Abstract
Early maturity is an important agronomic trait in most crops, because it can solve the problem of planting in stubble for multiple cropping as well as make full use of light and temperature resources in alpine regions, thereby avoiding damage from low temperatures in the early growth period and early frost damage in the late growth period to improve crop yield and quality. The expression of genes that determine flowering affects flowering time, which directly affects crop maturity and indirectly affects crop yield and quality. Therefore, it is important to analyze the regulatory network of flowering for the cultivation of early-maturing varieties. Foxtail millet (Setaria italica) is a reserve crop for future extreme weather and is also a model crop for functional gene research in C4 crops. However, there are few reports on the molecular mechanism regulating flowering in foxtail millet. A putative candidate gene, SiNF-YC2, was isolated based on quantitative trait loci (QTL) mapping analysis. Bioinformatics analysis showed that SiNF-YC2 has a conserved HAP5 domain, which indicates that it is a member of the NF-YC transcription factor family. The promoter of SiNF-YC2 contains light-response-, hormone-, and stress-resistance-related elements. The expression of SiNF-YC2 was sensitive to the photoperiod and was related to the regulation of biological rhythm. Expression also varied in different tissues and in response to drought and salt stress. In a yeast two-hybrid assay, SiNF-YC2 interacted with SiCO in the nucleus. Functional analysis suggested that SiNF-YC2 promotes flowering and improves resistance to salt stress.
1. Introduction
Early maturity is an important agronomic trait in most crops. It can solve the problem of planting in stubble for multiple cropping, improve the multiple cropping index, and increase the annual crop yield [1]. In particular, in high-latitude, high-altitude alpine regions with short frost-free periods, early maturity can make full use of light and temperature resources while also avoiding the damage caused by low temperatures in the early growth period and early frost in the late growth period to improve crop yield and quality [2]. Thus, in alpine regions, early maturity is important for safe crop production. The practices carried out in alpine regions of China are mostly a combination of agriculture and animal husbandry, with seriously deficient accumulated temperature and drought with little rain. The lack of effective crops with drought tolerance that provide both grain and grass is one of the main factors restricting the development of local economies. As a traditional characteristic crop in China, foxtail millet is drought-tolerant and can be used to provide both grain and grass, and therefore, it is suitable for development in arid and water-scarce alpine regions. However, because of the lack of early-maturing varieties that can mature normally in local areas, millet is difficult to plant in the alpine regions of China, restricting millet planting in those areas [3]. The cultivation of early-maturing millet varieties is the most suitable solution to the above problems. Thus, to expand the suitable planting range of millet, it is urgent to improve crop breeding and incorporate genes associated with early maturity. The discovery of early-maturity genes is vital in the breeding of early-maturity and high-yield crop varieties. An important index to evaluate the early maturity of crops is the time-of-flowering traits, and the expression of genes that determine flowering affects the time of flowering and directly controls the length of the crop growth period [4,5]. Therefore, it is essential to understand the network regulating foxtail millet flowering in order to develop early-maturing varieties and ultimately incorporate them into the planting structure of alpine regions.
The regulation of flowering time in plants is a self-adaptation to the environment and a major determinant of cereal crop yield. The photoperiod regulation of plant flowering has been extensively reported in Arabidopsis and rice [6,7,8,9]. To date, QTL mapping has been used to clone many genes involved in the photoperiod control of flowering pathways in rice, including Hd3a, Hd1, Ehd1, and Ghd7 [10,11,12]. Foxtail millet is widely regarded as a model C4 and energy crop [13,14], and the cloning and functional analysis of millet stress-related genes are increasing [15]. However, there are few studies on the photoperiod, mainly on the QTL mapping of photoperiod-sensitive correlations [16], and few reports on the screening, cloning, and functional identification of photoperiod-related genes.
The GI–CO–FT pathway is involved in photoperiodic responses in both dicots and monocots. The CO gene is a key gene in the photoperiodic response because it integrates light and clock signals, regulates interactions between various proteins, and thus plays a role in inhibiting or promoting flowering [10,17,18]. FLOWERING LOCUS T (FT), which belongs to the phosphatidylethanolamine binding protein (PEBP) family, controls flowering time by encoding a small protein called florigen. The CO-induced initiation of FT and other downstream genes must occur in combination with nuclear factor-Y (NF-Y) [19,20,21]. In an NF-Y transcription factor mutant, FT expression decreases [22]. Nuclear factor-Y is a class of transcription factors that are widely distributed in eukaryotes and are also known as heme-activator proteins (HAPs) or CCAAT-binding factors (CBFs). Nuclear factor-Y usually regulates the expression of downstream genes in the form of heterotrimers composed of three subunits: NF-YA (CBF-B/HAP2), NF-YB (CBF-A/HAP3), and NF-YC (CBF-C/HAP5) [23]. The transcription and protein levels of CO and NF-Y transcription factors change with time, and thus, the regulation of FT by NF-YA/B/C and NF-YB/C-CO trimers changes regularly to dynamically regulate the plant flowering process. The overexpression of a single NF-Y gene can alter plant flowering time [24], suggesting that NF-Y subunits are involved in regulating flowering time in a highly redundant and complex mechanism. However, how the CO gene regulates FT is not well understood.
In a previous study [25], 116 candidate genes were selected by mapping photoperiod-sensitive traits in millet, among which 4 were members of the NF-Y gene family. Studies on the functions of NF-Y family genes are limited to stress resistance [26,27], and little attention has been paid to the regulation of heading and flowering times in millet.
In this study, the candidate gene SiNF-YC2, associated with the regulation of flowering in foxtail millet, was cloned, and the biological characteristics and functions were examined. The SiNF-YC2 gene promoted early flowering and improved plant salt tolerance. The results of this study provide a foundation to study the mechanism regulating the early maturation of foxtail millet and also guidance to improve the molecular breeding of foxtail millet.
2. Results
2.1. Localization and Bioinformatics Analysis of SiNF-YC2
In our previous study [21], 116 candidate genes were predicted according to gene annotations. The SiNF-YC2 gene, which is located in the region of Block 63,401–63,475 on chromosome 9 (Figure 1), was identified as an important candidate gene based on the integrated results of gene annotation, cis-element analysis, and the expression patterns of candidate genes in different varieties that have different photoperiod sensitivities.
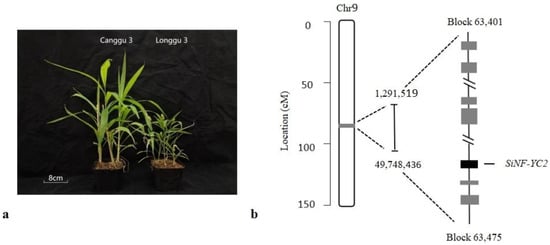
Figure 1.
QTL mapping and candidate gene screening for early-maturity traits in foxtail millet. (a) Heading phenotype of parent plants. (b) Location of SiNF-YC2 on the chromosome.
According to bioinformatics analysis, SiNF-YC2 is an unstable protein with an open coding frame of 738 bp and 245 amino acid residues. The content of hydrophilic amino acids in the SiNF-YC2 peptide chain is higher than that of hydrophobic amino acids, indicating that SiNF-YC2 is a hydrophilic protein (Supplementary Figure S1a). The whole SiNF-YC2 peptide chain does not have a transmembrane domain, indicating that it is not a transmembrane protein (Supplementary Figure S1b). According to signal peptide prediction, the SiNF-YC2 protein does not have a signal peptide, is a nonsecreted protein, and likely has multiple phosphorylation sites (Supplementary Figure S1c).
In the analysis of conserved SiNF-YC2 domains, a conserved HAP5 domain was predicted in the 49–148 amino acid region of the protein, indicating its membership in the CCAAT-box-bound NF-YC transcription factor family (Figure 2a). According to the analysis of the secondary structure, the SiNF-YC2 protein is composed of 28.98% α-helices (71 amino acids), 11.43% elongation links (28 amino acids), 5.71% β-angles (14 amino acids), and 53.88% random coils (132 amino acids) (Figure 2b). SWISS-MODEL (https://swissmodel.expasy.org/interactive, accessed on 6 October 2022) was used to predict the tertiary structure of the SiNF-YC2 protein. The similarity between the template sequence required for modeling and the target sequence reached 58.18%, indicating that the prediction results were close to the actual results (Figure 2e).
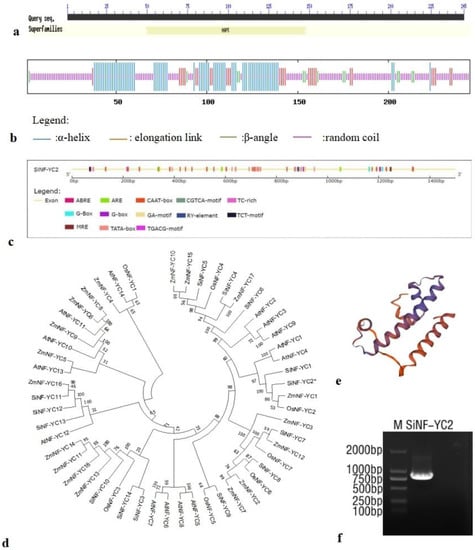
Figure 2.
Bioinformatics analysis of SiNF-YC2. (a) Conserved domain prediction. (b) Protein secondary structure analysis. (c) cis-Acting elements in SiNF-YC2 promoter. (d) Evolutionary analysis of SiNF-YC2 (At: Arabidopsis thaliana; Zm: Zea mays; Si: Setaria italica; Os: Oryza sativa). The asterisk indicates SiNF-YC2. (e) Protein tertiary structure prediction. (f) Cloning of SiNF-YC2 (M: 2 kb DNA marker).
According to the position of SiNF-YC2 in the millet genome, the promoter of SiNF-YC2 was analyzed using the online analysis software PlantCARE(plant cis-acting regulatory elements, http://bioinformatics.psd.ugent.be/welotools/plantcare/html/, accessed on 14 October 2022). In addition to the basic regulatory elements TATA box and CAAT box, the promoter of the millet SiNF-YC2 gene also contains several hormone-related action elements, such as ABRE, the CGTCA motif, and the TGACG motif; optical response elements, such as the G-box, G-Box, GA motif, MRE, and TCT motif; and anti-stress-response-related elements, such as TC-rich repeats. In addition, the promoter contains ARE action elements that respond to anaerobic induction and RY cis-acting elements that participate in seed-specific regulation (Figure 2c; Supplementary Table S2).
The sequence alignment of the SiNF-YC2 protein with the conserved domain of NF-YC proteins in other species showed that it also contains two β-linked domains separated by α 1, α 2, and α 3 helices and an α C structure, which are indicators of the NF-YC family (Supplementary Figure S1d). In the phylogenetic analysis of NF-YC family members in A. thaliana, maize, and rice, SiNF-YC2 of millet is closely related to ZmNF-YC1 of maize and OsNF-YC2 of rice (Figure 2d).
2.2. Analysis of Expression Characteristics of SiNF-YC2
To analyze circadian expression, fresh leaf samples from plants growing under SD and LD photoperiods were collected every 3 h starting at 10 a.m. (light onset) for 48 h. The SiNF-YC2 gene was rhythmically expressed in the leaves of Longgu 3 for 24 h under both long- and short-sunshine conditions (Figure 3a,b). Under the SD photoperiod, the expression level decreased with the onset of light, reached the lowest level at 3 h, then began to increase, and reached the first peak at 6 h. The expression level of SiNF-YC2 reached a second peak at the 10th hour of darkness, then decreased, and reached the lowest level at the 13th hour of darkness. At the end of darkness, the expression level of SiNF-YC2 reached a third peak. In the next cycle, 24 h later, the same trend was observed. Thus, the expression of SiNF-YC2 showed three peaks within one day, one in the light period and two in the dark period (Figure 3a). Under the LD photoperiod, the expression of SiNF-YC2 also began to decrease with the onset of light and began to increase at 3 h but reached a first peak at 9 h of light, with a second peak at 2 h of darkness (18 h). As was observed under the SD photoperiod, there was a third peak of SiNF-YC2 expression at the end of darkness. The expression pattern in the next cycle was similar to that in the previous 24 h (Figure 3b). The results showed that the expression pattern of the SiNF-YC2 gene was different under different photoperiods.
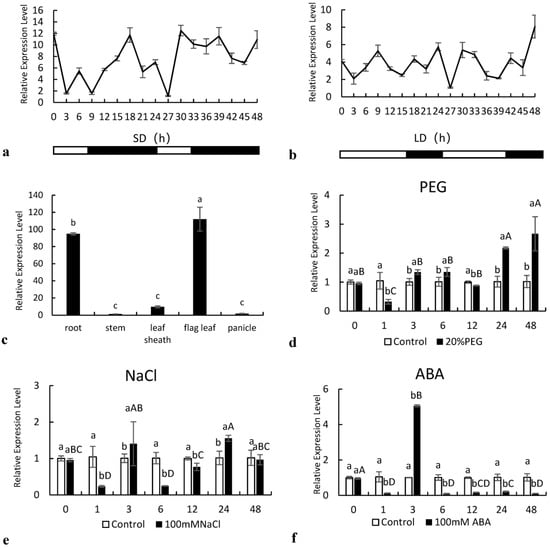
Figure 3.
Expression analysis of SiNF-YC2. (a) Short-day (SD) photoperiod. (b) Long-day (LD) photoperiod. (c) Relative expression of SiNF-YC2 in different tissues of foxtail millet. Expression of SiNF-YC2 under (d) 20% PEG6000, (e) 200 mM NaCl, and (f) 100 μM ABA treatments. Different letters on columns indicate significant differences at the level of 0.05. p < 0.05 indicates significant differences between statistical data, p > 0.05 indicates insignificant differences between statistical data, and p < 0.01 indicates extremely significant differences between statistical data. Different lowercase letters indicate differences between the control and treatment at the same time point; different capital letters indicated differences in SiNF-YC2 expression in millet exposed to stress at different time points.
The SiNF-YC2 gene was expressed in the different tissues of Longgu 3, but the relative expression was significantly different (p < 0.05). The relative expression of the SiNF-YC2 gene was the highest in the flag leaf, followed by that in roots, with the lowest expression in the stem (Figure 3c).
The expression SiNF-YC2 was examined under different stress treatments (Figure 3d–f). In the 20% PEG6000 treatment, the expression of the SiNF-YC2 gene was significantly different from that in the control at different times, and the highest expression was at 48 h, which was approximately 2.5 times higher than that in the control. In the NaCl treatment, the expression of the SiNF-YC2 gene was significantly different from that in the control group, first decreasing and then increasing, then decreasing and then increasing, and finally decreasing. A first peak appeared at 3 h after treatment, and a second peak appeared at 24 h after treatment. The expression of SiNF-YC2 in the ABA treatment was significantly lower than that in the control, except at 3 h.
2.3. SiNF-YC2 Interacts with SiCO in the Nucleus
The localization results of SiNF-YC2 showed that the p35S-SiNF-YC2-GFP fusion protein only emitted a green fluorescence signal in the nucleus, indicating that the SiNF-YC2 protein was primarily localized in the nucleus (Figure 4a). In the Y2H assays, the experimental group was able to grow on the four-deficient SD/-Ade-His-Leu-Trp medium containing X-α-gal and was stained blue, which indicated that SiNF-YC2 interacted with SiCO (Figure 4b).
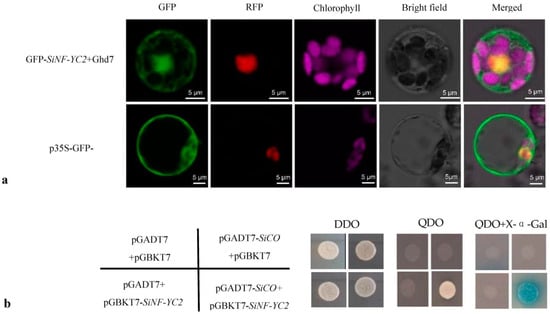
Figure 4.
SiNF-YC2 and SiCO interact in the nucleus. (a) Subcellular localization of SiNF-YC2 protein. (b) Interaction of SiNF-YC2 and SiCO. DDO: two-deficient SD/-Leu-Trp medium; QDO: four-deficient SD/-Ade-His-Leu-Trp medium.
2.4. Functional Analysis of SiNF-YC2
Leaf DNA was extracted from plants screened on hygromycin medium, and then PCR was performed. Nine of ten transgenic Arabidopsis (OE-1 to OE-10) plants contained the target genes (Figure 5a). Then, three lines with high expression, which were OE-1, OE-3, and OE-5, were screened from the other six lines by RT-qPCR and used for subsequent functional analysis (Figure 5b).
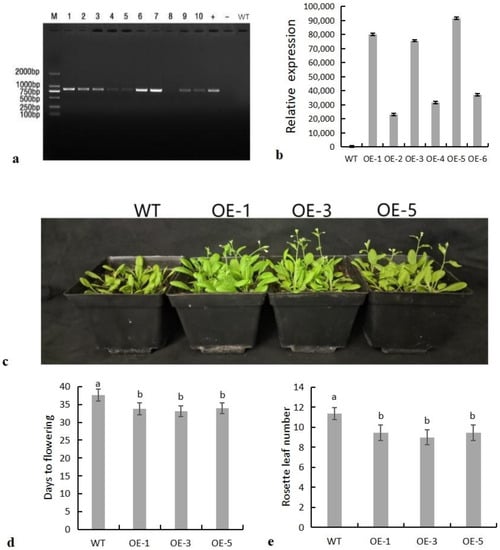
Figure 5.
SiNF-YC2 positively regulates flowering time. (a) PCR detection of transgenic Arabidopsis (M: 2 kb DNA marker; +: positive control; −: negative control; WT: wild-type Arabidopsis; 1–10: transgenic Arabidopsis OE-1 to OE-10). (b) Quantitative screening of SiNF-YC2. (c) Flowering phenotypes of wild-type and transgenic Arabidopsis. (d) Flowering time statistics. (e) Rosette leaf phenotypes of wild-type and transgenic Arabidopsis. Different letters on columns indicate significant differences at the level of 0.05. p < 0.05 indicates significant differences between statistical data, p > 0.05 indicates insignificant differences between statistical data, and p < 0.01 indicates extremely significant differences between statistical data. Different lowercase letters indicate differences between different strains.
The SiNF-YC2 gene showed circadian expression patterns in both LD and SD photoperiods (Figure 3a,b), indicating that SiNF-YC2 was sensitive to the photoperiod and regulated by biological rhythms. Therefore, SiNF-YC2 was overexpressed in wild-type Arabidopsis plants to study its function. Compared with wild-type Arabidopsis, transgenic Arabidopsis lines overexpressing SiNF-YC2 produced fewer rosette leaves (Figure 5e) and flowered earlier (Figure 5c,d), indicating that SiNF-YC2 positively regulated flowering time in Arabidopsis.
Transgenic Arabidopsis performed differently under different stress treatments (Figure 6). At different concentrations of NaCl, the germination rate of transgenic Arabidopsis was slightly higher than that of the wild type, but the difference was not significant (Figure 6g). After seven days of vertical culture, the root length of transgenic SiNF-YC2 Arabidopsis was not significantly different from that of the wild type on normal medium, whereas the root length of SiNF-YC2 transgenic Arabidopsis was significantly higher than that of the wild type under 125 mM and 150 mM NaCl stress (Figure 6h,i). After transplanting to soil, Arabidopsis was treated with salt. The results further indicated that SiNF-YC2 played a role in promoting the response of transgenic Arabidopsis to salt stress (Figure 6c–j,k). At different concentrations of mannitol and ABA, the phenotypic results of transgenic Arabidopsis indicated that there was no difference in sensitivity between SiNF-YC2 transgenic Arabidopsis and the wild type at either the germination or seedling stage (Figure 6a–f). Thus, the results indicated that SiNF-YC2 did not have an important role in promoting the response to osmotic stress in Arabidopsis.
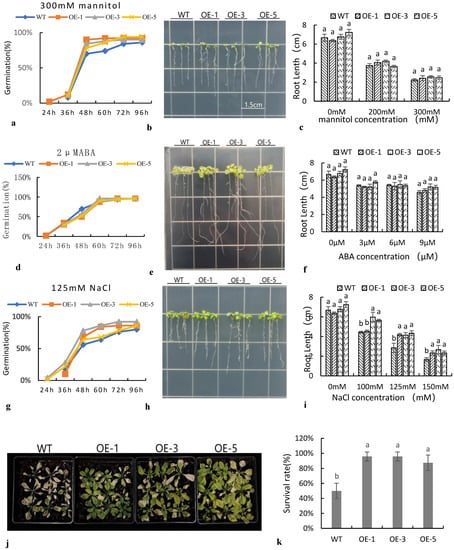
Figure 6.
Comparison of transgenic Arabidopsis germination under different stress treatments. Response of transgenic Arabidopsis treated with (a–c) mannitol, (d–f) ABA, and (g–k) salt. (a,d,g) Germination rate, (b,e,h) root length, (c,f,i) root length data analysis, (j) salt stress at seedling stage, and (k) survival rate. WT: wild type. OE: SiNF-YC2 transgenic Arabidopsis. Bar = 1.5 cm. Different letters on columns indicate significant differences at the level of 0.05. p < 0.05 indicates significant differences between statistical data, p > 0.05 indicates insignificant differences between statistical data, and p < 0.01 indicates extremely significant differences between statistical data. Different lowercase letters indicate differences between different strains.
3. Discussion
A relatively long crop growth period affects the cultivation of the resulting crops, and the yield and quality of late-maturing varieties can also be adversely affected by climatic factors such as high temperatures or heavy rains. In cold areas with cultivation, low-temperature injury is a major threat to crops, primarily affecting the heading, grouting, and seed-setting rate of late-ripening varieties. Although late-ripening varieties can produce high and stable yields, it is difficult to optimize the agricultural industry structure with such varieties. However, early-maturing varieties can effectively avoid the effects of cold, dew, and wind and often achieve high and stable yields [28]. Therefore, early-maturing varieties with an appropriate maturity stage need to be selected. The discovery of early-maturity genes was an important step in selecting crop varieties with early maturity and high yields [28]. OsNF-YB11 is very important for the heading date, plant height, and yield of rice [29]. OsNF-YB1 is specifically expressed in the aleurone layer of developing endosperm and regulates grain filling and endosperm development, and it affects grain yield and quality [30,31]. Flowering time is an important index to determine the maturity stage of crops [32]. However, the molecular mechanism regulating flowering in millet remains unclear. The NF-YC protein is an important component of the NF-Y transcription factor complex. In previous studies [25], SiNF-YC2 was isolated, but its function remained unclear. In this study, the SiNF-YC2 protein was cloned and analyzed, and it was found to contain DNA-binding domains, NF-YA and NF-YB interaction domains, and CCAAT-binding domains. This study confirmed that the SiNF-YC2 gene is associated with early maturity, which provides genetic resources and theoretical support for the cultivation of early-maturity millet varieties.
In the present study, our results showed that SiNF-YC2 may promote early flowering, which is similar to the function of AtNF-YC4 in Arabidopsis thaliana. This means that the function of homologous genes may be conserved; however, this conclusion is not always valid: for example, Hd1, which is the homologous gene of the Arabidopsis CO gene in rice, negatively regulates the FT co-orthologs HD3A and RFT1 under long-day conditions, which means that the function of homologous genes in different plants may vary. In addition, the function of different members of the NF-YC gene family may vary. In Arabidopsis thaliana, overexpression of the AtNF-YC2/3 gene can promote the flowering of plants and increase the transcription level of FT [33]. In rice, OsNF-YC2 and OsNF-YC6 can interact with DHD1 (delayed heading date1), a member of the GRAS family, to synergically inhibit the expression of Ehd1, thus delaying the heading date [34]. The above research showed that the overexpression of the NF-Y gene can alter flowering time via a highly redundant and complex mechanism in various plants [35]; therefore, there is still much work to be completed on the function of NF-Y genes in flowering regulation in various plants.
In the present study, our result showed that SiNF-YC2 interacted with SiCO in the nucleus. Previous research showed that both NF-YA/B/C trimers and NF-YB/C-CO (CONSTANS) trimers can be targeted to the promoter region of FT [35,36]. An NF-Y protein can recognize the CCAAT site in the promoter. The CCAAT-acting element in the distal promoter is close to the CORE site, and through the interaction between the NF-Y complex and CO protein, the CO-binding CORE is recruited. The recruitment makes the FT promoter form a ring, which then positively regulates the transcription of FT to promote plant flowering. So, we speculate that SiNF-YC2 may participate in the flowering regulation of foxtail millet by forming a dimer with the CO protein and then inducing the expression of florin by the FT gene [22]. However, the detailed molecular mechanism of how SiNF-YC2 binds to FT promoters is not yet clear, so further verification is required in foxtail millet.
According to previous studies, NF-YC functions in the abiotic stress response [27,37,38]. AtNF-YC1 regulates the response of Arabidopsis to cold stress by binding to the CCAAT box in the promoter region of AtXTH21 [39]. In rice, OsNF-YC1 is involved in the response to salt stress, and the salt tolerance of overexpressed OsNF-YC1 plants increases significantly compared with that of the wild type [40]. In addition, TaNF-YC5 is induced by drought and salt stress and is important in the resistance to drought and salt stress [41], and the overexpression of CdtNF-YC1 in rice increases plant sensitivity to salt stress [42]. The cis-element in the SiNF-YC2 promoter contains ABRE elements that can increase gene expression under drought and high-salt stress, and RT-qPCR results indicated that it was responsive to drought, salt stress, and ABA. In this study, Arabidopsis plants were treated with different concentrations of NaCl, and the salt tolerance of transgenic SiNF-YC2 plants was higher than that of the control, indicating that SiNF-YC2 could increase plant salt tolerance to a certain extent. However, the overexpression of SiNF-YC2 did not increase the resistance of transgenic plants to drought and ABA. The absence of a response to drought and ABA may be because SiNF-YC2 needs to bind to NF-YB and NF-YA to form a polymer in order to perform its function. Therefore, further studies are needed to determine whether SiNF-YC2 is involved in regulating plant responses to drought and ABA stresses and identify the possible regulatory mechanisms.
4. Materials and Methods
4.1. Bioinformatics Analysis of SiNF-YC2
Several candidate genes were screened by QTL mapping in the early stage [21]. Among the candidate genes, SiNF-YC2 (Seita. 9G468100) was selected as an important gene for the photoperiod response on the basis of the integrated results of gene annotation, cis-element analysis, and expression analyses. The gene and protein sequences of SiNF-YC2 were downloaded from the Phytozome database (http://www.phytozome.com/, accessed on 1 October 2022). The physical and chemical properties of the SiNF-YC2 protein were analyzed in Protparam (https://web.express.org/ProtParam/, accessed on 8 October 2022) [43,44]. ProtScale (https://web.expasy.org/protscale/, accessed on 8 October 2022) was used for hydrophilic/hydrophobic analysis. The online software TMHMM (http://www.cbs.dtu.dk/services/TMHMM/, accessed on 8 October 2022) was used for transmembrane structural domain analysis. The amino acid sequence was input to SignalP (http://www.cbs.dtu.dk/services/SignalP/, accessed on 8 October 2022) to predict whether there was a signal peptide structure. The online CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 10 October 2022) was used to conduct conserved domain analysis. The software Netphos (http://www.cbs.dtu.dk/services/NetPhos/, accessed on 12 October 2022) was used to analyze protein phosphorylation sites. The secondary structure was predicted using PBIL’s SOMPA online website (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html, accessed on 12 October 2022). The tertiary structure was predicted using the SWISS-MODEL of Expasy (https://swissmodel.expasy.org/interactive, accessed on 6 October 2022) with amino acid sequence input. According to the rules of amino acid arrangement, the protein tertiary structure model that had the highest overall quality estimation score was built.
From the Phytozome database, the 1500 bp region upstream of the SiNF-YC2 gene was identified as the promoter. The plant promoter online analysis software PlantCARE (plant cis-acting regulatory elements, http://bioinformatics.psd.ugent.be/welotools/plantcare/html/, accessed on 14 October 2022) was used to analyze the gene promoter, predict cis-acting elements, and visually analyze them in TBtools.
The protein sequence of SiNF-YC2 was submitted to the Uniprot database (http://www.uniprot.org, accessed on 8 October 2022) for protein sequence BLAST, and corresponding homologous protein sequences of different species were selected. A phylogenetic tree was constructed using the neighbor-joining method in MEGA 11.0 [45], and the obtained genes were compared by analyzing multiple sequences using DNAMAN8.
4.2. Plant Materials and Growth Conditions
The foxtail millet variety Longgu 3 was used for the cloning and expression analysis of the SiNF-YC2 gene. Colombian zero-type (Col-0) wild Arabidopsis thaliana was used as a transgenic receptor and control material.
Longgu 3 foxtail millet was sown in flowerpots (10 cm × 10 cm) with field soil and nutrient soil mixed in a 1:1 ratio and cultured in a light incubator. The temperature was 25 °C, the light intensity was 8000 Lx, and the light time was adjusted according to the test treatment. Wild-type or transgenic Arabidopsis plants (ecotype Columbia (Col-0)) were grown in soil or Murashige and Skoog (MS) medium at 25 °C and 70% humidity with a 16 h light/8 h dark photoperiod at a light intensity of 8000 Lx.
4.3. Cloning of SiNF-YC2 and Generation of Transgenic Plants
The gene sequence of SiNF-YC2 was downloaded from the Phytozome database, and the specific primers C2-F1 and C2-R1 were designed with Primer Premier 5.0 (Premier, Canada) (Supplementary Table S1). Total RNA of Longgu 3 was extracted and reverse-transcribed into cDNA. SiNF-YC2 was amplified using cDNA as a template, and then the PCR product was purified and inserted into a pLB vector. Then, the pLB SiNF-YC2 plasmid was used as a template, and SiNF-YC2 was amplified by the primers C2-F2 and C2-R2 (Supplementary Table S1), inserted into a pCAMBIA1300 vector (abcam, Catalog No. ab275754), and digested by Kpn I and Sma I. The recombination vector was introduced into Agrobacterium strain GV3101 (Biomed, Beijing, China).
Agrobacterium tumefaciens strain GV3101 (Biomed, Beijing, China) containing the SiNF-YC2 recombinant vector was transformed into Arabidopsis by using the floral dip method [46]. Transgenic plants were selected by culturing them on plates supplemented with hygromycin (30 mg/L). Transgenic plants were detected at the DNA and transcription levels by PCR and reverse-transcription quantitative PCR (RT-qPCR), respectively. Homozygous transgenic lines of the T3 generation were screened and propagated for further functional analysis.
4.4. Expression Analysis of SiNF-YC2
In each pot, 8 to 10 Longgu 3 were planted, for a total of 28 pots. Under a 12 h light/12 h dark photoperiod, seedlings were grown to the second-leaf stage, and then four seedlings with the same level of growth were retained in each pot. For different photoperiod treatments, when foxtail millet reached the three-leaf stage, 18 pots were placed in an incubator with a short-day photoperiod (8 h light/16 h dark), and 10 pots were placed in an incubator with a long-day photoperiod (16 h light/8 h dark). Two pots of foxtail millet were cultured in the short-day incubator for one week, and then tender leaves were used to extract total RNA and clone SiNF-YC2. To characterize the expression of SiNF-YC2, six pots of foxtail millet in the short-day incubator were cultured to the heading stage, and the roots, stem, leaf sheath, top leaf, secondary top leaf, and spike were collected, with three replicates of each. Foxtail millet in the different photoperiod treatments was sampled after three weeks of culture. To analyze circadian expression, fresh leaf samples were collected at 3 h intervals over a 48 h period from growing plants. All samples were immediately frozen with liquid nitrogen and stored at –80 °C.
Foxtail millet at the 4-leaf stage under the short-day photoperiod was exposed to salt (100 mM NaCl), drought (20% PEG6000), and ABA (100 µM) stress. Drought and salt treatments were applied by irrigation, the ABA treatment was applied by spraying, and the control group received normal watering. There were 20 pots per treatment. After 0, 1, 3, 6, 12, 24, and 48 h of treatment, fresh leaves were collected, and the expression of SiNF-YC2 was analyzed under different stress treatments. Three biological replicates were collected at each time point. Samples were immediately frozen with liquid nitrogen and stored at –80 °C.
Total RNA was isolated using a Fast Pure®Plant Total RNA Isolation Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. First-strand cDNA was reverse-transcribed from total RNA using HiScript II Reverse Transcriptase (Vazyme, Nanjing, China) with oligo (dT) as the primer. PCR was performed in a total volume of 10 µL containing 5 µL of 2× ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China), 0.2 µL of each gene-specific primer (Supplementary Table S1), 1 µL of cDNA, and 3.6 µL of ddH2O on a Roche LightCycler 480 real-time PCR machine. Reactions were conducted using the following program: 95 °C for 2 min and 40 cycles of 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 30 s, and fluorescent signals were detected. The standard procedure for the dissolution curve was 95 °C for 5 s; 60 °C for 1 min; 95 °C continuously; and 50 °C for 30 s. Gene expression was calculated by the 2−∆∆Ct method [47], and data were collated and analyzed. Each experiment was performed with three technical replicates. Student’s t-tests were used to determine significant differences.
4.5. Subcellular Localization
To determine the subcellular location of the SiNF-YC2 protein, the fusion expression vector p35S-SiNF-YC2-GFP was generated by using homologous recombination. Protoplasts were isolated from rice leaf cells and were transformed with the fusion expression vector p35S-SiNF-YC2-GFP+p35S-OsGhd7-GFP and the control vector GFP+p35S-OsGhd7-GFP. Transformed protoplasts were incubated for 16 to 18 h in the dark. Signals from the green fluorescent protein (GFP) were observed using a confocal laser-scanning microscope (FLV1200, Olympus, Tokyo, Japan), which uses lasers to excite fluorescent dyes in the sample and captures the emitted light to produce high-resolution images.
4.6. Yeast Two-Hybrid Assays
To analyze the function and regulatory network of SiNF-YC2, yeast two-hybrid assays were conducted. Fusion expression vectors pGADT7-SiCO and pGBKT7-SiNF-YC2 were constructed, and then the fusion expression vector pGADT7-SiCO+pGBKT7-SiNF-YC2 and negative control vectors pGADT7+pGBKT7, pGADT7-SiCO+pGBKT7, and pGADT7+pGBKT7-SiNF-YC2 were transformed separately into yeast (Saccharomyces cerevisiae) strain AH109 by using the polyethylene glycol/lithium acetate method. Transformed yeast was first coated on two-deficient SD/-Leu-Trp medium and then on four-deficient SD/-Ade-His-Leu-Trp medium to observe colony growth. Last, yeast in the test and control groups was applied to media containing X-α-gal. Plates with four-deficient SD/-Ade-His-Leu-Trp medium with gal were cultured upside down at 30 °C for three to four days, and then the change in colony color was observed.
4.7. Function Analysis of SiNF-YC2
To analyze the germination rate of transgenic Arabidopsis seeds under stress, 50 plump wild-type and 50 T3 generation transgenic Arabidopsis seeds were selected and, after disinfection, were evenly spot-sown in MS solid medium containing 0, 0.5, 1.0, or 2.0 μM ABA; 0, 75, 100, or 125 mM NaCl; or 0, 100, 200, or 300 mM mannitol. After vernalization, seeds were put into a light-temperature incubator for culture, and germination rates were determined at 1, 1.5, 2, 2.5, 3, and 4 days.
To analyze changes in the root length of transgenic Arabidopsis plants under stress, wild-type and T3-generation transgenic Arabidopsis grown to the second-leaf stage on MS solid medium were transferred to MS solid medium containing 0, 3.0, 6.0, or 9.0 μM ABA; 0, 100, 125, or 150 mM NaCl; or 0, 150, 200, or 300 mM mannitol. Plates were placed vertically, and culturing continued. After differential phenotypes appeared, root lengths were analyzed using a root scanner (WINRHIZO proLA2400). Each experiment was performed with three replicates.
To observe the stress resistance of transgenic Arabidopsis in the seedling stage, wild-type and transgenic Arabidopsis grown on MS solid medium for approximately 10 days were transplanted into soil. After two weeks of normal growth, some of the plants were subjected to drought stress by stopping watering. After differential phenotypes appeared, the water supply was restored, and the survival rate of each line was determined one week after the recovery of growth. The other plants were sprayed with 200 mM NaCl solution. After differential phenotypes appeared, the survival rate of each line was determined.
To observe the flowering phenotype of transgenic Arabidopsis, wild-type and transgenic Arabidopsis grown on MS solid medium for approximately 10 days were transplanted to soil and cultured, with 20 to 25 plants per line. The number of days was recorded from the first day of sowing to the first flower, and the number of rosette leaves was counted.
5. Conclusions
SiNF-YC2 was cloned based on QTL mapping. The expression of the SiNF-YC2 gene exhibited a circadian rhythm. The gene was highly expressed in millet leaf and root tissues and was involved in responses to abiotic stresses such as drought, salt, and ABA. The overexpression of the SiNF-YC2 gene increased the resistance of transgenic Arabidopsis to salt stress. The overexpression of SiNF-YC2 also led to the early flowering of transgenic Arabidopsis, suggesting that the gene is involved in regulating plant flowering by promoting early flowering. The results of this study not only lay a foundation for elucidating the flowering regulation network of foxtail millet but also provide excellent genetic resources for the breeding of early-maturing foxtail millet varieties.
Supplementary Materials
The supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms24087217/s1.
Author Contributions
F.L. and Y.G. coordinated the project and conceived and designed the experiments. J.N. and X.Y. conducted the experiments. J.N. and F.L. wrote the first draft. L.Q., E.C., Y.Y. and R.W. edited the manuscript. H.Z. and H.W. contributed valuable discussions. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the Key Research and Development Project of Shandong Province (2021LZGC025), China Agriculture Research System of MOF and MARA (CARS-06-14.5-A19), Agricultural Fine Seed Project of Shandong Province (2021LZGC006), and the Open Project of the National Engineering Research Center of Wheat and Maize.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
qRT-PCR: quantitative real-time; WT: wild type; GFP: green fluorescent protein; QTL: quantitative trait locus; Y2H: yeast two-hybrid.
References
- Fang, J.; Zhang, F.T.; Wang, H.R.; Wang, W.; Zhao, F.; Li, Z.J.; Sun, C.H.; Chen, F.M.; Xu, F.; Chang, S.Q.; et al. Ef-cd locus shortens rice maturity duration without yield penalty. Proc. Natl. Acad. Sci. USA 2019, 116, 18717–18722. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.Y.; Jin, F.R.; Dong, G.Y.; Guan, M.; Tan, T.L. Exploring the growth and development properties of early variety of winter rapeseed. Strateg. Study CAE 2012, 14, 4–12. [Google Scholar]
- Liu, Z.L.; Cheng, R.H.; Shi, Z.G.; Xia, X.Y.; Zhang, Y.Z.; Hou, S.L. Innovation in Jigu 28 of Super Prematurity and High Quality New Millet Variety with Flexible Growth Period and Study on Its Related Physiological Mechanism. Sci. Agric. Sin. 2009, 42, 1145–1151. [Google Scholar]
- Jie, S.; Andrew, A.; Martin, H.; Caroline, D. Vernalization—A cold-induced epigenetic switch. J. Cell Sci. 2012, 125, 3723–3731. [Google Scholar]
- Richard, A. Seasonal and developmental timing of flowering. Plant J. Cell Mol. Biol. 2010, 61, 1001–1013. [Google Scholar]
- Mariko, S.; Kay, S.A. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 11698–11703. [Google Scholar]
- Roshi, S.; Jorge, G.-A.; Vittoria, B.; Fabio, F. Molecular control of seasonal flowering in rice, Arabidopsis and temperate cereals. Ann. Bot. 2014, 114, 1445–1458. [Google Scholar]
- Shengjie, B.; Changmei, H.; Lisha, S.; Hao, Y. New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 118–131. [Google Scholar]
- Koo, B.-H.; Yoo, S.-C.; Park, J.-W.; Kwon, C.-T.; Lee, B.-D.; An, G.; Zhang, Z.; Li, J.; Li, Z.; Paek, N.-C. Natural Variation in OsPRR37 Regulates Heading Date and Contributes to Rice Cultivation at a Wide Range of Latitudes. Molecular Plant. 2013, 6, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2484. [Google Scholar] [CrossRef]
- Kazuyuki, D.; Takeshi, I.; Takuichi, F.; Utako, Y.; Takahiko, K.; Zenpei, S.; Masahiro, Y.; Atsushi, Y. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar]
- Tsuji, H.; Tamaki, S.; Komiya, R.; Shimamoto, K. Florigen and the Photoperiodic Control of Flowering in Rice. Rice. 2008, 1, 25–35. [Google Scholar] [CrossRef]
- Li, P.; Brutnell, T.P. Setaria viridis and Setaria italica, model genetic systems for the Panicoid grasses. J. Exp. Bot. 2011, 62, 3031–3037. [Google Scholar] [CrossRef] [PubMed]
- Periyasamy, R.; Subramani, P.; Kasinathan, R.; Manoharan, B.; Rajaiah, A.; Lakkakula, S.; Ramakrishnan, R.; Manikandan, R. The protective effects of polyamines on salinity stress tolerance in foxtail millet (Setaria italica L.), an important C4 model crop. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2020, 26, 1815–1829. [Google Scholar]
- Yuan, X.M.; He, L.; Zhang, K.Y.; Wu, X.; Ma, F.F.; Wang, J.; Han, Y.H. Analysis of XTH genes that related to drought stress in foxtail millet. J. Shanxi Agric. Univ. Nat. Sci. Ed. 2017, 37, 1–6. [Google Scholar]
- Margarita, M.-H.; Wang, X.W.; Barbier, H.; Brutnell, T.P.; Devos, K.M.; Doust, A.N. Genetic control and comparative genomic analysis of flowering time in Setaria (Poaceae). G3 2013, 3, 283–295. [Google Scholar]
- Laura, L.; Martín, K.; Maximiliano, S.-L.; Elizabeth, K.; Cerdan, P.D.; Casal, J.J. CONSTANS delays Arabidopsis flowering under short days. Plant J. Cell Mol. Biol. 2019, 97, 923–932. [Google Scholar]
- Takeshi, K.; Samia, S.; Elli, K.; Katriina, M.; Timo, H. Fragaria vesca CONSTANS controls photoperiodic flowering and vegetative development. J. Exp. Bot. 2017, 68, 4839–4850. [Google Scholar]
- Lee, Y.-S.; An, G. OsGI controls flowering time by modulating rhythmic flowering time regulators preferentially under short day in rice. J. Plant Biol. 2015, 58, 137–145. [Google Scholar] [CrossRef]
- Sim, S.A.; Woo, S.G.; Hwang, D.Y.; Kim, J.-H.; Lee, S.S.; Lim, C.O.; Hong, J.C.; Song, Y.H. FLOWERING HTH1 is involved in CONSTANS-mediated flowering regulation in Arabidopsis. Appl. Biol. Chem. 2019, 62, 56. [Google Scholar] [CrossRef]
- Usman, A.M.; Amanda, D.; Jon, D.S.; Marcel, Q. Photoperiod sensing of the circadian clock is controlled by EARLY FLOWERING 3 and GIGANTEA. Plant J. Cell Mol. Biol. 2020, 101, 1397–1410. [Google Scholar]
- Kumimoto, R.W.; Zhang, Y.; Siefers, N.; Holt, B.F., 3rd. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 2010, 63, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Gusmaroli, G.; Tonelli, C.; Mantovani, R. Regulation of the CCAAT-Binding NF-Y subunits in Arabidopsis thaliana. Gene 2001, 264, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Li, T.; Xu, P.B.; Li, L.; Du, S.S.; Lian, H.L.; Yang, H.Q. DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett. 2016, 590, 541–549. [Google Scholar] [CrossRef]
- Li, F.F.; Niu, J.H.; Yu, X.; Kong, Q.H.; Wang, R.F.; Qin, L.; Chen, E.Y.; Yang, Y.B.; Liu, Z.Y.; Lang, L.N.; et al. Isolation and identification of SiCOL5, which is involved in photoperiod response, based on the quantitative trait locus mapping of Setaria italica. Front. Plant Sci. 2022, 13, 969604. [Google Scholar] [CrossRef]
- Huang, S.; Hu, L.Q.; Xu, D.B.; Li, W.W.; Xu, Z.S.; Li, L.C.; Zhou, Y.B.; Diao, X.M.; Jia, G.Q.; Ma, Z.Y.; et al. Transcription Factor SiNF-YA5 from Foxtail Millet (Setaria italica) Conferred Tolerance to High-salt Stress through ABA-independent Pathway in Transgenic Arabidopsis. Acta Agron. Sin. 2016, 42, 1787–1797. [Google Scholar] [CrossRef]
- Swain, S.; Myers, Z.A.; Siriwardana, C.L.; Holt, B.F. The multifaceted roles of NUCLEAR FACTOR-Y in Arabidopsis thaliana development and stress responses. BBA Gene Regul. Mech. 2016, 1860, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Rahul, C.; Changsoo, K.; Patel, J.D.; Hui, G.; Tariq, S.; Wallace, J.G.; Douhua, H.; Zhengsheng, Z.; Jeevan, A.; Sameer, K.; et al. Identification of small effect quantitative trait loci of plant architectural, flowering, and early maturity traits in reciprocal interspecific introgression population in cotton. Front. Plant Sci. 2022, 13, 981682. [Google Scholar]
- Manjari, M.; Singh, R.R.; Rohit, J.; Ashwani, P.; Singla-Pareek, S.-L. DTH8 overexpression induces early flowering, boosts yield, and improves stress recovery in rice cv IR64. Physiol. Plant. 2022, 174, e13691. [Google Scholar]
- Xu, J.J.; Zhang, X.F.; Xue, H.W. Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor. J. Exp. Bot. 2016, 67, 757–758. [Google Scholar] [CrossRef] [PubMed]
- Martin, B. Filling the grain: Transcription factor OsNF-YB1 triggers auxin biosynthesis to boost rice grain size. Plant Physiol. 2021, 185, 6399–6411. [Google Scholar]
- Aidyn, M.; Frédéric, C.; George, C. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 2002, 14, S111. [Google Scholar]
- Hackenberg, D.; Wu, Y.; Voigt, A.; Adams, R.; Schramm, P.; Grimm, B. Studies on Differential Nuclear Translocation Mechanism and Assembly of the Three Subunits of the Arabidopsis thaliana Transcription Factor NF-Y. Molecular Plant. 2012, 5, 876–888. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, S.S.; Liu, T.Z.; Wang, C.M.; Cheng, Z.J.; Zhang, X.; Chen, L.P.; Sheng, P.K.; Cai, M.H.; Li, C.N.; et al. DELAYED HEADING DATE1 interacts with OsHAP5C/D, delays flowering time and enhances yield in rice. Plant Biotechnol. J. 2019, 17, 531–539. [Google Scholar] [CrossRef]
- Xu, M.Y.; Zhu, J.X.; Zhang, M.; Wang, L. Advances on plant miR169/NF-YA regulation modules. Yi Chuan Hered. 2016, 38, 700–706. [Google Scholar]
- Myers, Z.A.; Kumimoto, R.W.; Siriwardana, C.L.; Gayler, K.K.; Risinger, J.R.; Pezzetta, D.; Iii, B.F.H. NUCLEAR FACTOR Y, Subunit C (NF-YC) Transcription Factors Are Positive Regulators of Photomorphogenesis in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006333. [Google Scholar] [CrossRef]
- Palmeros-Suárez, P.A.; Massange-Sánchez, J.A.; Martínez-Gallardo, N.A.; Montero-Vargas, J.M.; Gómez-Leyva, J.F.; Délano-Frier, J.P. The overexpression of an Amaranthus hypochondriacus NF-YC gene modifies growth and confers water deficit stress resistance in Arabidopsis. Plant Sci. 2015, 240, 25–40. [Google Scholar] [CrossRef]
- Hackenberg, D.; Grimm, B.; Keetman, U. Homologous NF-YC2 Subunit from Arabidopsis and Tobacco Is Activated by Photooxidative Stress and Induces Flowering. Int. J. Mol. Sci. 2012, 13, 3458–3477. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.T.; Ye, T.T.; Zhong, B.; Liu, X.; Jin, R.; Chan, Z.L. AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol. 2014, 203, 554–567. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, C.J.; Liu, Y.J.; Huang, G.X.; Zhang, L.Y. Prokaryotic expression and polyclonal antibody preparation of Arabidopsis transcription factor NF-YC. Biotechnol. Bull. 2012, 3, 57–62. [Google Scholar]
- Ma, Z.F. Study on Wheat NF-Y Type Transcription Factor Gene TaNFYB-A13 and TaNF-YC1 Mediate the Function of Plant Resisting Osmotic Stress; Hebei Agricultural University: Baoding, China, 2020. [Google Scholar]
- Chen, M.; Zhao, Y.J.; Zhuo, C.L.; Lu, S.Y.; Guo, Z.F. Overexpression of a NF-YC transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol. J. 2015, 13, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Bjellqvist, B.; Hughes, G.J.; Pasquali, C.; Paquet, N.; Ravier, F.; Sanchez, J.C.; Frutiger, S.; Hochstrasser, D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 1993, 14, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. Proteom. Protoc. Handb. 2005, 53, 571–607. [Google Scholar]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Xiuren, Z.; Rossana, H.; Shih-Shun, L.; Qi-Wen, N.; Nam-Hai, C. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).