubMed]
- Pogorelko, G.; Lionetti, V.; Fursova, O.; Sundaram, R.M.; Qi, M.; Whitham, S.A.; Bogdanove, A.J.; Bellincampi, D.; Zabotina, O.A. Arabidopsis and Brachypodium distachyon transgenic plants expressing Aspergillus nidulans acetylesterases have decreased degree of polysaccharide acetylation and increased resistance to pathogens. Plant Physiol. 2013, 162, 9–23. [Google Scholar] [CrossRef]
- Osorio, S.; Castillejo, C.; Quesada, M.A.; Medina-Escobar, N.; Brownsey, G.J.; Suau, R.; Heredia, A.; Botella, M.A.; Valpuesta, V. Partial demethylation of oligogalacturonides by pectin methyl esterase 1 is required for eliciting defence responses in wild strawberry (Fragaria vesca). Plant J. 2008, 54, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Bethke, G.; Thao, A.; Xiong, G.; Li, B.; Soltis, N.E.; Hatsugai, N.; Hillmer, R.A.; Katagiri, F.; Kliebenstein, D.J.; Pauly, M. Pectin biosynthesis is critical for cell wall integrity and immunity in Arabidopsis thaliana. Plant Cell 2016, 28, 537–556. [Google Scholar] [CrossRef]
- Lionetti, V.; Fabri, E.; De Caroli, M.; Hansen, A.R.; Willats, W.G.; Piro, G.; Bellincampi, D. Three pectin methylesterase inhibitors protect cell wall integrity for Arabidopsis immunity to Botrytis. Plant Physiol. 2017, 173, 1844–1863. [Google Scholar] [CrossRef]
- Del Corpo, D.; Fullone, M.R.; Miele, R.; Lafond, M.; Pontiggia, D.; Grisel, S.; Kieffer-Jaquinod, S.; Giardina, T.; Bellincampi, D.; Lionetti, V. AtPME17 is a functional Arabidopsis thaliana pectin methylesterase regulated by its PRO region that triggers PME activity in the resistance to Botrytis cinerea. Mol. Plant Pathol. 2020, 21, 1620–1633. [Google Scholar] [CrossRef] [PubMed]
- Spadoni, S.; Zabotina, O.; Di Matteo, A.; Mikkelsen, J.D.; Cervone, F.; De Lorenzo, G.; Mattei, B.; Bellincampi, D. Polygalacturonase-inhibiting protein interacts with pectin through a binding site formed by four clustered residues of arginine and lysine. Plant Physiol. 2006, 141, 557–564. [Google Scholar] [CrossRef]
- Agueero, C.B.; Uratsu, S.L.; Greve, C.; Powell, A.L.T.; Labavitch, J.M.; Meredith, C.P.; Dandekar, A.M. Evaluation of tolerance to Pierce’s disease and Botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol. Plant Pathol. 2005, 6, 43–51. [Google Scholar] [CrossRef]
- Ferrari, S.; Sella, L.; Janni, M.; De Lorenzo, G.; Favaron, F.; D’ovidio, R. Transgenic expression of polygalacturonase-inhibiting proteins in Arabidopsis and wheat increases resistance to the flower pathogen Fusarium graminearum. Plant Biol. 2012, 14, 31–38. [Google Scholar] [CrossRef]
- Lionetti, V.; Raiola, A.; Camardella, L.; Giovane, A.; Obel, N.; Pauly, M.; Favaron, F.; Cervone, F.; Bellincampi, D. Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol. 2007, 143, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Valente, M.T.; Infantino, A.; Aragona, M. Molecular and functional characterization of an endoglucanase in the phytopathogenic fungus Pyrenochaeta lycopersici. Curr. Genet. 2011, 57, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Jagadeeswaran, G.; Veale, L.; Mort, A.J. Do lytic polysaccharide monooxygenases aid in plant pathogenesis and herbivory? Trends Plant Sci. 2021, 26, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Cairo, J.P.L.F.; Cannella, D.; Oliveira, L.C.; Gonçalves, T.A.; Rubio, M.V.; Terrasan, C.R.; Tramontina, R.; Mofatto, L.S.; Carazzolle, M.F.; Garcia, W. On the roles of AA15 lytic polysaccharide monooxygenases derived from the termite Coptotermes gestroi. J. Inorg. Biochem. 2021, 216, 111316. [Google Scholar] [CrossRef] [PubMed]
- Langston, J.A.; Shaghasi, T.; Abbate, E.; Xu, F.; Vlasenko, E.; Sweeney, M.D. Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl. Environ. Microbiol. 2011, 77, 7007–7015. [Google Scholar]
- Vaaje-Kolstad, G.; Westereng, B.; Horn, S.J.; Liu, Z.; Zhai, H.; Sørlie, M.; Eijsink, V.G. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 2010, 330, 219–222. [Google Scholar] [CrossRef]
- Forsberg, Z.; Vaaje-Kolstad, G.; Westereng, B.; Bunæs, A.C.; Stenstrøm, Y.; MacKenzie, A.; Sørlie, M.; Horn, S.J.; Eijsink, V.G. Cleavage of cellulose by a CBM33 protein. Protein Sci. 2011, 20, 1479–1483. [Google Scholar] [CrossRef]
- Vermaas, J.V.; Crowley, M.F.; Beckham, G.T.; Payne, C.M. Effects of lytic polysaccharide monooxygenase oxidation on cellulose structure and binding of oxidized cellulose oligomers to cellulases. J. Phys. Chem. B 2015, 119, 6129–6143. [Google Scholar] [CrossRef]
- Johansen, K.S. Lytic polysaccharide monooxygenases: The microbial power tool for lignocellulose degradation. Trends Plant Sci. 2016, 21, 926–936. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Calderaro, F.; Bevers, L.E.; van den Berg, M.A. Oxidative power: Tools for assessing LPMO activity on cellulose. Biomolecules 2021, 11, 1098. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, Y.; Zheng, Y.; Hsieh, Y.S. Recent advances in screening methods for the functional investigation of lytic polysaccharide monooxygenases. Front. Chem. 2021, 9, 653754. [Google Scholar] [CrossRef] [PubMed]
- Sagarika, M.S.; Parameswaran, C.; Senapati, A.; Barala, J.; Mitra, D.; Prabhukarthikeyan, S.; Kumar, A.; Nayak, A.K.; Panneerselvam, P. Lytic polysaccharide monooxygenases (LPMOs) producing microbes: A novel approach for rapid recycling of agricultural wastes. Sci. Total Environ. 2022, 806, 150451. [Google Scholar] [CrossRef]
- Pontiggia, D.; Benedetti, M.; Costantini, S.; De Lorenzo, G.; Cervone, F. Dampening the DAMPs: How plants maintain the homeostasis of cell wall molecular patterns and avoid hyper-immunity. Front. Plant Sci. 2020, 11, 613259. [Google Scholar] [CrossRef]
- Benedetti, M.; Verrascina, I.; Pontiggia, D.; Locci, F.; Mattei, B.; De Lorenzo, G.; Cervone, F. Four Arabidopsis berberine bridge enzyme-like proteins are specific oxidases that inactivate the elicitor-active oligogalacturonides. Plant J. 2018, 94, 260–273. [Google Scholar] [CrossRef]
- Messenlehner, J.; Hetman, M.; Tripp, A.; Wallner, S.; Macheroux, P.; Gruber, K.; Daniel, B. The catalytic machinery of the FAD-dependent AtBBE-like protein 15 for alcohol oxidation: Y193 and Y479 form a catalytic base, Q438 and R292 an alkoxide binding site. Arch. Biochem. Biophys. 2021, 700, 108766. [Google Scholar] [CrossRef] [PubMed]
- Daniel, B.; Wallner, S.; Steiner, B.; Oberdorfer, G.; Kumar, P.; van der Graaff, E.; Roitsch, T.; Sensen, C.W.; Gruber, K.; Macheroux, P. Structure of a berberine bridge enzyme-like enzyme with an active site specific to the plant family Brassicaceae. PLoS ONE 2016, 11, e0156892. [Google Scholar] [CrossRef]
- Daniel, B.; Konrad, B.; Toplak, M.; Lahham, M.; Messenlehner, J.; Winkler, A.; Macheroux, P. The family of berberine bridge enzyme-like enzymes: A treasure-trove of oxidative reactions. Arch. Biochem. Biophys. 2017, 632, 88–103. [Google Scholar] [CrossRef]
- Liu, M.; Hu, J.; Zhang, A.; Dai, Y.; Chen, W.; He, Y.; Zhang, H.; Zheng, X.; Zhang, Z. Auxilin-like protein MoSwa2 promotes effector secretion and virulence as a clathrin uncoating factor in the rice blast fungus Magnaporthe oryzae. New Phytol. 2021, 230, 720–736. [Google Scholar] [CrossRef]
- Engelsdorf, T.; Gigli-Bisceglia, N.; Veerabagu, M.; McKenna, J.F.; Vaahtera, L.; Augstein, F.; Van der Does, D.; Zipfel, C.; Hamann, T. The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci. Signal. 2018, 11, eaao3070. [Google Scholar] [CrossRef]
- De Lorenzo, G.; Brutus, A.; Savatin, D.V.; Sicilia, F.; Cervone, F. Engineering plant resistance by constructing chimeric receptors that recognize damage-associated molecular patterns (DAMPs). Febs Lett. 2011, 585, 1521–1528. [Google Scholar] [CrossRef]
- Denoux, C.; Galletti, R.; Mammarella, N.; Gopalan, S.; Werck, D.; De Lorenzo, G.; Ferrari, S.; Ausubel, F.M.; Dewdney, J. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 2008, 1, 423–445. [Google Scholar] [CrossRef] [PubMed]
- Ngou, B.P.M.; Ding, P.; Jones, J.D. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 2022, 34, 1447–1478. [Google Scholar] [CrossRef] [PubMed]
- Del Hierro, I.; Melida, H.; Broyart, C.; Santiago, J.; Molina, A. Computational prediction method to decipher receptor–glycoligand interactions in plant immunity. Plant J. 2021, 105, 1710–1726. [Google Scholar] [CrossRef]
- Franck, C.M.; Westermann, J.; Boisson-Dernier, A. Plant malectin-like receptor kinases: From cell wall integrity to immunity and beyond. Annu. Rev. Plant Biol. 2018, 69, 301–328. [Google Scholar] [CrossRef]
- De Coninck, T.; Van Damme, E.J. Plant lectins: Handymen at the cell surface. Cell Surf. 2022, 8, 100091. [Google Scholar] [CrossRef] [PubMed]
- Bellande, K.; Bono, J.-J.; Savelli, B.; Jamet, E.; Canut, H. Plant lectins and lectin receptor-like kinases: How do they sense the outside? Int. J. Mol. Sci. 2017, 18, 1164. [Google Scholar] [CrossRef]
- Ortiz-Morea, F.A.; Liu, J.; Shan, L.; He, P. Malectin-like receptor kinases as protector deities in plant immunity. Nat. Plants 2022, 8, 27–37. [Google Scholar] [CrossRef]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. Bioessays 2006, 28, 799–808. [Google Scholar] [CrossRef]
- Klarzynski, O.; Plesse, B.; Joubert, J.-M.; Yvin, J.-C.; Kopp, M.; Kloareg, B.; Fritig, B. Linear β-1, 3 glucans are elicitors of defense responses in tobacco. Plant Physiol. 2000, 124, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, D.; Guo, L.; Pan, H.; Yvon, R.; Garman, S.; Wu, H.-M.; Cheung, A.Y. Malectin/Malectin-like domain-containing proteins: A repertoire of cell surface molecules with broad functional potential. Cell Surf. 2021, 7, 100056. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Biswas, R.; Lenz, H.D.; Rauhut, T.; Ranf, S.; Kemmerling, B.; Götz, F.; Glawischnig, E.; Lee, J.; Felix, G. Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 2007, 282, 32338–32348. [Google Scholar] [CrossRef] [PubMed]
- Versluys, M.; Toksoy Öner, E.; Van den Ende, W. Fructan oligosaccharide priming alters apoplastic sugar dynamics and improves resistance against Botrytis cinerea in chicory. J. Exp. Bot. 2022, 73, 4214–4235. [Google Scholar] [CrossRef] [PubMed]
- Brutus, A.; Sicilia, F.; Macone, A.; Cervone, F.; De Lorenzo, G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 2010, 107, 9452–9457. [Google Scholar] [CrossRef]
- Tang, W.; Lin, W.; Zhou, X.; Guo, J.; Dang, X.; Li, B.; Lin, D.; Yang, Z. Mechano-transduction via the pectin-FERONIA complex activates ROP6 GTPase signaling in Arabidopsis pavement cell morphogenesis. Curr. Biol. 2022, 32, 508–517.e503. [Google Scholar] [CrossRef]
- Ridley, B.L.; O’Neill, M.A.; Mohnen, D. Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 2001, 57, 929–967. [Google Scholar] [CrossRef]
- Ferrari, S.; Savatin, D.V.; Sicilia, F.; Gramegna, G.; Cervone, F.; Lorenzo, G.D. Oligogalacturonides: Plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 2013, 4, 49. [Google Scholar] [CrossRef]
- Savatin, D.V.; Bisceglia, N.G.; Marti, L.; Fabbri, C.; Cervone, F.; De Lorenzo, G. The Arabidopsis NUCLEUS-AND PHRAGMOPLAST-LOCALIZED KINASE1-related protein kinases are required for elicitor-induced oxidative burst and immunity. Plant Physiol. 2014, 165, 1188–1202. [Google Scholar] [CrossRef]
- Vallarino, J.G.; Osorio, S. Signaling role of oligogalacturonides derived during cell wall degradation. Plant Signal. Behav. 2012, 7, 1447–1449. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Pontiggia, D.; Raggi, S.; Cheng, Z.; Scaloni, F.; Ferrari, S.; Ausubel, F.M.; Cervone, F.; De Lorenzo, G. Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc. Natl. Acad. Sci. USA 2015, 112, 5533–5538. [Google Scholar] [CrossRef]
- Silva-Sanzana, C.; Estevez, J.M.; Blanco-Herrera, F. Influence of cell wall polymers and their modifying enzymes during plant–aphid interactions. J. Exp. Bot. 2020, 71, 3854–3864. [Google Scholar] [CrossRef]
- Ochoa-Meza, L.C.; Quintana-Obregón, E.A.; Vargas-Arispuro, I.; Falcón-Rodríguez, A.B.; Aispuro-Hernández, E.; Virgen-Ortiz, J.J.; Martínez-Téllez, M.Á. Oligosaccharins as elicitors of defense responses in wheat. Polymers 2021, 13, 3105. [Google Scholar] [CrossRef] [PubMed]
- Kohorn, B.D.; Johansen, S.; Shishido, A.; Todorova, T.; Martinez, R.; Defeo, E.; Obregon, P. Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 2009, 60, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Kohorn, B.D.; Kobayashi, M.; Johansen, S.; Friedman, H.P.; Fischer, A.; Byers, N. Wall-associated kinase 1 (WAK1) is crosslinked in endomembranes, and transport to the cell surface requires correct cell-wall synthesis. J. Cell Sci. 2006, 119, 2282–2290. [Google Scholar] [CrossRef]
- Cabrera, J.C.; Boland, A.; Messiaen, J.; Cambier, P.; Van Cutsem, P. Egg box conformation of oligogalacturonides: The time-dependent stabilization of the elicitor-active conformation increases its biological activity. Glycobiology 2008, 18, 473–482. [Google Scholar] [CrossRef]
- Verica, J.A.; He, Z.-H. The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 2002, 129, 455–459. [Google Scholar] [CrossRef]
- Verica, J.A.; Chae, L.; Tong, H.; Ingmire, P.; He, Z.-H. Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall-associated kinase-like kinase genes in Arabidopsis. Plant Physiol. 2003, 133, 1732–1746. [Google Scholar] [CrossRef]
- Diener, A.C.; Ausubel, F.M. RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 2005, 171, 305–321. [Google Scholar] [CrossRef]
- Chen, S.; Cui, L.; Wang, X. A plant cell wall-associated kinase encoding gene is dramatically downregulated during nematode infection of potato. Plant Signal. Behav. 2022, 17, 2004026. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, W.; Long, S.; Zhao, C. Maintenance of cell wall integrity under high salinity. Int. J. Mol. Sci. 2021, 22, 3260. [Google Scholar] [CrossRef]
- Feng, H.; Li, C.; Zhou, J.; Yuan, Y.; Feng, Z.; Shi, Y.; Zhao, L.; Zhang, Y.; Wei, F.; Zhu, H. A cotton WAKL protein interacted with a DnaJ protein and was involved in defense against Verticillium dahliae. Int. J. Biol. Macromol. 2021, 167, 633–643. [Google Scholar] [CrossRef]
- Li, H.; Zhou, S.-Y.; Zhao, W.-S.; Su, S.-C.; Peng, Y.-L. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 2009, 69, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Hurni, S.; Scheuermann, D.; Krattinger, S.G.; Kessel, B.; Wicker, T.; Herren, G.; Fitze, M.N.; Breen, J.; Presterl, T.; Ouzunova, M. The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. USA 2015, 112, 8780–8785. [Google Scholar] [CrossRef]
- Zuo, W.; Chao, Q.; Zhang, N.; Ye, J.; Tan, G.; Li, B.; Xing, Y.; Zhang, B.; Liu, H.; Fengler, K.A. A maize wall-associated kinase confers quantitative resistance to head smut. Nat. Genet. 2015, 47, 151–157. [Google Scholar] [CrossRef]
- Zhang, N.; Pombo, M.A.; Rosli, H.G.; Martin, G.B. Tomato wall-associated kinase SlWak1 depends on Fls2/Fls3 to promote apoplastic immune responses to Pseudomonas syringae. Plant Physiol. 2020, 183, 1869–1882. [Google Scholar] [CrossRef] [PubMed]
- Kanneganti, V.; Gupta, A.K. RNAi mediated silencing of a wall associated kinase, OsiWAK1 in Oryza sativa results in impaired root development and sterility due to anther indehiscence. Physiol. Mol. Biol. Plants 2011, 17, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Tong, H.; Selby, J.; DeWitt, J.; Peng, X.; He, Z.-H. Involvement of a cell wall-associated kinase, WAKL4, in Arabidopsis mineral responses. Plant Physiol. 2005, 139, 1704–1716. [Google Scholar] [CrossRef]
- Tripathi, R.K.; Aguirre, J.A.; Singh, J. Genome-wide analysis of wall associated kinase (WAK) gene family in barley. Genomics 2021, 113, 523–530. [Google Scholar] [CrossRef]
- Zhang, Z.; Ma, W.; Ren, Z.; Wang, X.; Zhao, J.; Pei, X.; Liu, Y.; He, K.; Zhang, F.; Huo, W. Characterization and expression analysis of wall-associated kinase (WAK) and WAK-like family in cotton. Int. J. Biol. Macromol. 2021, 187, 867–879. [Google Scholar] [CrossRef]
- Park, A.R.; Cho, S.K.; Yun, U.J.; Jin, M.Y.; Lee, S.H.; Sachetto-Martins, G.; Park, O.K. Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J. Biol. Chem. 2001, 276, 26688–26693. [Google Scholar] [CrossRef] [PubMed]
- Gramegna, G.; Modesti, V.; Savatin, D.V.; Sicilia, F.; Cervone, F.; De Lorenzo, G. GRP-3 and KAPP, encoding interactors of WAK1, negatively affect defense responses induced by oligogalacturonides and local response to wounding. J. Exp. Bot. 2016, 67, 1715–1729. [Google Scholar] [CrossRef]
- Mangeon, A.; Menezes-Salgueiro, A.D.; Sachetto-Martins, G. Start me up: Revision of evidences that AtGRP3 acts as a potential switch for AtWAK1. Plant Signal. Behav. 2017, 12, e1191733. [Google Scholar] [CrossRef]
- Mangeon, A.; Pardal, R.; Menezes-Salgueiro, A.D.; Duarte, G.L.; de Seixas, R.; Cruz, F.P.; Cardeal, V.; Magioli, C.; Ricachenevsky, F.K.; Margis, R. AtGRP3 is implicated in root size and aluminum response pathways in Arabidopsis. PLoS ONE 2016, 11, e0150583. [Google Scholar] [CrossRef]
- Sivaguru, M.; Ezaki, B.; He, Z.-H.; Tong, H.; Osawa, H.; Baluška, F.; Volkmann, D.; Matsumoto, H. Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 2003, 132, 2256–2266. [Google Scholar] [CrossRef]
- Lou, H.Q.; Fan, W.; Jin, J.F.; Xu, J.M.; Chen, W.W.; Yang, J.L.; Zheng, S.J. A NAC-type transcription factor confers aluminium resistance by regulating cell wall-associated receptor kinase 1 and cell wall pectin. Plant Cell Environ. 2020, 43, 463–478. [Google Scholar] [CrossRef]
- Schallus, T.; Jaeckh, C.; Fehér, K.; Palma, A.S.; Liu, Y.; Simpson, J.C.; Mackeen, M.; Stier, G.; Gibson, T.J.; Feizi, T. Malectin: A novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol. Biol. Cell 2008, 19, 3404–3414. [Google Scholar] [CrossRef] [PubMed]
- Schallus, T.; Fehér, K.; Sternberg, U.; Rybin, V.; Muhle-Goll, C. Analysis of the specific interactions between the lectin domain of malectin and diglucosides. Glycobiology 2010, 20, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.N.; Muhle-Goll, C.; Biskup, M.B. The Glc 2 Man 2-fragment of the N-glycan precursor–a novel ligand for the glycan-binding protein malectin? Org. Biomol. Chem. 2010, 8, 3294–3299. [Google Scholar] [CrossRef] [PubMed]
- Galli, C.; Bernasconi, R.; Soldà, T.; Calanca, V.; Molinari, M. Malectin participates in a backup glycoprotein quality control pathway in the mammalian ER. PLoS ONE 2011, 6, e16304. [Google Scholar] [CrossRef]
- Takeda, K.; Qin, S.-Y.; Matsumoto, N.; Yamamoto, K. Association of malectin with ribophorin I is crucial for attenuation of misfolded glycoprotein secretion. Biochem. Biophys. Res. Commun. 2014, 454, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Tannous, A.; Pisoni, G.B.; Hebert, D.N.; Molinari, M. N-linked sugar-regulated protein folding and quality control in the ER. Semin. Cell Dev. Biol. 2015, 41, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Qiu, T.; Yin, C.; Zhao, X.; Xu, G.; Qi, L.; Zhang, Y.; Peng, Y.; Zhao, W. The Rice Malectin Regulates Plant Cell Death and Disease Resistance by Participating in Glycoprotein Quality Control. Int. J. Mol. Sci. 2022, 23, 5819. [Google Scholar] [CrossRef]
- Cooper, G.M.; Hausman, R.E. The Cell: A Molecular Approach; ASM Press: Washington, DC, USA, 2007. [Google Scholar]
- Hématy, K.; Sado, P.-E.; Van Tuinen, A.; Rochange, S.; Desnos, T.; Balzergue, S.; Pelletier, S.; Renou, J.-P.; Höfte, H. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 2007, 17, 922–931. [Google Scholar] [CrossRef]
- Escobar-Restrepo, J.-M.; Huck, N.; Kessler, S.; Gagliardini, V.; Gheyselinck, J.; Yang, W.-C.; Grossniklaus, U. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 2007, 317, 656–660. [Google Scholar] [CrossRef]
- Hématy, K.; Höfte, H. Novel receptor kinases involved in growth regulation. Curr. Opin. Plant Biol. 2008, 11, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.Y.; Wu, H.-M. THESEUS 1, FERONIA and relatives: A family of cell wall-sensing receptor kinases? Curr. Opin. Plant Biol. 2011, 14, 632–641. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Kessler, S.A.; Grossniklaus, U. The walls have ears: The role of plant CrRLK1Ls in sensing and transducing extracellular signals. J. Exp. Bot. 2011, 62, 1581–1591. [Google Scholar] [CrossRef]
- Galindo-Trigo, S.; Gray, J.E.; Smith, L.M. Conserved roles of CrRLK1L receptor-like kinases in cell expansion and reproduction from algae to angiosperms. Front. Plant Sci. 2016, 7, 1269. [Google Scholar] [CrossRef]
- Doblas, V.G.; Gonneau, M.; Höfte, H. Cell wall integrity signaling in plants: Malectin-domain kinases and lessons from other kingdoms. Cell Surf. 2018, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S. Plant cell wall signalling and receptor-like kinases. Biochem. J. 2017, 474, 471–492. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Kessler, S.A.; Lindner, H.; Jones, D.S.; Grossniklaus, U. Functional analysis of related Cr RLK 1L receptor-like kinases in pollen tube reception. EMBO Rep. 2015, 16, 107–115. [Google Scholar] [CrossRef]
- Nissen, K.S.; Willats, W.G.; Malinovsky, F.G. Understanding CrRLK1L function: Cell walls and growth control. Trends Plant Sci. 2016, 21, 516–527. [Google Scholar] [CrossRef]
- Gonneau, M.; Desprez, T.; Martin, M.; Doblas, V.G.; Bacete, L.; Miart, F.; Sormani, R.; Hématy, K.; Renou, J.; Landrein, B. Receptor kinase THESEUS1 is a rapid alkalinization factor 34 receptor in Arabidopsis. Curr. Biol. 2018, 28, 2452–2458.e2454. [Google Scholar] [CrossRef]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef]
- Ge, Z.; Bergonci, T.; Zhao, Y.; Zou, Y.; Du, S.; Liu, M.-C.; Luo, X.; Ruan, H.; García-Valencia, L.E.; Zhong, S. Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 2017, 358, 1596–1600. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef]
- Huck, N.; Moore, J.M.; Federer, M.; Grossniklaus, U. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 2003, 130, 2149–2159. [Google Scholar] [CrossRef]
- Deslauriers, S.D.; Larsen, P.B. FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Mol. Plant 2010, 3, 626–640. [Google Scholar] [CrossRef]
- Ngo, Q.A.; Vogler, H.; Lituiev, D.S.; Nestorova, A.; Grossniklaus, U. A calcium dialog mediated by the FERONIA signal transduction pathway controls plant sperm delivery. Dev. Cell 2014, 29, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Li, J.; Huang, Y.; Liu, L.; Li, D.; Chen, L.; Luan, S. FERONIA receptor kinase controls seed size in Arabidopsis thaliana. Mol. Plant 2014, 7, 920–922. [Google Scholar] [CrossRef] [PubMed]
- Masachis, S.; Segorbe, D.; Turrà, D.; Leon-Ruiz, M.; Fürst, U.; El Ghalid, M.; Leonard, G.; López-Berges, M.S.; Richards, T.A.; Felix, G. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 2016, 1, 16043. [Google Scholar] [CrossRef]
- Feng, W.; Kita, D.; Peaucelle, A.; Cartwright, H.N.; Doan, V.; Duan, Q.; Liu, M.-C.; Maman, J.; Steinhorst, L.; Schmitz-Thom, I. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 2018, 28, 666–675.e665. [Google Scholar] [CrossRef]
- Li, L.; Chen, H.; Alotaibi, S.S.; Pěnčík, A.; Adamowski, M.; Novák, O.; Friml, J. RALF1 peptide triggers biphasic root growth inhibition upstream of auxin biosynthesis. Proc. Natl. Acad. Sci. USA 2022, 119, e2121058119. [Google Scholar] [CrossRef] [PubMed]
- Merino, M.C.; Guidarelli, M.; Negrini, F.; De Biase, D.; Pession, A.; Baraldi, E. Induced expression of the Fragaria× ananassa Rapid alkalinization factor-33-like gene decreases anthracnose ontogenic resistance of unripe strawberry fruit stages. Mol. Plant Pathol. 2019, 20, 1252–1263. [Google Scholar] [CrossRef]
- Kou, X.; Sun, J.; Wang, P.; Wang, D.; Cao, P.; Lin, J.; Chang, Y.; Zhang, S.; Wu, J. PbrRALF2-elicited reactive oxygen species signaling is mediated by the PbrCrRLK1L13-PbrMPK18 module in pear pollen tubes. Hortic. Res. 2021, 8, 222. [Google Scholar] [CrossRef]
- Xie, Y.-H.; Zhang, F.-J.; Sun, P.; Li, Z.-Y.; Zheng, P.-F.; Gu, K.-D.; Hao, Y.-J.; Zhang, Z.; You, C.-X. Apple receptor-like kinase FERONIA regulates salt tolerance and ABA sensitivity in Malus domestica. J. Plant Physiol. 2022, 270, 153616. [Google Scholar] [CrossRef]
- Gao, Q.; Wang, C.; Xi, Y.; Shao, Q.; Li, L.; Luan, S. A receptor–channel trio conducts Ca2+ signalling for pollen tube reception. Nature 2022, 607, 534–539. [Google Scholar] [CrossRef]
- Feng, H.; Liu, C.; Fu, R.; Zhang, M.; Li, H.; Shen, L.; Wei, Q.; Sun, X.; Xu, L.; Ni, B. LORELEI-LIKE GPI-ANCHORED PROTEINS 2/3 regulate pollen tube growth as chaperones and coreceptors for ANXUR/BUPS receptor kinases in Arabidopsis. Mol. Plant 2019, 12, 1612–1623. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Qu, L.J.; Xiao, J. Crystal structures of the extracellular domains of the CrRLK1L receptor-like kinases ANXUR1 and ANXUR2. Protein Sci. 2018, 27, 886–892. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liang, X.; Bao, Y.; Yang, S.; Zhang, X.; Yu, H.; Zhang, Q.; Xu, G.; Feng, X.; Dou, D. A malectin-like receptor kinase regulates cell death and pattern-triggered immunity in soybean. EMBO Rep. 2020, 21, e50442. [Google Scholar] [CrossRef] [PubMed]
- Sussholz, O.; Pizarro, L.; Schuster, S.; Avni, A. SlRLK-like is a malectin-like domain protein affecting localization and abundance of LeEIX2 receptor resulting in suppression of EIX-induced immune responses. Plant J. 2020, 104, 1369–1381. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yin, C.; Liu, J.; Feng, B.; Ge, D.; Kong, L.; Ortiz-Morea, F.A.; Richter, J.; Hauser, M.-T.; Wang, W.-M. A trimeric Cr RLK1L-LLG1 complex genetically modulates SUMM2-mediated autoimmunity. Nat. Commun. 2020, 11, 4859. [Google Scholar] [CrossRef]
- Liu, J.; Huang, Y.; Kong, L.; Yu, X.; Feng, B.; Liu, D.; Zhao, B.; Mendes, G.C.; Yuan, P.; Ge, D. The malectin-like receptor-like kinase LETUM1 modulates NLR protein SUMM2 activation via MEKK2 scaffolding. Nat. Plants 2020, 6, 1106–1115. [Google Scholar] [CrossRef] [PubMed]
- Hok, S.; Danchin, E.G.; Allasia, V.; Panabières, F.; Attard, A.; Keller, H. An Arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease. Plant Cell Environ. 2011, 34, 1944–1957. [Google Scholar] [CrossRef]
- Yeh, Y.-H.; Panzeri, D.; Kadota, Y.; Huang, Y.-C.; Huang, P.-Y.; Tao, C.-N.; Roux, M.; Chien, H.-C.; Chin, T.-C.; Chu, P.-W. The Arabidopsis malectin-like/LRR-RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 2016, 28, 1701–1721. [Google Scholar] [CrossRef]
- Yuan, N.; Yuan, S.; Li, Z.; Zhou, M.; Wu, P.; Hu, Q.; Mendu, V.; Wang, L.; Luo, H. STRESS INDUCED FACTOR 2, a leucine-rich repeat kinase regulates basal plant pathogen defense. Plant Physiol. 2018, 176, 3062–3080. [Google Scholar] [CrossRef]
- Chan, C.; Panzeri, D.; Okuma, E.; Tõldsepp, K.; Wang, Y.-Y.; Louh, G.-Y.; Chin, T.-C.; Yeh, Y.-H.; Yeh, H.-L.; Yekondi, S. STRESS INDUCED FACTOR 2 regulates Arabidopsis stomatal immunity through phosphorylation of the anion channel SLAC1. Plant Cell 2020, 32, 2216–2236. [Google Scholar] [CrossRef]
- Hajný, J.; Prát, T.; Rydza, N.; Rodriguez, L.; Tan, S.; Verstraeten, I.; Domjan, D.; Mazur, E.; Smakowska-Luzan, E.; Smet, W. Receptor kinase module targets PIN-dependent auxin transport during canalization. Science 2020, 370, 550–557. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhuang, Y.; Li, C.; Sun, X.; Zhao, S.; Ma, C.; Lin, H.; Zhou, H. SIMP1 modulates salt tolerance by elevating ERAD efficiency through UMP1A-mediated proteasome maturation in plants. New Phytol. 2021, 232, 625–641. [Google Scholar] [CrossRef]
- Jing, X.-Q.; Li, W.-Q.; Zhou, M.-R.; Shi, P.-T.; Zhang, R.; Shalmani, A.; Muhammad, I.; Wang, G.-F.; Liu, W.-T.; Chen, K.-M. Rice Carbohydrate-Binding Malectn-Like Protein, OsCBM1, Contributes to Drought-Stress Tolerance by Participating in NADPH Oxidase-Mediated ROS Production. Rice 2021, 14, 100. [Google Scholar] [CrossRef] [PubMed]
- Won, S.-K.; Lee, Y.-J.; Lee, H.-Y.; Heo, Y.-K.; Cho, M.; Cho, H.-T. Cis-element-and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 2009, 150, 1459–1473. [Google Scholar] [CrossRef] [PubMed]
- Antolín-Llovera, M.; Ried, M.K.; Parniske, M. Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr. Biol. 2014, 24, 422–427. [Google Scholar] [CrossRef]
- Antolín-Llovera, M.; Petutsching, E.K.; Ried, M.K.; Lipka, V.; Nürnberger, T.; Robatzek, S.; Parniske, M. Knowing your friends and foes–plant receptor-like kinases as initiators of symbiosis or defence. New Phytol. 2014, 204, 791–802. [Google Scholar] [CrossRef]
- Li, H.; Chen, M.; Duan, L.; Zhang, T.; Cao, Y.; Zhang, Z. Domain swap approach reveals the critical roles of different domains of SYMRK in root nodule symbiosis in Lotus japonicus. Front. Plant Sci. 2018, 9, 697. [Google Scholar] [CrossRef]
- Lerouge, P.; Roche, P.; Faucher, C.; Maillet, F.; Truchet, G.; Promé, J.C.; Dénarié, J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 1990, 344, 781–784. [Google Scholar] [CrossRef]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Grønlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef]
- Broghammer, A.; Krusell, L.; Blaise, M.; Sauer, J.; Sullivan, J.T.; Maolanon, N.; Vinther, M.; Lorentzen, A.; Madsen, E.B.; Jensen, K.J. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA 2012, 109, 13859–13864. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, W.; Sun, J.; Feng, F.; Deng, Y.; He, Z.; Oldroyd, G.E.; Wang, E. The receptor kinase CERK 1 has dual functions in symbiosis and immunity signalling. Plant J. 2015, 81, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Stracke, S.; Kistner, C.; Yoshida, S.; Mulder, L.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; Stougaard, J.; Szczyglowski, K. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 2002, 417, 959–962. [Google Scholar] [CrossRef]
- Mendy, B.; Wang’ombe, M.W.; Radakovic, Z.S.; Holbein, J.; Ilyas, M.; Chopra, D.; Holton, N.; Zipfel, C.; Grundler, F.M.; Siddique, S. Arabidopsis leucine-rich repeat receptor–like kinase NILR1 is required for induction of innate immunity to parasitic nematodes. PLoS Pathog. 2017, 13, e1006284. [Google Scholar] [CrossRef] [PubMed]
- Le, M.H.; Cao, Y.; Zhang, X.-C.; Stacey, G. LIK1, a CERK1-interacting kinase, regulates plant immune responses in Arabidopsis. PLoS ONE 2014, 9, e102245. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sanagi, M.; Lu, Y.; Nomura, Y.; Stolze, S.C.; Yasuda, S.; Saijo, Y.; Schulze, W.X.; Feil, R.; Stitt, M. Protein phosphorylation dynamics under carbon/nitrogen-nutrient stress and identification of a cell death-related receptor-like kinase in Arabidopsis. Front. Plant Sci. 2020, 11, 377. [Google Scholar] [CrossRef]
- Lee, H.K.; Goring, D.R. Two subgroups of receptor-like kinases promote early compatible pollen responses in the Arabidopsis thaliana pistil. J. Exp. Bot. 2021, 72, 1198–1211. [Google Scholar] [CrossRef]
- Onelli, E.; Idilli, A.I.; Moscatelli, A. Emerging roles for microtubules in angiosperm pollen tube growth highlight new research cues. Front. Plant Sci. 2015, 6, 51. [Google Scholar] [CrossRef]
- de Oliveira, H.C.; Rossi, S.A.; García-Barbazán, I.; Zaragoza, Ó.; Trevijano-Contador, N. Cell wall integrity pathway involved in morphogenesis, virulence and antifungal susceptibility in Cryptococcus neoformans. J. Fungi 2021, 7, 831. [Google Scholar] [CrossRef]
- Rodicio, R.; Heinisch, J.J. Together we are strong—Cell wall integrity sensors in yeasts. Yeast 2010, 27, 531–540. [Google Scholar] [CrossRef]
- Philip, B.; Levin, D.E. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 2001, 21, 271–280. [Google Scholar] [CrossRef]
- Jendretzki, A.; Wittland, J.; Wilk, S.; Straede, A.; Heinisch, J.J. How do I begin? Sensing extracellular stress to maintain yeast cell wall integrity. Eur. J. Cell Biol. 2011, 90, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Arroyo, J. The CWI pathway: Regulation of the transcriptional adaptive response to cell wall stress in yeast. J. Fungi 2017, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Jung, U.S.; Levin, D.E. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 1999, 34, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.N.; Jane, W.-N.; Verslues, P.E. Role of the putative osmosensor Arabidopsis histidine kinase1 in dehydration avoidance and low-water-potential response. Plant Physiol. 2013, 161, 942–953. [Google Scholar] [CrossRef] [PubMed]
- Engelsdorf, T.; Hamann, T. An update on receptor-like kinase involvement in the maintenance of plant cell wall integrity. Ann. Bot. 2014, 114, 1339–1347. [Google Scholar] [CrossRef]
- Tran, L.-S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef] [PubMed]
- Wormit, A.; Butt, S.M.; Chairam, I.; McKenna, J.F.; Nunes-Nesi, A.; Kjaer, L.; O’Donnelly, K.; Fernie, A.R.; Woscholski, R.; Barter, M.L. Osmosensitive changes of carbohydrate metabolism in response to cellulose biosynthesis inhibition. Plant Physiol. 2012, 159, 105–117. [Google Scholar] [CrossRef]
- Tojo, H.; Nakamura, A.; Ferjani, A.; Kazama, Y.; Abe, T.; Iida, H. A Method Enabling Comprehensive Isolation of Arabidopsis Mutants Exhibiting Unusual Root Mechanical Behavior. Front. Plant Sci. 2021, 12, 646404. [Google Scholar] [CrossRef]
- Burri, J.T.; Munglani, G.; Nelson, B.J.; Grossniklaus, U.; Vogler, H. Quantification of mechanical forces and physiological processes involved in pollen tube growth using microfluidics and microrobotics. In Pollen and Pollen Tube Biology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 275–292. [Google Scholar]
- Hartmann, F.P.; Tinturier, E.; Julien, J.-L.; Leblanc-Fournier, N. Between Stress and Response: Function and Localization of Mechanosensitive Ca2+ Channels in Herbaceous and Perennial Plants. Int. J. Mol. Sci. 2021, 22, 11043. [Google Scholar] [CrossRef]
- Mousavi, S.A.; Dubin, A.E.; Zeng, W.-Z.; Coombs, A.M.; Do, K.; Ghadiri, D.A.; Keenan, W.T.; Ge, C.; Zhao, Y.; Patapoutian, A. PIEZO ion channel is required for root mechanotransduction in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2102188118. [Google Scholar] [CrossRef]
- Moulia, B.; Bastien, R.; Chauvet-Thiry, H.; Leblanc-Fournier, N. Posture control in land plants: Growth, position sensing, proprioception, balance, and elasticity. J. Exp. Bot. 2019, 70, 3467–3494. [Google Scholar] [CrossRef] [PubMed]
- Shigematsu, H.; Iida, K.; Nakano, M.; Chaudhuri, P.; Iida, H.; Nagayama, K. Structural characterization of the mechanosensitive channel candidate MCA2 from Arabidopsis thaliana. PLoS ONE 2014, 9, e87724. [Google Scholar] [CrossRef] [PubMed]
- Kamano, S.; Kume, S.; Iida, K.; Lei, K.-J.; Nakano, M.; Nakayama, Y.; Iida, H. Transmembrane topologies of Ca2+-permeable mechanosensitive channels MCA1 and MCA2 in Arabidopsis thaliana. J. Biol. Chem. 2015, 290, 30901–30909. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, T.; Nakagawa, Y.; Mori, K.; Nakano, M.; Imamura, T.; Kataoka, H.; Terashima, A.; Iida, K.; Kojima, I.; Katagiri, T. MCA1 and MCA2 that mediate Ca2+ uptake have distinct and overlapping roles in Arabidopsis. Plant Physiol. 2010, 152, 1284–1296. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, K.; Iida, K.; Iida, H. MCAs in Arabidopsis are Ca2+-permeable mechanosensitive channels inherently sensitive to membrane tension. Nat. Commun. 2021, 12, 6074. [Google Scholar] [CrossRef]
- Hattori, T.; Otomi, Y.; Nakajima, Y.; Soga, K.; Wakabayashi, K.; Iida, H.; Hoson, T. MCA1 and MCA2 are involved in the response to hypergravity in Arabidopsis hypocotyls. Plants 2020, 9, 590. [Google Scholar] [CrossRef]
- Iida, H.; Furuichi, T.; Nakano, M.; Toyota, M.; Sokabe, M.; Tatsumi, H. New candidates for mechano-sensitive channels potentially involved in gravity sensing in Arabidopsis thaliana. Plant Biol. 2014, 16, 39–42. [Google Scholar] [CrossRef]
- Mori, K.; Renhu, N.; Naito, M.; Nakamura, A.; Shiba, H.; Yamamoto, T.; Suzaki, T.; Iida, H.; Miura, K. Ca2+-permeable mechanosensitive channels MCA1 and MCA2 mediate cold-induced cytosolic Ca2+ increase and cold tolerance in Arabidopsis. Sci. Rep. 2018, 8, 550. [Google Scholar] [CrossRef]
- Nakano, M.; Samejima, R.; Iida, H. Mechanosensitive channel candidate MCA2 is involved in touch-induced root responses in Arabidopsis. Front. Plant Sci. 2014, 5, 421. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Katagiri, T.; Shinozaki, K.; Qi, Z.; Tatsumi, H.; Furuichi, T.; Kishigami, A.; Sokabe, M.; Kojima, I.; Sato, S. Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc. Natl. Acad. Sci. USA 2007, 104, 3639–3644. [Google Scholar] [CrossRef]
- Furuichi, T.; Iida, H.; Sokabe, M.; Tatsumi, H. Expression of Arabidopsis MCA1 enhanced mechanosensitive channel activity in the Xenopus laevis oocyte plasma membrane. Plant Signal. Behav. 2012, 7, 1022–1026. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, T.; Takatani, S.; Motose, H.; Iida, H.; Takahashi, T. The root growth reduction in response to mechanical stress involves ethylene-mediated microtubule reorganization and transmembrane receptor-mediated signal transduction in Arabidopsis. Plant Cell Rep. 2021, 40, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Procko, C.; Murthy, S.; Keenan, W.T.; Mousavi, S.A.R.; Dabi, T.; Coombs, A.; Procko, E.; Baird, L.; Patapoutian, A.; Chory, J. Stretch-activated ion channels identified in the touch-sensitive structures of carnivorous Droseraceae plants. Elife 2021, 10, e64250. [Google Scholar] [CrossRef]
- Zhai, Y.; Wen, Z.; Han, Y.; Zhuo, W.; Wang, F.; Xi, C.; Liu, J.; Gao, P.; Zhao, H.; Wang, Y. Heterogeneous expression of plasma-membrane-localised OsOSCA1. 4 complements osmotic sensing based on hyperosmolality and salt stress in Arabidopsis osca1 mutant. Cell Calcium 2020, 91, 102261. [Google Scholar] [CrossRef]
- Zhai, Y.; Wen, Z.; Fang, W.; Wang, Y.; Xi, C.; Liu, J.; Zhao, H.; Wang, Y.; Han, S. Functional analysis of rice OSCA genes overexpressed in the arabidopsis osca1 mutant due to drought and salt stresses. Transgenic Res. 2021, 30, 811–820. [Google Scholar] [CrossRef]
- Maity, K.; Heumann, J.M.; McGrath, A.P.; Kopcho, N.J.; Hsu, P.-K.; Lee, C.-W.; Mapes, J.H.; Garza, D.; Krishnan, S.; Morgan, G.P. Cryo-EM structure of OSCA1. 2 from Oryza sativa elucidates the mechanical basis of potential membrane hyperosmolality gating. Proc. Natl. Acad. Sci. USA 2019, 116, 14309–14318. [Google Scholar] [CrossRef]
- Zhou, X.; Lu, J.; Zhang, Y.; Guo, J.; Lin, W.; Van Norman, J.M.; Qin, Y.; Zhu, X.; Yang, Z. Membrane receptor-mediated mechano-transduction maintains cell integrity during pollen tube growth within the pistil. Dev. Cell 2021, 56, 1030–1042.e1036. [Google Scholar] [CrossRef]
- Chesler, A.T.; Szczot, M. Portraits of a pressure sensor. Elife 2018, 7, e34396. [Google Scholar] [CrossRef]
- Zhang, Z.; Tong, X.; Liu, S.-Y.; Chai, L.-X.; Zhu, F.-F.; Zhang, X.-P.; Zou, J.-Z.; Wang, X.-B. Genetic analysis of a Piezo-like protein suppressing systemic movement of plant viruses in Arabidopsis thaliana. Sci. Rep. 2019, 9, 3187. [Google Scholar] [CrossRef]
- Fang, X.; Liu, B.; Shao, Q.; Huang, X.; Li, J.; Luan, S.; He, K. AtPiezo plays an important role in root cap mechanotransduction. Int. J. Mol. Sci. 2021, 22, 467. [Google Scholar] [CrossRef] [PubMed]
- Radin, I.; Richardson, R.A.; Coomey, J.H.; Weiner, E.R.; Bascom, C.S.; Li, T.; Bezanilla, M.; Haswell, E.S. Plant PIEZO homologs modulate vacuole morphology during tip growth. Science 2021, 373, 586–590. [Google Scholar] [CrossRef] [PubMed]
- Haswell, E.S.; Peyronnet, R.; Barbier-Brygoo, H.; Meyerowitz, E.M.; Frachisse, J.-M. Two MscS homologs provide mechanosensitive channel activities in the Arabidopsis root. Curr. Biol. 2008, 18, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, A.M.; Haswell, E.S. Charged pore-lining residues are required for normal channel kinetics in the eukaryotic mechanosensitive ion channel MSL1. Channels 2020, 14, 310–325. [Google Scholar] [CrossRef]
- Dünser, K.; Gupta, S.; Herger, A.; Feraru, M.I.; Ringli, C.; Kleine-Vehn, J. Extracellular matrix sensing by FERONIA and Leucine-Rich Repeat Extensins controls vacuolar expansion during cellular elongation in Arabidopsis thaliana. EMBO J. 2019, 38, e100353. [Google Scholar] [CrossRef]
- Basu, D.; Tian, L.; Debrosse, T.; Poirier, E.; Emch, K.; Herock, H.; Travers, A.; Showalter, A.M. Glycosylation of a fasciclin-like arabinogalactan-protein (SOS5) mediates root growth and seed mucilage adherence via a cell wall receptor-like kinase (FEI1/FEI2) pathway in Arabidopsis. PLoS ONE 2016, 11, e0145092. [Google Scholar] [CrossRef]
- Seifert, G.J. The FLA4-FEI pathway: A unique and mysterious signaling module related to cell wall structure and stress signaling. Genes 2021, 12, 145. [Google Scholar] [CrossRef]
- Van der Does, D.; Boutrot, F.; Engelsdorf, T.; Rhodes, J.; McKenna, J.F.; Vernhettes, S.; Koevoets, I.; Tintor, N.; Veerabagu, M.; Miedes, E. The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet. 2017, 13, e1006832. [Google Scholar] [CrossRef]
- Spari, D.; Beldi, G. Extracellular ATP as an Inter-Kingdom signaling molecule: Release mechanisms by bacteria and its implication on the host. Int. J. Mol. Sci. 2020, 21, 5590. [Google Scholar] [CrossRef]
- Oelmüller, R. Threat at one end of the plant: What travels to inform the other parts? Int. J. Mol. Sci. 2021, 22, 3152. [Google Scholar] [CrossRef]
- Pietrowska-Borek, M.; Dobrogojski, J.; Sobieszczuk-Nowicka, E.; Borek, S. New insight into plant signaling: Extracellular ATP and uncommon nucleotides. Cells 2020, 9, 345. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.; Tanaka, K. A crosstalk between extracellular ATP and jasmonate signaling pathways for plant defense. Plant Signal. Behav. 2018, 13, e1432229. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-Y.; Sivaguru, M.; Stacey, G. Extracellular ATP in plants. Visualization, localization, and analysis of physiological significance in growth and signaling. Plant Physiol. 2006, 142, 984–992. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-J.; Liu, Y.-S.; Wu, J.-Y. The signaling role of extracellular ATP and its dependence on Ca2+ flux in elicitation of Salvia miltiorrhiza hairy root cultures. Plant Cell Physiol. 2008, 49, 617–624. [Google Scholar] [CrossRef]
- Weerasinghe, R.R.; Swanson, S.J.; Okada, S.F.; Garrett, M.B.; Kim, S.-Y.; Stacey, G.; Boucher, R.C.; Gilroy, S.; Jones, A.M. Touch induces ATP release in Arabidopsis roots that is modulated by the heterotrimeric G-protein complex. FEBS Lett. 2009, 583, 2521–2526. [Google Scholar] [CrossRef]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef]
- Choi, J.; Tanaka, K.; Liang, Y.; Cao, Y.; Lee, S.Y.; Stacey, G. Extracellular ATP, a danger signal, is recognized by DORN1 in Arabidopsis. Biochem. J. 2014, 463, 429–437. [Google Scholar] [CrossRef]
- Song, C.J.; Steinebrunner, I.; Wang, X.; Stout, S.C.; Roux, S.J. Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in Arabidopsis. Plant Physiol. 2006, 140, 1222–1232. [Google Scholar] [CrossRef]
- Thomas, C.; Rajagopal, A.; Windsor, B.; Dudler, R.; Lloyd, A.; Roux, S.J. A role for ectophosphatase in xenobiotic resistance. Plant Cell 2000, 12, 519–533. [Google Scholar] [CrossRef]
- Rieder, B.; Neuhaus, H.E. Identification of an Arabidopsis plasma membrane–located ATP transporter important for anther development. Plant Cell 2011, 23, 1932–1944. [Google Scholar] [CrossRef]
- Lim, M.H.; Wu, J.; Yao, J.; Gallardo, I.F.; Dugger, J.W.; Webb, L.J.; Huang, J.; Salmi, M.L.; Song, J.; Clark, G. Apyrase suppression raises extracellular ATP levels and induces gene expression and cell wall changes characteristic of stress responses. Plant Physiol. 2014, 164, 2054–2067. [Google Scholar] [CrossRef] [PubMed]
- Clark, G.; Roux, S.J. Apyrases, extracellular ATP and the regulation of growth. Curr. Opin. Plant Biol. 2011, 14, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Clark, G.; Roux, S.J. Role of Ca2+ in mediating plant responses to extracellular ATP and ADP. Int. J. Mol. Sci. 2018, 19, 3590. [Google Scholar] [CrossRef]
- Feng, H.; Guan, D.; Bai, J.; Sun, K.; Jia, L. Extracellular ATP: A potential regulator of plant cell death. Mol. Plant Pathol. 2015, 16, 633–639. [Google Scholar] [CrossRef]
- Parsons, H.T.; Christiansen, K.; Knierim, B.; Carroll, A.; Ito, J.; Batth, T.S.; Smith-Moritz, A.M.; Morrison, S.; McInerney, P.; Hadi, M.Z. Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 2012, 159, 12–26. [Google Scholar] [CrossRef]
- Chiu, T.-Y.; Christiansen, K.; Moreno, I.; Lao, J.; Loqué, D.; Orellana, A.; Heazlewood, J.L.; Clark, G.; Roux, S.J. AtAPY1 and AtAPY2 function as Golgi-localized nucleoside diphosphatases in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 1913–1925. [Google Scholar] [CrossRef] [PubMed]
- Schiller, M.; Massalski, C.; Kurth, T.; Steinebrunner, I. The Arabidopsis apyrase AtAPY1 is localized in the Golgi instead of the extracellular space. BMC Plant Biol. 2012, 12, 123. [Google Scholar] [CrossRef]
- Wu, J.; Steinebrunner, I.; Sun, Y.; Butterfield, T.; Torres, J.; Arnold, D.; Gonzalez, A.; Jacob, F.; Reichler, S.; Roux, S.J. Apyrases (nucleoside triphosphate-diphosphohydrolases) play a key role in growth control in Arabidopsis. Plant Physiol. 2007, 144, 961–975. [Google Scholar] [CrossRef]
- Liu, X.; Wu, J.; Clark, G.; Lundy, S.; Lim, M.; Arnold, D.; Chan, J.; Tang, W.; Muday, G.K.; Gardner, G. Role for apyrases in polar auxin transport in Arabidopsis. Plant Physiol. 2012, 160, 1985–1995. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, C.; Zhang, X.; Deng, S.; Zhao, R.; Shen, X.; Chen, S. Extracellular ATP signaling and homeostasis in plant cells. Plant Signal. Behav. 2012, 7, 566–569. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, C.L.; Deng, S.R.; Lu, C.F.; Shen, X.; Zhou, X.Y.; Zheng, X.J.; Hu, Z.M.; Chen, S.L. An ATP signalling pathway in plant cells: Extracellular ATP triggers programmed cell death in Populus euphratica. Plant Cell Environ. 2012, 35, 893–916. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Dong, X.; Hao, W.; Gao, W.; Zhang, W.; Xia, S.; Liu, T.; Shang, Z. Heterotrimeric G protein-regulated Ca2+ influx and PIN2 asymmetric distribution are involved in Arabidopsis thaliana roots’ avoidance response to extracellular ATP. Front. Plant Sci. 2017, 8, 1522. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; De Sain, M.; Weide, R.; Gouget, A.; Klamer, S.; Canut, H.; Govers, F. The lectin receptor kinase LecRK-I. 9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 2011, 7, e1001327. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.-H.; Choi, J.; Stacey, G. Molecular mechanism of plant recognition of extracellular ATP. Protein Rev. 2017, 19, 233–253. [Google Scholar]
- Jewell, J.B.; Sowders, J.M.; He, R.; Willis, M.A.; Gang, D.R.; Tanaka, K. Extracellular ATP shapes a defense-related transcriptome both independently and along with other defense signaling pathways. Plant Physiol. 2019, 179, 1144–1158. [Google Scholar] [CrossRef]
- Jewell, J.B.; Tanaka, K. Transcriptomic perspective on extracellular ATP signaling: A few curious trifles. Plant Signal. Behav. 2019, 14, 1659079. [Google Scholar] [CrossRef]
- Nizam, S.; Qiang, X.; Wawra, S.; Nostadt, R.; Getzke, F.; Schwanke, F.; Dreyer, I.; Langen, G.; Zuccaro, A. Serendipita indica E5′ NT modulates extracellular nucleotide levels in the plant apoplast and affects fungal colonization. EMBO Rep. 2019, 20, e47430. [Google Scholar] [CrossRef]
- Tanaka, K.; Tóth, K.; Stacey, G. Role of ectoapyrases in nodulation. In Biological Nitrogen Fixation; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 517–524. [Google Scholar]
- Balagué, C.; Gouget, A.; Bouchez, O.; Souriac, C.; Haget, N.; Boutet-Mercey, S.; Govers, F.; Roby, D.; Canut, H. The Arabidopsis thaliana lectin receptor kinase LecRK-I. 9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol. Plant Pathol. 2017, 18, 937–948. [Google Scholar] [CrossRef]
- Wang, C.; Huang, X.; Li, Q.; Zhang, Y.; Li, J.-L.; Mou, Z. Extracellular pyridine nucleotides trigger plant systemic immunity through a lectin receptor kinase/BAK1 complex. Nat. Commun. 2019, 10, 4810. [Google Scholar] [CrossRef]
- Mou, Z. Extracellular pyridine nucleotides as immune elicitors in Arabidopsis. Plant Signal. Behav. 2017, 12, e25474. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, X.; Mou, Z. Comparison of nicotinamide adenine dinucleotide phosphate-induced immune responses against biotrophic and necrotrophic pathogens in Arabidopsis thaliana. Plant Signal. Behav. 2016, 11, e1169358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Mou, Z. Extracellular pyridine nucleotides induce PR gene expression and disease resistance in Arabidopsis. Plant J. 2009, 57, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, Y.; Kaya, H.; Hiraoka, G.; Yumoto, F.; Kimura, S.; Kadota, Y.; Hishinuma, H.; Senzaki, E.; Yamagoe, S.; Nagata, K. Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 2008, 283, 8885–8892. [Google Scholar] [CrossRef] [PubMed]
- Dubiella, U.; Seybold, H.; Durian, G.; Komander, E.; Lassig, R.; Witte, C.-P.; Schulze, W.X.; Romeis, T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 2013, 110, 8744–8749. [Google Scholar] [CrossRef]
- Kadota, Y.; Shirasu, K.; Zipfel, C. Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 2015, 56, 1472–1480. [Google Scholar] [CrossRef]
- Gandhi, A.; Tseng, Y.-H.; Oelmüller, R. The damage-associated molecular pattern cellotriose alters the phosphorylation pattern of proteins involved in cellulose synthesis and trans-Golgi trafficking in Arabidopsis Thaliana. Plant Signal. Behav. 2023, 18, 2184352. [Google Scholar] [CrossRef]
- Mangano, S.; Pacheco, J.M.; Marino-Buslje, C.; Estevez, J.M. How does pH fit in with oscillating polar growth? Trends Plant Sci. 2018, 23, 479–489. [Google Scholar] [CrossRef]
- Dora, S.; Terrett, O.M.; Sánchez-Rodríguez, C. Plant–microbe interactions in the apoplast: Communication at the plant cell wall. Plant Cell 2022, 34, 1532–1550. [Google Scholar] [CrossRef]
- Gámez-Arjona, F.M.; Sánchez-Rodríguez, C.; Montesinos, J.C. The root apoplastic pH as an integrator of plant signaling. Front. Plant Sci. 2022, 13, 931979. [Google Scholar] [CrossRef]
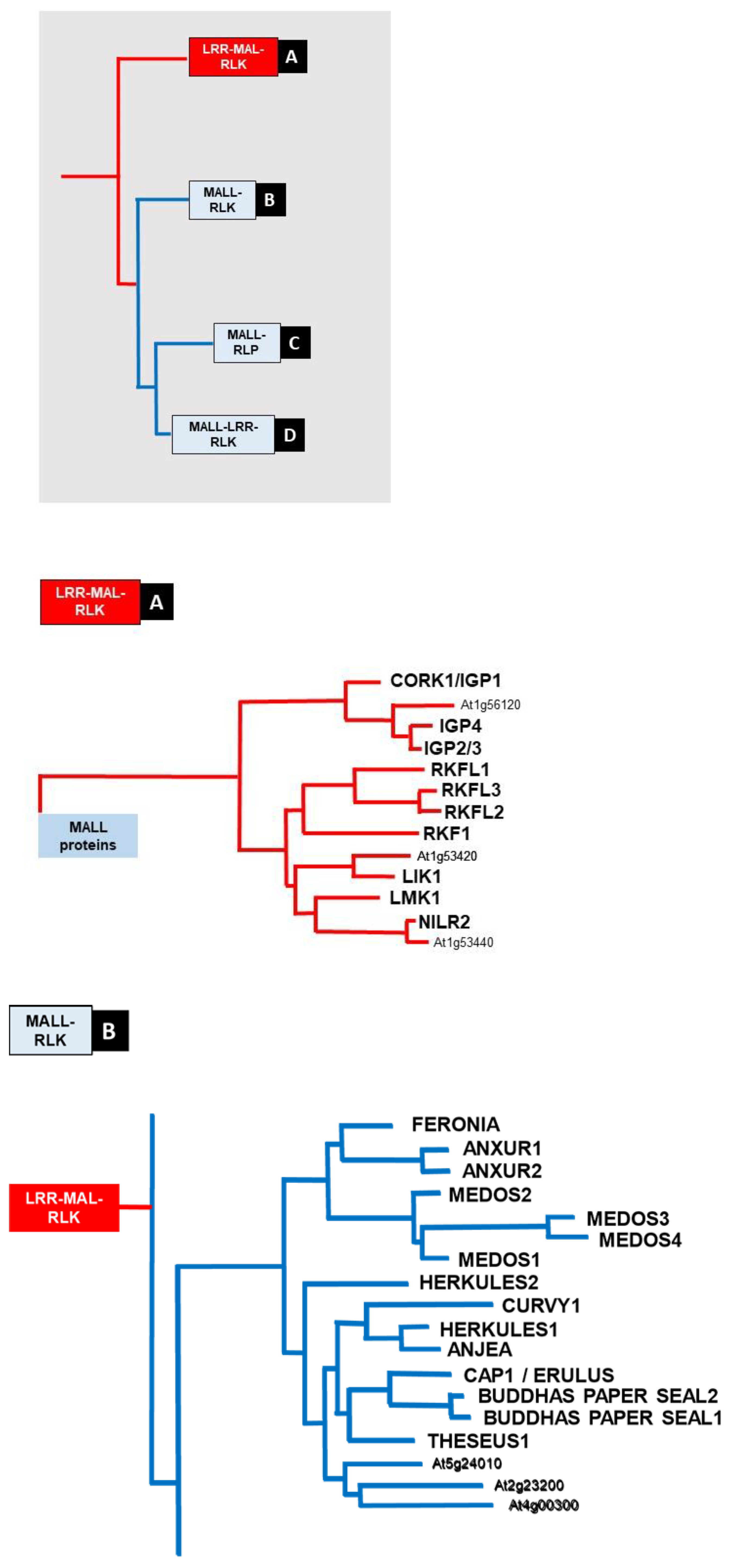

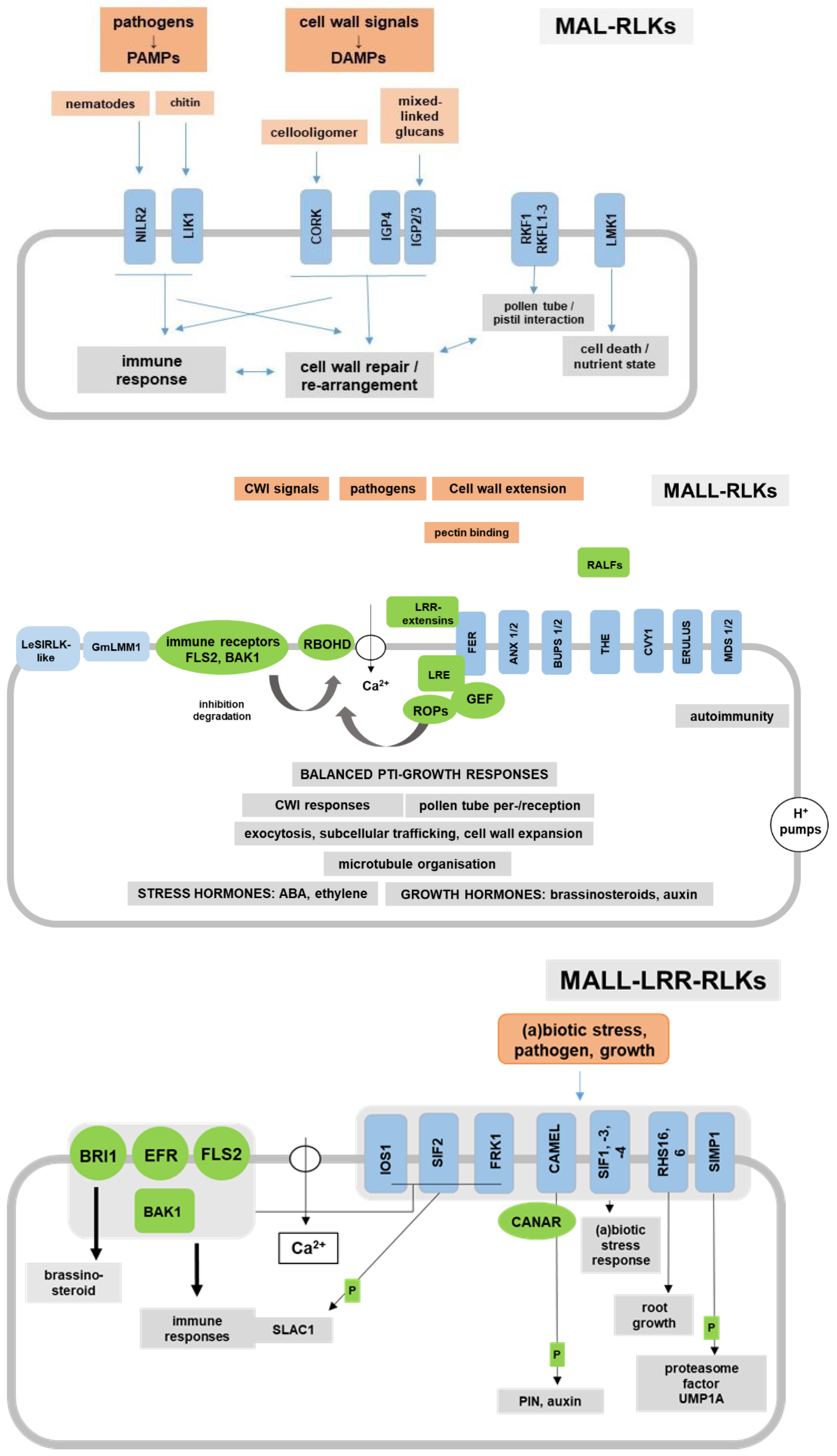
| Oligosaccharides | Origin | Receptor | References |
|---|---|---|---|
| cello-oligomer, n 2–7 (β-1,4-glucans) | cellulose | CORK1/IGP1 | [6,7,8,9,10,11,12] |
| β-1,3-glucans, non-branched | callose | CERK1 (for short-chain β-1,3-glucans), LYK4, LYK5 | [13,14] |
| α-1,4-oligogalacturonides (OGs) | pectin | WAKs | [12,13,15,16] |
| arabinoxylan | xylan | [17] | |
| mixed-linkage β-1,3/1,4 glucan oligosaccharides | mixed-linkage glucan polysaccharides | IGP2/3, IGP4 | [7,18,19,20] |
| xyloglucan oligosaccharides | xyloglucan | [21] | |
| mannan oligosaccharides, n = 2–6 | mannan polysaccharides | [22] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oelmüller, R.; Tseng, Y.-H.; Gandhi, A. Signals and Their Perception for Remodelling, Adjustment and Repair of the Plant Cell Wall. Int. J. Mol. Sci. 2023, 24, 7417. https://doi.org/10.3390/ijms24087417
Oelmüller R, Tseng Y-H, Gandhi A. Signals and Their Perception for Remodelling, Adjustment and Repair of the Plant Cell Wall. International Journal of Molecular Sciences. 2023; 24(8):7417. https://doi.org/10.3390/ijms24087417
Chicago/Turabian StyleOelmüller, Ralf, Yu-Heng Tseng, and Akanksha Gandhi. 2023. "Signals and Their Perception for Remodelling, Adjustment and Repair of the Plant Cell Wall" International Journal of Molecular Sciences 24, no. 8: 7417. https://doi.org/10.3390/ijms24087417
APA StyleOelmüller, R., Tseng, Y.-H., & Gandhi, A. (2023). Signals and Their Perception for Remodelling, Adjustment and Repair of the Plant Cell Wall. International Journal of Molecular Sciences, 24(8), 7417. https://doi.org/10.3390/ijms24087417





