Extracellular Vesicles in Breast Cancer: From Biology and Function to Clinical Diagnosis and Therapeutic Management
Abstract
1. Introduction
2. Extracellular Vesicle Nature, Structure, and Properties
2.1. Extracellular Vesicle Biogenesis
2.1.1. SEVs Biogenesis
2.1.2. MLEVs Biogenesis
2.2. Extracellular Vesicle Composition
2.3. Extracellular Vesicle Fate
3. Extracellular Vesicle’s Role in Normal Breast Tissue
3.1. Extracellular Vesicles Production in Normal Mammary Tissue
3.2. Exosome Role in the Maintenance of the Mammary Stem Cell
3.3. Milk Is an Essential Source of Extracellular Vesicles
4. Extracellular Vesicles Deregulation in Breast Cancer
4.1. Extracellular Vesicle and Cancer Stem Cells
4.2. Bidirectional Contributions of Extracellular Vesicles from Breast Tumor and Microenvironmental Cells to Breast Cancer Changes
4.2.1. Breast Cancer Cells-Derived Exosomes Transfer to Local Microenvironment
4.2.2. The Microenvironment Produces Exosomes That Could Transform Breast Cancer Cells
4.2.3. Local Inflammation at the Tumor Site and Extracellular Vesicles
4.3. Promotion of Tumor Expansion
4.4. Cancer Metabolism Reprogramming
4.5. Angiogenesis Induction
4.6. Immune Evasion
4.7. Metastatic Spread Induction and Secondary Settlement
4.7.1. Extracellular Vesicles and Epithelial to Mesenchymal Transition of BC Cells
4.7.2. Extracellular Vesicles Impact on Extracellular Matrix Disruption
4.7.3. Extracellular Vesicles and BC Cells Spread
4.7.4. Extracellular Vesicles, Pre-Metastatic Niche, and Secondary Organ Settlement
4.8. Cancer Cells Dormancy
4.9. Resistance to Therapy
4.9.1. Resistance to Hormone Therapy
4.9.2. Resistance to Chemotherapy
4.9.3. Resistance to Radiotherapy
4.9.4. Resistance to Targeted Therapy
5. Exosomes as Relevant Breast Cancer Biological Markers
5.1. Exosome Nucleic Acid Cargo as Biomarkers
5.1.1. Exosome mRNAs as Interesting BC Markers
5.1.2. SEVs miRNAs as Relevant BC Biological Markers
5.1.3. SEVs lncRNAs as Interesting Emerging Biomarkers
5.1.4. SEVs Circular Nucleic Acids as New Potential Diagnostic Tool
5.2. SEVs Protein Cargo as a Source of New Cancer Biomarkers
6. SEVs as Attractive Targets to Inhibit BC
6.1. Inhibition of SEV Uptake by Target Cells
6.2. Inhibition of SEV Biogenesis
SEV Release Inhibition
7. SEVs as Nanovectors to Drive Therapy in BC
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer Incidence and Mortality Patterns in Europe: Estimates for 40 Countries and 25 Major Cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef]
- Gennari, A.; André, F.; Barrios, C.H.; Cortés, J.; de Azambuja, E.; DeMichele, A.; Dent, R.; Fenlon, D.; Gligorov, J.; Hurvitz, S.A.; et al. ESMO Clinical Practice Guideline for the Diagnosis, Staging and Treatment of Patients with Metastatic Breast Cancer. Ann. Oncol. 2021, 32, 1475–1495. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast Cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef]
- Luque-Bolivar, A.; Pérez-Mora, E.; Villegas, V.E.; Rondón-Lagos, M. Resistance and Overcoming Resistance in Breast Cancer. Breast Cancer Targets Ther. 2020, 12, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Saha, T.; Lukong, K.E. Breast Cancer Stem-Like Cells in Drug Resistance: A Review of Mechanisms and Novel Therapeutic Strategies to Overcome Drug Resistance. Front. Oncol. 2022, 12, 856974. [Google Scholar] [CrossRef]
- Calaf, G.M.; Zepeda, A.B.; Castillo, R.L.; Figueroa, C.A.; Arias, C.; Figueroa, E.; Farías, J.G. Molecular Aspects of Breast Cancer Resistance to Drugs (Review). Int. J. Oncol. 2015, 47, 437–445. [Google Scholar] [CrossRef]
- Nazemi, M.; Rainero, E. Cross-Talk Between the Tumor Microenvironment, Extracellular Matrix, and Cell Metabolism in Cancer. Front. Oncol. 2020, 10, 239. [Google Scholar] [CrossRef]
- Pernot, S.; Evrard, S.; Khatib, A.-M. The Give-and-Take Interaction Between the Tumor Microenvironment and Immune Cells Regulating Tumor Progression and Repression. Front. Immunol. 2022, 13, 850856. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.R. The Matrix in Cancer. Nat. Rev. Cancer 2021, 21, 217–238. [Google Scholar] [CrossRef]
- Naito, Y.; Yoshioka, Y.; Yamamoto, Y.; Ochiya, T. How Cancer Cells Dictate Their Microenvironment: Present Roles of Extracellular Vesicles. Cell Mol. Life Sci. 2016, 74, 697–713. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Margolis, L.; Sadovsky, Y. The Biology of Extracellular Vesicles: The Known Unknowns. PLoS Biol. 2019, 17, e3000363. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular Vesicles in Cancer—Implications for Future Improvements in Cancer Care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef]
- Latifkar, A.; Cerione, R.A.; Antonyak, M.A. Probing the Mechanisms of Extracellular Vesicle Biogenesis and Function in Cancer. Biochem. Soc. Trans. 2018, 46, 1137–1146. [Google Scholar] [CrossRef]
- Rahbarghazi, R.; Jabbari, N.; Sani, N.A.; Asghari, R.; Salimi, L.; Kalashani, S.A.; Feghhi, M.; Etemadi, T.; Akbariazar, E.; Mahmoudi, M.; et al. Tumor-Derived Extracellular Vesicles: Reliable Tools for Cancer Diagnosis and Clinical Applications. Cell Commun. Signal. 2019, 17, 73. [Google Scholar] [CrossRef] [PubMed]
- Nazarenko, I. Recent Developments in Technology and Perspectives for Cancer Liquid Biopsy. In Tumor Liquid Biopsies; Recent Results in Cancer Research; Springer: Cham, Switzerland, 2020; Volume 215, pp. 319–344. [Google Scholar] [CrossRef]
- Liu, J.; Chen, Y.; Pei, F.; Zeng, C.; Yao, Y.; Liao, W.; Zhao, Z. Extracellular Vesicles in Liquid Biopsies: Potential for Disease Diagnosis. Biomed. Res. Int. 2021, 2021, 6611244. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Witwer, K.W.; Théry, C. Extracellular Vesicles or Exosomes? On Primacy, Precision, and Popularity Influencing a Choice of Nomenclature. J. Extracell. Vesicles 2019, 8, 1648167. [Google Scholar] [CrossRef]
- Caruso, S.; Poon, I.K.H. Apoptotic Cell-Derived Extracellular Vesicles: More Than Just Debris. Front. Immunol. 2018, 9, 1486. [Google Scholar] [CrossRef]
- Cocucci, E.; Meldolesi, J. Ectosomes and Exosomes: Shedding the Confusion between Extracellular Vesicles. Trends Cell Biol. 2015, 25, 364–372. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Alli, A.A. Extracellular Vesicles Mechanisms of Extracellular Vesicle Biogenesis, Cargo Loading, and Release. Physiology 2021, 13. [Google Scholar] [CrossRef]
- Sherman, C.D.; Lodha, S.; Sahoo, S. EV Cargo Sorting in Therapeutic Development for Cardiovascular Disease. Cells 2021, 10, 1500. [Google Scholar] [CrossRef]
- Mammes, A.; Pasquier, J.; Mammes, O.; Conti, M.; Douard, R.; Loric, S. Extracellular Vesicles: General Features and Usefulness in Diagnosis and Therapeutic Management of Colorectal Cancer. World J. Gastrointest. Oncol. 2021, 13, 1561–1598. [Google Scholar] [CrossRef]
- Nakano, A. The Golgi Apparatus and Its Next-Door Neighbors. Front. Cell Dev. Biol. 2022, 10, 884360. [Google Scholar] [CrossRef]
- Wang, S.; Thibault, G.; Ng, D.T.W. Routing Misfolded Proteins through the Multivesicular Body (MVB) Pathway Protects against Proteotoxicity. J. Biol. Chem. 2011, 286, 29376–29387. [Google Scholar] [CrossRef]
- Christ, L.; Raiborg, C.; Wenzel, E.M.; Campsteijn, C.; Stenmark, H. Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem. Sci. 2017, 42, 42–56. [Google Scholar] [CrossRef]
- Olmos, Y. The ESCRT Machinery: Remodeling, Repairing, and Sealing Membranes. Membranes 2022, 12, 633. [Google Scholar] [CrossRef]
- Kajimoto, T.; Okada, T.; Miya, S.; Zhang, L.; Nakamura, S. Ongoing activation of sphingosine 1-phosphate receptors mediates maturation of exosomal multivesicular endosomes. Nat. Commun. 2013, 4, 2712. [Google Scholar] [CrossRef]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT Pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef]
- Peng, X.; Yang, L.; Ma, Y.; Li, Y.; Li, H. Focus on the Morphogenesis, Fate and the Role in Tumor Progression of Multivesicu-lar Bodies. Cell Commun. Signal. 2020, 18, 122. [Google Scholar] [CrossRef]
- Scott, C.C.; Gruenberg, J. Ion Flux and the Function of Endosomes and Lysosomes: PH Is Just the Start. Bioessays 2011, 33, 103–110. [Google Scholar] [CrossRef]
- Savina, A.; Furlán, M.; Vidal, M.; Colombo, M.I. Exosome Release Is Regulated by a Calcium-dependent Mechanism in K562 Cells. J. Biol. Chem. 2003, 278, 20083–20090. [Google Scholar] [CrossRef]
- Kumar, A.; Deep, G. Hypoxia in Tumor Microenvironment Regulates Exosome Biogenesis: Molecular Mechanisms and Translational Opportunities. Cancer Lett. 2020, 479, 23–30. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, 1–40. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Eskelinen, E.-L.; Deretic, V. Autophagosomes, Phagosomes, Autolysosomes, Phagolysosomes, Autophagoly-sosomes… Wait, I’m Confused. Autophagy 2014, 10, 549–551. [Google Scholar] [CrossRef]
- Xing, H.; Tan, J.; Miao, Y.; Lv, Y.; Zhang, Q. Crosstalk between Exosomes and Autophagy: A Review of Molecular Mechanisms and Therapies. J. Cell Mol. Med. 2021, 25, 2297–2308. [Google Scholar] [CrossRef]
- Falguières, T.; Luyet, P.-P.; Gruenberg, J. Molecular Assemblies and Membrane Domains in Multivesicular Endosome Dynamics. Exp. Cell Res. 2009, 315, 1567–1573. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current Knowledge on Exosome Biogenesis and Release. Cell Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Anand, S.; Samuel, M.; Kumar, S.; Mathivanan, S. Ticket to a Bubble Ride: Cargo Sorting into Exosomes and Extracellular Vesicles. Biochim. Biophys. Acta BBA-Proteins Proteom. 2019, 1867, 140203. [Google Scholar] [CrossRef]
- Granger, E.; McNee, G.; Allan, V.; Woodman, P. The Role of the Cytoskeleton and Molecular Motors in Endosomal Dynamics. Semin. Cell Dev. Biol. 2014, 31, 20–29. [Google Scholar] [CrossRef]
- Titus, M.A. Myosin-Driven Intracellular Transport. Cold Spring Harb. Perspect. Biol. 2018, 10, a021972. [Google Scholar] [CrossRef]
- Langemeyer, L.; Fröhlich, F.; Ungermann, C. Rab GTPase Function in Endosome and Lysosome Biogenesis. Trends Cell Biol. 2018, 28, 957–970. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef]
- Song, L.; Tang, S.; Han, X.; Jiang, Z.; Dong, L.; Liu, C.; Liang, X.; Dong, J.; Qiu, C.; Wang, Y.; et al. KIBRA Controls Exosome Secretion via Inhibiting the Proteasomal Degradation of Rab27a. Nat. Commun. 2019, 10, 1639. [Google Scholar] [CrossRef]
- Hong, W.; Lev, S. Tethering the Assembly of SNARE Complexes. Trends Cell Biol. 2014, 24, 35–43. [Google Scholar] [CrossRef]
- Südhof, T.C.; Rothman, J.E. Membrane Fusion: Grappling with SNARE and SM Proteins. Science 2009, 323, 474–477. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, D.; Jin, F.; Bian, Z.; Li, L.; Liang, H.; Li, M.; Shi, L.; Pan, C.; Zhu, D.; et al. Pyruvate Kinase Type M2 Promotes Tumour Cell Exosome Release via Phosphorylating Synaptosome-Associated Protein 23. Nat. Commun. 2017, 8, 14041. [Google Scholar] [CrossRef]
- Xu, M.; Ji, J.; Jin, D.; Wu, Y.; Wu, T.; Lin, R.; Zhu, S.; Jiang, F.; Ji, Y.; Bao, B.; et al. The Biogenesis and Secretion of Exosomes and Multivesicular Bodies (MVBs): Intercellular Shuttles and Implications in Human Diseases. Genes Dis. 2022, in press. [Google Scholar] [CrossRef]
- Nabhan, J.F.; Hu, R.; Oh, R.S.; Cohen, S.N.; Lu, Q. Formation and Release of Arrestin Domain-Containing Protein 1-Mediated Microvesicles (ARMMs) at Plasma Membrane by Recruitment of TSG101 Protein. Proc. Natl. Acad. Sci. USA 2012, 109, 4146–4151. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, A.; Ochiya, T. Exosomes and Extracellular Vesicles: Rethinking the Essential Values in Cancer Biology. Semin. Cancer Biol. 2021, 74, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Juan, T.; Fürthauer, M. Biogenesis and Function of ESCRT-Dependent Extracellular Vesicles. Semin. Cell Dev. Biol. 2018, 74, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Andrews, A.M.; Rizzo, V. Microparticle-Induced Activation of the Vascular Endothelium Requires Caveolin-1/Caveolae. PLoS ONE 2016, 11, e0149272. [Google Scholar] [CrossRef]
- Li, S.; Lisanti, M.; Puszkin, S. Purification and Molecular Characterization of NP185, a Neuronal-Specific and Syn-apse-Enriched Clathrin Assembly Polypeptide. Bioquim. Y Patol. Clin. Bypc. Rev. Asoc. Bioquim. Argent. 1998, 62, 5–17. [Google Scholar]
- Sun, M.; Xue, X.; Li, L.; Xu, D.; Li, S.; Li, S.C.; Su, Q. Ectosome Biogenesis and Release Processes Observed by Using Live-Cell Dynamic Imaging in Mammalian Glial Cells. Quant. Imaging Med. Surg. 2021, 11, 4604–4616. [Google Scholar] [CrossRef] [PubMed]
- Hugel, B.; Martínez, M.C.; Kunzelmann, C.; Freyssinet, J.-M. Membrane Microparticles: Two Sides of the Coin. Physiology 2005, 20, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, J.; Wysoczynski, M.; Hayek, F.; Janowska-Wieczorek, A.; Ratajczak, M.Z. Membrane-derived microvesicles: Im-portant and underappreciated mediators of cell-to-cell communication. Leukemia 2006, 20, 1487–1495. [Google Scholar] [CrossRef]
- Bevers, E.M.; Comfurius, P.; Dekkers, D.W.C.; Zwaal, R.F.A. Lipid Translocation across the Plasma Membrane of Mammalian Cells. Biochim. Biophys. Acta BBA-Mol. Cell Biol. Lipids 1999, 1439, 317–330. [Google Scholar] [CrossRef]
- MacKenzie, A.; Wilson, H.L.; Kiss-Toth, E.; Dower, S.K.; North, R.A.; Surprenant, A. Rapid Secretion of Interleukin-1beta by Microvesicle Shedding. Immunity 2001, 15, 825–835. [Google Scholar] [CrossRef]
- Mir, B.; Goettsch, C. Extracellular Vesicles as Delivery Vehicles of Specific Cellular Cargo. Cells 2020, 9, 1601. [Google Scholar] [CrossRef]
- Isaac, R.; Reis, F.C.G.; Ying, W.; Olefsky, J.M. Exosomes as Mediators of Intercellular Crosstalk in Metabolism. Cell Metab. 2021, 33, 1744–1762. [Google Scholar] [CrossRef] [PubMed]
- Freeman, D.W.; Hooten, N.N.; Eitan, E.; Green, J.; Mode, N.A.; Bodogai, M.; Zhang, Y.; Lehrmann, E.; Zonderman, A.B.; Biragyn, A.; et al. Altered Extracellular Vesicle Concentration, Cargo, and Function in Diabetes. Diabetes 2018, 67, 2377–2388. [Google Scholar] [CrossRef]
- Burbidge, K.; Zwikelmaier, V.; Cook, B.; Long, M.M.; Balva, B.; Lonigro, M.; Ispas, G.; Rademacher, D.J.; Campbell, E.M. Cargo and Cell-Specific Differences in Extracellular Vesicle Populations Identified by Multiplexed Immunofluorescent Analysis. J. Extracell. Vesicles 2020, 9, 1789326. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Du, Y.; Peng, L.; Qin, Y.; Liu, H.; Ma, X.; Wei, Y. Extracellular Vesicle MicroRNA Cargoes from Intermittent Hypoxia-Exposed Cardiomyocytes and Their Effect on Endothelium. Biochem. Bioph. Res. Commun. 2021, 548, 182–188. [Google Scholar] [CrossRef]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef] [PubMed]
- Vietri, M.; Radulovic, M.; Stenmark, H. The Many Functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 2020, 21, 25–42. [Google Scholar] [CrossRef]
- Tahir, T. Rabs Mediated Membrane Trafficking in Cancer Progression. Digit. Med. Health Technol. 2022, 2022, 1–11. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Pollet, H.; Conrard, L.; Cloos, A.-S.; Tyteca, D. Plasma Membrane Lipid Domains as Platforms for Vesicle Biogenesis and Shedding? Biomolecules 2018, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Escola, J.-M.; Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective Enrichment of Tetraspan Proteins on the Internal Vesicles of Multivesicular Endosomes and on Exosomes Secreted by Human B-Lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar] [CrossRef] [PubMed]
- Toribio, V.; Yáñez-Mó, M. Tetraspanins Interweave EV Secretion, Endosomal Network Dynamics and Cellular Metabolism. Eur. J. Cell Biol. 2022, 101, 151229. [Google Scholar] [CrossRef] [PubMed]
- Hemler, M.E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 2003, 19, 397–422. [Google Scholar] [CrossRef] [PubMed]
- Perez-Hernandez, D.; Gutiérrez-Vázquez, C.; Jorge, I.; López-Martín, S.; Ursa, A.; Sánchez-Madrid, F.; Vázquez, J.; Yáñez-Mó, M. The Intracellular Interactome of Tetraspanin-Enriched Microdomains Reveals Their Function as Sorting Machineries toward Exosomes. J. Biol. Chem. 2013, 288, 11649–11661. [Google Scholar] [CrossRef] [PubMed]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in Extracellular Vesicle Formation and Function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The Exosome Journey: From Biogenesis to Uptake and Intracellular Signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Sahu, R.; Kaushik, S.; Clement, C.C.; Cannizzo, E.S.; Scharf, B.; Follenzi, A.; Potolicchio, I.; Nieves, E.; Cuervo, A.M.; Santambrogio, L. Microautophagy of Cytosolic Proteins by Late Endosomes. Dev. Cell 2011, 20, 131–139. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA Delivery by Extracellular Vesicles in Mammalian Cells and Its Applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Turchinovich, A.; Drapkina, O.; Tonevitsky, A. Transcriptome of Extracellular Vesicles: State-of-the-Art. Front. Immunol. 2019, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-Stranded DNA in Exosomes: A Novel Biomarker in Cancer Detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and Transfer of Mitochondrial DNA via Exosomes Regulate Escape from Dormancy in Hormonal Therapy-Resistant Breast Cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef] [PubMed]
- Vagner, T.; Spinelli, C.; Minciacchi, V.R.; Balaj, L.; Zandian, M.; Conley, A.; Zijlstra, A.; Freeman, M.R.; Demichelis, F.; De, S.; et al. Large Extracellular Vesicles Carry Most of the Tumour DNA Circulating in Prostate Cancer Patient Plasma. J. Extracell. Vesicles 2018, 7, 1505403. [Google Scholar] [CrossRef]
- Shelke, G.; Jang, S.C.; Yin, Y.; Lässer, C.; Lötvall, J. Human Mast Cells Release Extracellular Vesicle-Associated DNA. Matters 2016, 2, e201602000034. [Google Scholar] [CrossRef]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New Evidence That a Large Proportion of Human Blood Plasma Cell-Free DNA Is Localized in Exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef] [PubMed]
- Kujala, J.; Hartikainen, J.M.; Tengström, M.; Sironen, R.; Kosma, V.; Mannermaa, A. High Mutation Burden of Circulating Cell-free DNA in Early-stage Breast Cancer Patients Is Associated with a Poor Relapse-free Survival. Cancer Med. 2020, 9, 5922–5931. [Google Scholar] [CrossRef]
- Alimirzaie, S.; Bagherzadeh, M.; Akbari, M.R. Liquid Biopsy in Breast Cancer: A Comprehensive Review. Clin. Genet. 2019, 95, 643–660. [Google Scholar] [CrossRef]
- Hall, C.; Sarli, V.; Meas, S.; Lucci, A. Role of Liquid Biopsy in Clinical Decision-Making for Breast Cancer. Curr. Breast Cancer Rep. 2019, 11, 52–66. [Google Scholar] [CrossRef]
- Zebrowska, A.; Skowronek, A.; Wojakowska, A.; Widlak, P.; Pietrowska, M. Metabolome of Exosomes: Focus on Vesicles Released by Cancer Cells and Present in Human Body Fluids. Int. J. Mol. Sci. 2019, 20, 3461. [Google Scholar] [CrossRef]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in Exosomes: Current Knowledge and the Way Forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef]
- Fonseca, P.; Vardaki, I.; Occhionero, A.; Panaretakis, T. Chapter Five Metabolic and Signaling Functions of Cancer Cell-Derived Extracellular Vesicles. Int. Rev. Cell Mol. Biol. 2016, 326, 175–199. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Kowal, J.; Théry, C. Why the Need and How to Approach the Functional Diversity of Extracellular Vesicles. Phil-Osophical Trans. R. Soc. B Biol. Sci. 2018, 373, 20160479. [Google Scholar] [CrossRef] [PubMed]
- Maisano, D.; Mimmi, S.; Russo, R.; Fioravanti, A.; Fiume, G.; Vecchio, E.; Nisticò, N.; Quinto, I.; Iaccino, E. Uncovering the Exosomes Diversity: A Window of Opportunity for Tumor Progression Monitoring. Pharmaceuticals 2020, 13, 180. [Google Scholar] [CrossRef]
- Zabeo, D.; Cvjetkovic, A.; Lässer, C.; Schorb, M.; Lötvall, J.; Höög, J.L. Exosomes Purified from a Single Cell Type Have Di-verse Morphology. J. Extracell. Vesicles 2017, 6, 1329476. [Google Scholar] [CrossRef] [PubMed]
- Vagner, T.; Chin, A.; Mariscal, J.; Bannykh, S.; Engman, D.M.; Vizio, D.D. Protein Composition Reflects Extracellular Vesicle Heterogeneity. Proteomics 2019, 19, e1800167. [Google Scholar] [CrossRef]
- Montecchi, T.; Shaba, E.; Tommaso, D.D.; Giuseppe, F.D.; Angelucci, S.; Bini, L.; Landi, C.; Baldari, C.T.; Ulivieri, C. Differen-tial Proteomic Analysis of Astrocytes and Astrocytes-Derived Extracellular Vesicles from Control and Rai Knockout Mice: In-sights into the Mechanisms of Neuroprotection. Int. J. Mol. Sci. 2021, 22, 7933. [Google Scholar] [CrossRef]
- Sork, H.; Corso, G.; Krjutskov, K.; Johansson, H.J.; Nordin, J.Z.; Wiklander, O.P.B.; Lee, Y.X.F.; Westholm, J.O.; Lehtiö, J.; Wood, M.J.A.; et al. Heterogeneity and Interplay of the Extracellular Vesicle Small RNA Transcriptome and Proteome. Sci. Rep. 2018, 8, 10813. [Google Scholar] [CrossRef]
- Shaba, E.; Vantaggiato, L.; Governini, L.; Haxhiu, A.; Sebastiani, G.; Fignani, D.; Grieco, G.E.; Bergantini, L.; Bini, L.; Landi, C. Multi-Omics Integrative Approach of Extracellular Vesicles: A Future Challenging Milestone. Proteomes 2022, 10, 12. [Google Scholar] [CrossRef]
- Greening, D.W.; Simpson, R.J. Understanding Extracellular Vesicle Diversity—Current Status. Expert Rev. Proteomic 2018, 15, 887–910. [Google Scholar] [CrossRef] [PubMed]
- Kolonics, F.; Szeifert, V.; Timár, C.I.; Ligeti, E.; Lőrincz, Á.M. The Functional Heterogeneity of Neutrophil-Derived Extra-cellular Vesicles Reflects the Status of the Parent Cell. Cells 2020, 9, 2718. [Google Scholar] [CrossRef] [PubMed]
- Carnino, J.M.; Ni, K.; Jin, Y. Post-Translational Modification Regulates Formation and Cargo-Loading of Extracellular Vesicles. Front. Immunol. 2020, 11, 948. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Chen, Q.; Lin, L.; Sha, C.; Li, T.; Liu, Y.; Yin, X.; Xu, Y.; Chen, L.; Gao, W.; et al. Regulation of Exosome Production and Cargo Sorting. Int. J. Biol. Sci. 2021, 17, 163–177. [Google Scholar] [CrossRef]
- Riches, A.; Campbell, E.; Borger, E.; Powis, S. Regulation of Exosome Release from Mammary Epithelial and Breast Cancer Cells—A New Regulatory Pathway. Eur. J. Cancer 2014, 50, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Muntasell, A.; Berger, A.C.; Roche, P.A. T cell-induced secretion of MHC class II–peptide complexes on B cell exosomes. EMBO J. 2007, 26, 4263–4272. [Google Scholar] [CrossRef]
- Takahashi, Y.; Nishikawa, M.; Shinotsuka, H.; Matsui, Y.; Ohara, S.; Imai, T.; Takakura, Y. Visualization and in Vivo Tracking of the Exosomes of Murine Melanoma B16-BL6 Cells in Mice after Intravenous Injection. J. Biotechnol. 2013, 165, 77–84. [Google Scholar] [CrossRef]
- Zech, D.; Rana, S.; Büchler, M.W.; Zöller, M. Tumor-Exosomes and Leukocyte Activation: An Ambivalent Crosstalk. Cell Commun. Signal. 2012, 10, 37. [Google Scholar] [CrossRef]
- Horibe, S.; Tanahashi, T.; Kawauchi, S.; Murakami, Y.; Rikitake, Y. Mechanism of Recipient Cell-Dependent Differences in Exosome Uptake. BMC Cancer 2018, 18, 47. [Google Scholar] [CrossRef]
- Joshi, B.S.; de Beer, M.A.; Giepmans, B.N.G.; Zuhorn, I.S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14, 4444–4455. [Google Scholar] [CrossRef]
- Gonda, A.; Kabagwira, J.; Senthil, G.N.; Wall, N.R. Internalization of Exosomes through Receptor-Mediated Endocytosis. Mol. Cancer Res. 2018, 17, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Kwok, Z.H.; Wang, C.; Jin, Y. Extracellular Vesicle Transportation and Uptake by Recipient Cells: A Critical Process to Regulate Human Diseases. Process 2021, 9, 273. [Google Scholar] [CrossRef] [PubMed]
- Dzobo, K.; Senthebane, D.A.; Ganz, C.; Thomford, N.E.; Wonkam, A.; Dandara, C. Advances in Therapeutic Targeting of Cancer Stem Cells within the Tumor Microenvironment: An Updated Review. Cells 2020, 9, 1896. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, D.; Gao, Y.; Lin, C.; An, Q.; Feng, Y.; Liu, Y.; Liu, D.; Luo, H.; Wang, D. The Biology and Function of Extracellular Vesicles in Cancer Development. Front. Cell Dev. Biol. 2021, 9, 777441. [Google Scholar] [CrossRef]
- Bissell, M.J.; Rizki, A.; Mian, I.S. Tissue Architecture: The Ultimate Regulator of Breast Epithelial Function. Curr. Opin. Cell Biol. 2003, 15, 753–762. [Google Scholar] [CrossRef]
- Daniel, C.W.; Smith, G.H. The Mammary Gland: A Model for Development. J. Mammary Gland. Biol. 1999, 4, 3–8. [Google Scholar] [CrossRef]
- Krause, S.; Maffini, M.V.; Soto, A.M.; Sonnenschein, C. A Novel 3D In Vitro Culture Model to Study StromalEpithelial Interactions in the Mammary Gland. Tissue Eng. Part C Methods 2008, 14, 261–271. [Google Scholar] [CrossRef]
- Howard, B.A.; Lu, P. Stromal Regulation of Embryonic and Postnatal Mammary Epithelial Development and Differentiation. Semin. Cell Dev. Biol. 2014, 25, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Schedin, P.; Hovey, R.C. Editorial: The Mammary Stroma in Normal Development and Function. J. Mammary Gland. Biol. 2010, 15, 275–277. [Google Scholar] [CrossRef]
- Ronnov-Jessen, L.; Petersen, O.W.; Bissell, M.J. Cellular Changes Involved in Conversion of Normal to Malignant Breast: Importance of the Stromal Reaction. Physiol. Rev. 1996, 76, 69–125. [Google Scholar] [CrossRef] [PubMed]
- Welsch, U.; Oppermann, T.; Mortezza, M.; Höfter, E.; Unterberger, P. Secretory Phenomena in the Non-Lactating Human Mammary Gland. Ann. Anat.-Anat. Anz. 2007, 189, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, H.; Aoki, N.; Nakagawa, Y.; Jin-No, S.; Aoyama, K.; Oshima, K.; Ohira, S.; Sato, C.; Nadano, D.; Matsuda, T. Weaning-Induced Expression of a Milk-Fat Globule Protein, MFG-E8, in Mouse Mammary Glands, as Demonstrated by the Analyses of Its MRNA, Protein and Phosphatidylserine-Binding Activity. Biochem. J. 2006, 395, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Conklin, M.W.; Keely, P.J. Why the Stroma Matters in Breast Cancer: Insights into Breast Cancer Patient Outcomes through the Examination of Stromal Biomarkers. Cell Adhes. Migr. 2012, 6, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Pujuguet, P.; Simian, M.; Liaw, J.; Timpl, R.; Werb, Z.; Bissell, M.J. Nidogen-1 Regulates Laminin-1-Dependent Mammary-Specific Gene Expression. J. Cell Sci. 2000, 113, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, A.; Hume, A.N. Exosome Signaling in Mammary Gland Development and Cancer. Int. J. Dev. Biol. 2011, 55, 879–887. [Google Scholar] [CrossRef]
- Lin, M.-C.; Chen, S.-Y.; He, P.-L.; Luo, W.-T.; Li, H.-J. Transfer of Mammary Gland-Forming Ability Between Mammary Basal Epithelial Cells and Mammary Luminal Cells via Extracellular Vesicles/Exosomes. J. Vis. Exp. 2017, 124, e55736. [Google Scholar] [CrossRef]
- Lakkaraju, A.; Rodriguez-Boulan, E. Itinerant exosomes: Emerging roles in cell and tissue polarity. Trends Cell Biol. 2008, 18, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.R.; Yan, W.; Cao, M.; Liu, X.; Wang, S.E. Polarized Secretion of Extracellular Vesicles by Mammary Epithelia. J. Mammary Gland. Biol. 2018, 23, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Takada, R.; Noda, C.; Kobayashi, S.; Takada, S. Different Populations of Wnt-Containing Vesicles Are Individually Released from Polarized Epithelial Cells. Sci. Rep. 2016, 6, 35562. [Google Scholar] [CrossRef]
- Colombo, F.; Casella, G.; Podini, P.; Finardi, A.; Racchetti, G.; Norton, E.G.; Cocucci, E.; Furlan, R. Polarized Cells Display Asymmetric Release of Extracellular Vesicles. Traffic 2021, 22, 98–110. [Google Scholar] [CrossRef]
- Sreekumar, P.G.; Kannan, R.; Kitamura, M.; Spee, C.; Barron, E.; Ryan, S.J.; Hinton, D.R. αB Crystallin Is Apically Secreted within Exosomes by Polarized Human Retinal Pigment Epithelium and Provides Neuroprotection to Adjacent Cells. PLoS ONE 2010, 5, e12578. [Google Scholar] [CrossRef]
- Muschler, J.; Streuli, C.H. Cell–Matrix Interactions in Mammary Gland Development and Breast Cancer. Cold Spring Harb. Perspect. Biol. 2010, 2, a003202. [Google Scholar] [CrossRef]
- Sternlicht, M.D.; Kouros-Mehr, H.; Lu, P.; Werb, Z. Hormonal and Local Control of Mammary Branching Morphogenesis. Differentiation 2006, 74, 365–381. [Google Scholar] [CrossRef]
- Fu, N.Y.; Nolan, E.; Lindeman, G.J.; Visvader, J.E. Stem Cells and the Differentiation Hierarchy in Mammary Gland Development. Physiol. Rev. 2020, 100, 489–523. [Google Scholar] [CrossRef]
- Fu, N.Y.; Rios, A.C.; Pal, B.; Law, C.W.; Jamieson, P.; Liu, R.; Vaillant, F.; Jackling, F.; Liu, K.H.; Smyth, G.K.; et al. Identification of Quiescent and Spatially Restricted Mammary Stem Cells That Are Hormone Responsive. Nat. Cell Biol. 2017, 19, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Stingl, J.; Eirew, P.; Ricketson, I.; Shackleton, M.; Vaillant, F.; Choi, D.; Li, H.I.; Eaves, C.J. Purification and Unique Properties of Mammary Epithelial Stem Cells. Nature 2006, 439, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Kalisky, T.; Sahoo, D.; Dalerba, P.; Feng, W.; Lin, Y.; Qian, D.; Kong, A.; Yu, J.; Wang, F.; et al. A Quiescent Bcl11b High Stem Cell Population Is Required for Maintenance of the Mammary Gland. Cell Stem Cell 2017, 20, 247–260.e5. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.A.; Nusse, R. Wnt Proteins Are Self-Renewal Factors for Mammary Stem Cells and Promote Their Long-Term Expansion in Culture. Cell Stem Cell 2010, 6, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Cai, S.; Shin, K.; Lim, A.; Kalisky, T.; Lu, W.-J.; Clarke, M.F.; Beachy, P.A. Stromal Gli2 Activity Coordinates a Niche Signaling Program for Mammary Epithelial Stem Cells. Science 2017, 356. [Google Scholar] [CrossRef] [PubMed]
- Pardal, R.; Clarke, M.F.; Morrison, S.J. Applying the Principles of Stem-Cell Biology to Cancer. Nat. Rev. Cancer 2003, 3, 895–902. [Google Scholar] [CrossRef]
- Abdelhamed, S.; Butler, J.T.; Doron, B.; Halse, A.; Nemecek, E.; Wilmarth, P.A.; Marks, D.L.; Chang, B.H.; Horton, T.; Kurre, P. Extracellular Vesicles Impose Quiescence on Residual Hematopoietic Stem Cells in the Leukemic Niche. EMBO Rep. 2019, 20, e47546. [Google Scholar] [CrossRef]
- Victora, C.G.; Bahl, R.; Barros, A.J.D.; França, G.V.A.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st Century: Epidemiology, Mechanisms, and Lifelong Effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef]
- Liao, Y.; Du, X.; Li, J.; Lönnerdal, B. Human Milk Exosomes and Their MicroRNAs Survive Digestion in Vitro and Are Taken up by Human Intestinal Cells. Mol. Nutr. Food Res. 2017, 61, 1700082. [Google Scholar] [CrossRef] [PubMed]
- Kusuma, R.J.; Manca, S.; Friemel, T.; Sukreet, S.; Nguyen, C.; Zempleni, J. Human Vascular Endothelial Cells Transport Foreign Exosomes from Cow’s Milk by Endocytosis. Am. J. Physiol.-Cell Physiol. 2016, 310, C800–C807. [Google Scholar] [CrossRef]
- Jiang, X.; You, L.; Zhang, Z.; Cui, X.; Zhong, H.; Sun, X.; Ji, C.; Chi, X. Biological Properties of Milk-Derived Extracellular Vesicles and Their Physiological Functions in Infant. Front. Cell Dev. Biol. 2021, 9, 693534. [Google Scholar] [CrossRef] [PubMed]
- Paredes, P.T.; Gutzeit, C.; Johansson, S.; Admyre, C.; Stenius, F.; Alm, J.; Scheynius, A.; Gabrielsson, S. Differences in Exosome Populations in Human Breast Milk in Relation to Allergic Sensitization and Lifestyle. Allergy 2014, 69, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Zonneveld, M.I.; van Herwijnen, M.J.C.; Fernandez-Gutierrez, M.M.; Giovanazzi, A.; de Groot, A.M.; Kleinjan, M.; van Capel, T.M.M.; Sijts, A.J.A.M.; Taams, L.S.; Garssen, J.; et al. Human Milk Extracellular Vesicles Target Nodes in Interconnected Signalling Pathways That Enhance Oral Epithelial Barrier Function and Dampen Immune Responses. J. Extracell. Vesicles 2021, 10, e12071. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Chen, T.; Luo, J.-Y.; Zhang, L.; Xi, Q.-Y.; Jiang, Q.-Y.; Sun, J.-J.; Zhang, Y.-L. Biological Characteristics and Roles of Noncoding RNAs in Milk-Derived Extracellular Vesicles. Adv. Nutr. 2021, 12, 1006–1019. [Google Scholar] [CrossRef]
- Aguilar-Lozano, A.; Baier, S.; Grove, R.; Shu, J.; Giraud, D.; Leiferman, A.; Mercer, K.E.; Cui, J.; Badger, T.M.; Adamec, J.; et al. Concentrations of Purine Metabolites Are Elevated in Fluids from Adults and Infants and in Livers from Mice Fed Diets Depleted of Bovine Milk Exosomes and Their RNA Cargos. J. Nutr. 2018, 148, 1886–1894. [Google Scholar] [CrossRef]
- Nichols, H.B.; Schoemaker, M.J.; Cai, J.; Xu, J.; Wright, L.B.; Brook, M.N.; Jones, M.E.; Adami, H.-O.; Baglietto, L.; Bertrand, K.A.; et al. Breast Cancer Risk After Recent Childbirth: A Pooled Analysis of 15 Prospective Studies. Ann. Intern. Med. 2018, 170, 22. [Google Scholar] [CrossRef]
- Sauter, E.R.; Reidy, D. How Exosomes in Human Breast Milk May Influence Breast Cancer Risk. Transl. Cancer Res. 2017, 6, S1384–S1388. [Google Scholar] [CrossRef]
- Qin, W.; Tsukasaki, Y.; Dasgupta, S.; Mukhopadhyay, N.; Ikebe, M.; Sauter, E.R. Exosomes in Human Breast Milk Promote EMT. Clin. Cancer Res. 2016, 22, 4517–4524. [Google Scholar] [CrossRef] [PubMed]
- Mallini, P.; Lennard, T.; Kirby, J.; Meeson, A. Epithelial-to-Mesenchymal Transition: What Is the Impact on Breast Cancer Stem Cells and Drug Resistance. Cancer Treat. Rev. 2014, 40, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthi, G.; Kodumudi, K.; Gallen, C.; Zachariah, N.N.; Basu, A.; Albert, G.; Beyer, A.; Snyder, C.; Wiener, D.; Costa, R.L.B.; et al. Disseminated Cancer Cells in Breast Cancer: Mechanism of Dissemination and Dormancy and Emerging Insights on Therapeutic Opportunities. Semin. Cancer Biol. 2022, 78, 78–89. [Google Scholar] [CrossRef]
- Harmes, D.C.; DiRenzo, J. Cellular Quiescence in Mammary Stem Cells and Breast Tumor Stem Cells: Got Testable Hypotheses? J. Mammary Gland. Biol. 2009, 14, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Lin, H. Cell Biology of Stem Cells: An Enigma of Asymmetry and Self-Renewal. J. Cell Biol. 2008, 180, 257–260. [Google Scholar] [CrossRef]
- Guen, V.J.; Chavarria, T.E.; Kröger, C.; Ye, X.; Weinberg, R.A.; Lees, J.A. EMT Programs Promote Basal Mammary Stem Cell and Tumor-Initiating Cell Stemness by Inducing Primary Ciliogenesis and Hedgehog Signaling. Proc. Natl. Acad. Sci. USA 2017, 114, E10532–E10539. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Gires, O.; Zhu, L.; Liu, J.; Li, J.; Yang, H.; Ju, G.; Huang, J.; Ge, W.; Chen, Y.; et al. TSPAN8 Promotes Cancer Cell Stemness via Activation of Sonic Hedgehog Signaling. Nat. Commun. 2019, 10, 2863. [Google Scholar] [CrossRef]
- Giuli, M.V.; Giuliani, E.; Screpanti, I.; Bellavia, D.; Checquolo, S. Notch Signaling Activation as a Hallmark for Triple-Negative Breast Cancer Subtype. J. Oncol. 2019, 2019, 8707053. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective Identification of Tumorigenic Breast Cancer Cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Woodward, W.A.; Chen, M.S.; Behbod, F.; Rosen, J.M. On Mammary Stem Cells. J. Cell Sci. 2005, 118, 3585–3594. [Google Scholar] [CrossRef]
- Vuong, D.; Simpson, P.T.; Green, B.; Cummings, M.C.; Lakhani, S.R. Molecular Classification of Breast Cancer. Virchows Arch. 2014, 465, 1–14. [Google Scholar] [CrossRef]
- Parmar, H.; Cunha, G.R. Epithelial–Stromal Interactions in the Mouse and Human Mammary Gland in Vivo. Endocr.-Relat. Cancer 2004, 11, 437–458. [Google Scholar] [CrossRef]
- Ingthorsson, S.; Briem, E.; Bergthorsson, J.T.; Gudjonsson, T. Epithelial Plasticity During Human Breast Morphogenesis and Cancer Progression. J. Mammary Gland. Biol. 2016, 21, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Sansone, P.; Ceccarelli, C.; Berishaj, M.; Chang, Q.; Rajasekhar, V.K.; Perna, F.; Bowman, R.L.; Vidone, M.; Daly, L.; Nnoli, J.; et al. Self-Renewal of CD133(Hi) Cells by IL6/Notch3 Signalling Regulates Endocrine Resistance in Metastatic Breast Cancer. Nat. Commun. 2016, 7, 10442. [Google Scholar] [CrossRef]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Badve, S.; Nakshatri, H. Breast-Cancer Stem Cells—Beyond Semantics. Lancet Oncol. 2012, 13, e43–e48. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Chen, S.; Tsai, H.; He, P.; Lin, Y.; Herschman, H.; Li, H. PGE2/EP4 Signaling Controls the Transfer of the Mammary Stem Cell State by Lipid Rafts in Extracellular Vesicles. Stem Cells 2017, 35, 425–444. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, J.; Li, J.; Shao, J.; Fang, L. MiR-130a-3p Inhibits Migration and Invasion by Regulating RAB5B in Human Breast Cancer Stem Cell-like Cells. Biochem. Bioph. Res. Commun. 2018, 501, 486–493. [Google Scholar] [CrossRef]
- Visvader, J.E.; Lindeman, G.J. Cancer Stem Cells in Solid Tumours: Accumulating Evidence and Unresolved Questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef]
- Bliss, S.A.; Sinha, G.; Sandiford, O.A.; Williams, L.M.; Engelberth, D.J.; Guiro, K.; Isenalumhe, L.L.; Greco, S.J.; Ayer, S.; Bryan, M.; et al. Mesenchymal Stem Cell–Derived Exosomes Stimulate Cycling Quiescence and Early Breast Cancer Dormancy in Bone Marrow. Cancer Res. 2016, 76, 5832–5844. [Google Scholar] [CrossRef]
- Shen, M.; Dong, C.; Ruan, X.; Yan, W.; Cao, M.; Pizzo, D.; Wu, X.; Yang, L.; Liu, L.; Ren, X.; et al. Chemotherapy-Induced Extracellular Vesicle MiRNAs Promote Breast Cancer Stemness by Targeting ONECUT2. Cancer Res. 2019, 79, 3608–3621. [Google Scholar] [CrossRef]
- Ciardiello, C.; Leone, A.; Budillon, A. The Crosstalk between Cancer Stem Cells and Microenvironment Is Critical for Solid Tumor Progression: The Significant Contribution of Extracellular Vesicles. Stem Cells Int. 2018, 2018, 6392198. [Google Scholar] [CrossRef] [PubMed]
- Chou, J.; Shahi, P.; Werb, Z. MicroRNA-Mediated Regulation of the Tumor Microenvironment. Cell Cycle 2013, 12, 3262–3271. [Google Scholar] [CrossRef]
- Leal-Orta, E.; Ramirez-Ricardo, J.; Cortes-Reynosa, P.; Galindo-Hernandez, O.; Salazar, E.P. Role of PI3K/Akt on Migration and Invasion of MCF10A Cells Treated with Extracellular Vesicles from MDA-MB-231 Cells Stimulated with Linoleic Acid. J. Cell Commun. Signal. 2019, 13, 235–244. [Google Scholar] [CrossRef]
- Ozawa, P.M.M.; Alkhilaiwi, F.; Cavalli, I.J.; Malheiros, D.; Ribeiro, E.M.d.S.F.; Cavalli, L.R. Extracellular Vesicles from Triple-Negative Breast Cancer Cells Promote Proliferation and Drug Resistance in Non-Tumorigenic Breast Cells. Breast Cancer Res. Treat. 2018, 172, 713–723. [Google Scholar] [CrossRef]
- Ren, Z.; Lv, M.; Yu, Q.; Bao, J.; Lou, K.; Li, X. MicroRNA-370-3p Shuttled by Breast Cancer Cell-derived Extracellular Vesicles Induces Fibroblast Activation through the CYLD/Nf-κB Axis to Promote Breast Cancer Progression. FASEB J. 2021, 35, e21383. [Google Scholar] [CrossRef]
- Naito, Y.; Yamamoto, Y.; Sakamoto, N.; Shimomura, I.; Kogure, A.; Kumazaki, M.; Yokoi, A.; Yashiro, M.; Kiyono, T.; Yanagihara, K.; et al. Cancer Extracellular Vesicles Contribute to Stromal Heterogeneity by Inducing Chemokines in Cancer-Associated Fibroblasts. Oncogene 2019, 38, 5566–5579. [Google Scholar] [CrossRef]
- Baroni, S.; Romero-Cordoba, S.; Plantamura, I.; Dugo, M.; D’Ippolito, E.; Cataldo, A.; Cosentino, G.; Angeloni, V.; Rossini, A.; Daidone, M.G.; et al. Exosome-Mediated Delivery of MiR-9 Induces Cancer-Associated Fibroblast-like Properties in Human Breast Fibroblasts. Cell Death Dis. 2016, 7, e2312. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R.; et al. Cancer-Secreted MiR-105 Destroys Vascular Endothelial Barriers to Promote Metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Rabe, D.C.; Rustandy, F.D.; Lee, J.; Rosner, M.R. Tumor Extracellular Vesicles Are Required for Tumor-Associated Macrophage Programming. Biorxiv 2018, 375022. [Google Scholar] [CrossRef]
- Tkach, M.; Thalmensi, J.; Timperi, E.; Gueguen, P.; Névo, N.; Grisard, E.; Sirven, P.; Cocozza, F.; Gouronnec, A.; Martin-Jaular, L.; et al. Extracellular Vesicles from Triple Negative Breast Cancer Promote Pro-Inflammatory Macrophages Associated with Better Clinical Outcome. Proc. Natl. Acad. Sci. USA 2022, 119, e2107394119. [Google Scholar] [CrossRef]
- Xun, J.; Du, L.; Gao, R.; Shen, L.; Wang, D.; Kang, L.; Chen, C.; Zhang, Z.; Zhang, Y.; Yue, S.; et al. Cancer-Derived Exosomal MiR-138-5p Modulates Polarization of Tumor-Associated Macrophages through Inhibition of KDM6B. Theranostics 2021, 11, 6847–6859. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Song, X.; Li, Y.; Chen, B.; Zhao, W.; Wang, L.; Zhang, H.; Liu, Y.; Han, D.; Zhang, N.; et al. RETRACTED: LncRNA BCRT1 Promotes Breast Cancer Progression by Targeting MiR-1303/PTBP3 Axis. Mol. Cancer 2020, 19, 85. [Google Scholar] [CrossRef]
- Ham, S.; Lima, L.G.; Chai, E.P.Z.; Muller, A.; Lobb, R.J.; Krumeich, S.; Wen, S.W.; Wiegmans, A.P.; Möller, A. Breast Cancer-Derived Exosomes Alter Macrophage Polarization via Gp130/STAT3 Signaling. Front. Immunol. 2018, 9, 871. [Google Scholar] [CrossRef] [PubMed]
- Maji, S.; Chaudhary, P.; Akopova, I.; Nguyen, P.M.; Hare, R.J.; Gryczynski, I.; Vishwanatha, J.K. Exosomal Annexin II Promotes Angiogenesis and Breast Cancer Metastasis. Mol. Cancer Res. 2017, 15, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Dumas, J.-F.; Brisson, L. Interaction between Adipose Tissue and Cancer Cells: Role for Cancer Progression. Cancer Metast. Rev. 2021, 40, 31–46. [Google Scholar] [CrossRef]
- Lapeire, L.; Hendrix, A.; Lambein, K.; Bockstal, M.V.; Braems, G.; Broecke, R.V.D.; Limame, R.; Mestdagh, P.; Vandesompele, J.; Vanhove, C.; et al. Cancer-Associated Adipose Tissue Promotes Breast Cancer Progression by Paracrine Oncostatin M and Jak/STAT3 Signaling. Cancer Res. 2014, 74, 6806–6819. [Google Scholar] [CrossRef]
- Wu, Q.; Li, J.; Li, Z.; Sun, S.; Zhu, S.; Wang, L.; Wu, J.; Yuan, J.; Zhang, Y.; Sun, S.; et al. RETRACTED: Exosomes from the Tumour-Adipocyte Interplay Stimulate Beige/Brown Differentiation and Reprogram Metabolism in Stromal Adipocytes to Promote Tumour Progression. J. Exp. Clin. Cancer Res. 2019, 38, 223. [Google Scholar] [CrossRef]
- Cho, J.A.; Park, H.; Lim, E.H.; Lee, K.W. Exosomes from Breast Cancer Cells Can Convert Adipose Tissue-Derived Mesenchymal Stem Cells into Myofibroblast-like Cells. Int. J. Oncol. 2011, 40, 130–138. [Google Scholar] [CrossRef]
- Bao, Q.; Huang, Q.; Chen, Y.; Wang, Q.; Sang, R.; Wang, L.; Xie, Y.; Chen, W. Tumor-Derived Extracellular Vesicles Regulate Cancer Progression in the Tumor Microenvironment. Front. Mol. Biosci. 2022, 8, 796385. [Google Scholar] [CrossRef]
- Li, C.; Teixeira, A.F.; Zhu, H.-J.; Dijke, P. ten Cancer Associated-Fibroblast-Derived Exosomes in Cancer Progression. Mol. Cancer 2021, 20, 154. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wei, H.; Wang, J.; Li, L.; Chen, A.; Li, Z. MicroRNA-181d-5p-Containing Exosomes Derived from CAFs Promote EMT by Regulating CDX2/HOXA5 in Breast Cancer. Mol. Ther.-Nucleic Acids 2020, 19, 654–667. [Google Scholar] [CrossRef]
- Donnarumma, E.; Fiore, D.; Nappa, M.; Roscigno, G.; Adamo, A.; Iaboni, M.; Russo, V.; Affinito, A.; Puoti, I.; Quintavalle, C.; et al. Cancer-Associated Fibroblasts Release Exosomal MicroRNAs That Dictate an Aggressive Phenotype in Breast Cancer. Oncotarget 2017, 8, 19592–19608. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zeng, C.; Zhan, Y.; Wang, H.; Jiang, X.; Li, W. Aberrant Low Expression of P85α in Stromal Fibroblasts Promotes Breast Cancer Cell Metastasis through Exosome-Mediated Paracrine Wnt10b. Oncogene 2017, 36, 4692–4705. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Z.; Liu, W.; Li, X. SNHG3 Functions as MiRNA Sponge to Promote Breast Cancer Cells Growth Through the Metabolic Reprogramming. Appl. Biochem. Biotech. 2020, 191, 1084–1099. [Google Scholar] [CrossRef]
- Yang, S.-S.; Ma, S.; Dou, H.; Liu, F.; Zhang, S.-Y.; Jiang, C.; Xiao, M.; Huang, Y.-X. Breast Cancer-Derived Exosomes Regulate Cell Invasion and Metastasis in Breast Cancer via MiR-146a to Activate Cancer Associated Fibroblasts in Tumor Microenvironment. Exp. Cell Res. 2020, 391, 111983. [Google Scholar] [CrossRef]
- Rezaei, M.; Cavaco, A.C.M.; Stehling, M.; Nottebaum, A.; Brockhaus, K.; Caliandro, M.F.; Schelhaas, S.; Schmalbein, F.; Vestweber, D.; Eble, J.A. Extracellular Vesicle Transfer from Endothelial Cells Drives VE-Cadherin Expression in Breast Cancer Cells, Thereby Causing Heterotypic Cell Contacts. Cancers 2020, 12, 2138. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, G.; Gili, M.; Grange, C.; Cavallari, C.; Dentelli, P.; Togliatto, G.; Taverna, D.; Camussi, G.; Brizzi, M.F. IL-3R-Alpha Blockade Inhibits Tumor Endothelial Cell-Derived Extracellular Vesicle (EV)-Mediated Vessel Formation by Targeting the β-Catenin Pathway. Oncogene 2018, 37, 1175–1191. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, Q.; Zhang, X.; Han, Q.; Li, H.; Mao, Y.; Wang, X.; Guo, H.; Irwin, D.M.; Niu, G.; et al. Exosomes from Macrophages Exposed to Apoptotic Breast Cancer Cells Promote Breast Cancer Proliferation and Metastasis. J. Cancer 2019, 10, 2892–2906. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, Y.; Ma, L.; Chen, Y.; Liu, J.; Guo, Y.; Yu, T.; Zhang, L.; Zhu, L.; Shu, Y. Role of Exosomal Non-Coding RNAs from Tumor Cells and Tumor-Associated Macrophages in the Tumor Microenvironment. Mol. Ther. 2022, 30, 3133–3154. [Google Scholar] [CrossRef] [PubMed]
- Camera, G.L.; Gelsomino, L.; Malivindi, R.; Barone, I.; Panza, S.; Rose, D.D.; Giordano, F.; D’Esposito, V.; Formisano, P.; Bonofiglio, D.; et al. Adipocyte-Derived Extracellular Vesicles Promote Breast Cancer Cell Malignancy through HIF-1α Activity. Cancer Lett. 2021, 521, 155–168. [Google Scholar] [CrossRef]
- Wang, S.; Su, X.; Xu, M.; Xiao, X.; Li, X.; Li, H.; Keating, A.; Zhao, R.C. Exosomes Secreted by Mesenchymal Stromal/Stem Cell-Derived Adipocytes Promote Breast Cancer Cell Growth via Activation of Hippo Signaling Pathway. Stem Cell Res. Ther. 2019, 10, 117. [Google Scholar] [CrossRef]
- Ramos-Andrade, I.; Moraes, J.; Brandão-Costa, R.M.; da Silva, S.V.; de Souza, A.; da Silva, C.; Renovato-Martins, M.; Barja-Fidalgo, C. Obese Adipose Tissue Extracellular Vesicles Raise Breast Cancer Cell Malignancy. Endocr.-Relat. Cancer 2020, 27, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Wang, S.; Zhao, R.C. Exosomes from Human Adipose-Derived Mesenchymal Stem Cells Promote Migration through Wnt Signaling Pathway in a Breast Cancer Cell Model. Mol. Cell Biochem. 2013, 383, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Khanh, V.C.; Fukushige, M.; Moriguchi, K.; Yamashita, T.; Osaka, M.; Hiramatsu, Y.; Ohneda, O. Type 2 Diabetes Mellitus Induced Paracrine Effects on Breast Cancer Metastasis Through Extracellular Vesicles Derived from Human Mesenchymal Stem Cells. Stem Cells Dev. 2020, 29, 1382–1394. [Google Scholar] [CrossRef]
- Wu, S.; Wang, Y.; Yuan, Z.; Wang, S.; Du, H.; Liu, X.; Wang, Q.; Zhu, X. Human Adipose-Derived Mesenchymal Stem Cells Promote Breast Cancer MCF7 Cell Epithelial-Mesenchymal Transition by Cross Interacting with the TGF-β/Smad and PI3K/AKT Signaling Pathways. Mol. Med. Rep. 2019, 19, 177–186. [Google Scholar] [CrossRef]
- Moraes, J.A.; Encarnação, C.; Franco, V.A.; Botelho, L.G.X.; Rodrigues, G.P.; Ramos-Andrade, I.; Barja-Fidalgo, C.; Renovato-Martins, M. Adipose Tissue-Derived Extracellular Vesicles and the Tumor Microenvironment: Revisiting the Hallmarks of Cancer. Cancers 2021, 13, 3328. [Google Scholar] [CrossRef]
- Mantovani, A.; Garlanda, C.; Allavena, P. Molecular Pathways and Targets in Cancer-Related Inflammation. Ann. Med. 2010, 42, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Othman, N.; Jamal, R.; Abu, N. Cancer-Derived Exosomes as Effectors of Key Inflammation-Related Players. Front. Immunol. 2019, 10, 2103. [Google Scholar] [CrossRef]
- Chow, A.; Zhou, W.; Liu, L.; Fong, M.Y.; Champer, J.; Haute, D.V.; Chin, A.R.; Ren, X.; Gugiu, B.G.; Meng, Z.; et al. Macrophage Immunomodulation by Breast Cancer-Derived Exosomes Requires Toll-like Receptor 2-Mediated Activation of NF-ΚB. Sci. Rep. 2014, 4, 5750. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.M.; Zuppan, P.J.; Anderson, L.A.; Huey, B.; Carter, C.; King, M.C. Oncogenes and Human Breast Cancer. Am. J. Hum. Genet. 1989, 44, 577–584. [Google Scholar]
- Kalimutho, M.; Nones, K.; Srihari, S.; Duijf, P.H.G.; Waddell, N.; Khanna, K.K. Patterns of Genomic Instability in Breast Cancer. Trends Pharmacol. Sci. 2019, 40, 198–211. [Google Scholar] [CrossRef]
- Liu, B.; Wen, X.; Cheng, Y. Survival or Death: Disequilibrating the Oncogenic and Tumor Suppressive Autophagy in Cancer. Cell Death Dis. 2013, 4, e892. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e10. [Google Scholar] [CrossRef]
- Walerych, D.; Napoli, M.; Collavin, L.; Sal, G.D. The Rebel Angel: Mutant P53 as the Driving Oncogene in Breast Cancer. Carcinogenesis 2012, 33, 2007–2017. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.; Silva, J.; Herrera, A.; Herrera, M.; Peña, C.; Martín, P.; Gil-Calderón, B.; Larriba, M.J.; Coronado, M.J.; Soldevilla, B.; et al. Exosomes Enriched in Stemness/Metastatic-Related MRNAS Promote Oncogenic Potential in Breast Cancer. Oncotarget 2015, 6, 40575–40587. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Feng, Y.; Li, L.; Yao, J.; Zhou, M.; Zhao, P.; Huang, F.; Zeng, L.; Yuan, L. Long Non-Coding RNA SNHG1 Promotes Breast Cancer Progression by Regulation of LMO4. Oncol. Rep. 2020, 43, 1503–1515. [Google Scholar] [CrossRef]
- Dai, G.; Yang, Y.; Liu, S.; Liu, H. Hypoxic Breast Cancer Cell-Derived Exosomal SNHG1 Promotes Breast Cancer Growth and Angiogenesis via Regulating MiR-216b-5p/JAK2 Axis. Cancer Manag. Res. 2022, 14, 123–133. [Google Scholar] [CrossRef]
- Ma, S.; McGuire, M.H.; Mangala, L.S.; Lee, S.; Stur, E.; Hu, W.; Bayraktar, E.; Villar-Prados, A.; Ivan, C.; Wu, S.Y.; et al. Gain-of-Function P53 Protein Transferred via Small Extracellular Vesicles Promotes Conversion of Fibroblasts to a Cancer-Associated Phenotype. Cell Rep. 2021, 34, 108726. [Google Scholar] [CrossRef] [PubMed]
- Kilinc, S.; Paisner, R.; Camarda, R.; Gupta, S.; Momcilovic, O.; Kohnz, R.A.; Avsaroglu, B.; L’Etoile, N.D.; Perera, R.M.; Nomura, D.K.; et al. Oncogene-regulated release of extracellular vesicles. Dev Cell. 2021, 56, 1989–2006.e6. [Google Scholar] [CrossRef]
- Lehuédé, C.; Dupuy, F.; Rabinovitch, R.; Jones, R.G.; Siegel, P.M. Metabolic Plasticity as a Determinant of Tumor Growth and Metastasis. Cancer Res. 2016, 76, 5201–5208. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.M.R. Cancer Metabolism: The Warburg Effect Today. Exp. Mol. Pathol. 2010, 89, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does It Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Sciacovelli, M.; Frezza, C. Metabolic Reprogramming and Epithelial-to-mesenchymal Transition in Cancer. FEBS J. 2017, 284, 3132–3144. [Google Scholar] [CrossRef] [PubMed]
- Lunetti, P.; Giacomo, M.D.; Vergara, D.; Domenico, S.D.; Maffia, M.; Zara, V.; Capobianco, L.; Ferramosca, A. Metabolic Reprogramming in Breast Cancer Results in Distinct Mitochondrial Bioenergetics between Luminal and Basal Subtypes. FEBS J. 2019, 286, 688–709. [Google Scholar] [CrossRef] [PubMed]
- Vlashi, E.; Lagadec, C.; Vergnes, L.; Reue, K.; Frohnen, P.; Chan, M.; Alhiyari, Y.; Dratver, M.B.; Pajonk, F. Metabolic Differences in Breast Cancer Stem Cells and Differentiated Progeny. Breast Cancer Res. Treat. 2014, 146, 525–534. [Google Scholar] [CrossRef]
- Kim, M.-J.; Kim, D.-H.; Jung, W.-H.; Koo, J.-S. Expression of Metabolism-Related Proteins in Triple-Negative Breast Cancer. Int. J. Clin. Exp. Pathol. 2013, 7, 301–312. [Google Scholar]
- Evans, K.W.; Yuca, E.; Scott, S.S.; Zhao, M.; Arango, N.P.; Pico, C.X.C.; Saridogan, T.; Shariati, M.; Class, C.A.; Bristow, C.A.; et al. Oxidative Phosphorylation Is a Metabolic Vulnerability in Chemotherapy-Resistant Triple Negative Breast Cancer. Cancer Res. 2021, 81, 5572–5581. [Google Scholar] [CrossRef]
- Hirschhaeuser, F.; Sattler, U.G.A.; Mueller-Klieser, W. Lactate: A Metabolic Key Player in Cancer. Cancer Res. 2011, 71, 6921–6925. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.S.; Almeida, C.R.; Helguero, L.A.; Duarte, I.F. Metabolic Crosstalk in the Breast Cancer Microenvironment. Eur. J. Cancer 2019, 121, 154–171. [Google Scholar] [CrossRef]
- Martinez-Outschoorn, U.E.; Pavlides, S.; Howell, A.; Pestell, R.G.; Tanowitz, H.B.; Sotgia, F.; Lisanti, M.P. Stromal–Epithelial Metabolic Coupling in Cancer: Integrating Autophagy and Metabolism in the Tumor Microenvironment. Int. J. Biochem. Cell Biol. 2011, 43, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Bonuccelli, G.; Whitaker-Menezes, D.; Castello-Cros, R.; Pavlides, S.; Pestell, R.G.; Fatatis, A.; Witkiewicz, A.K.; Heiden, M.G.V.; Migneco, G.; Chiavarina, B.; et al. The Reverse Warburg Effect: Glycolysis Inhibitors Prevent the Tumor Promoting Effects of Caveolin-1 Deficient Cancer Associated Fibroblasts. Cell Cycle 2010, 9, 1960–1971. [Google Scholar] [CrossRef] [PubMed]
- Zaoui, M.; Morel, M.; Ferrand, N.; Fellahi, S.; Bastard, J.-P.; Lamazière, A.; Larsen, A.K.; Béréziat, V.; Atlan, M.; Sabbah, M. Breast-Associated Adipocytes Secretome Induce Fatty Acid Uptake and Invasiveness in Breast Cancer Cells via CD36 Independently of Body Mass Index, Menopausal Status and Mammary Density. Cancers 2019, 11, 2012. [Google Scholar] [CrossRef] [PubMed]
- Carracedo, A.; Cantley, L.C.; Pandolfi, P.P. Cancer Metabolism: Fatty Acid Oxidation in the Limelight. Nat. Rev. Cancer 2013, 13, 227–232. [Google Scholar] [CrossRef]
- Hensley, C.T.; Wasti, A.T.; DeBerardinis, R.J. Glutamine and Cancer: Cell Biology, Physiology, and Clinical Opportunities. J. Clin. Investig. 2013, 123, 3678–3684. [Google Scholar] [CrossRef]
- Risha, Y.; Minic, Z.; Ghobadloo, S.M.; Berezovski, M.V. The Proteomic Analysis of Breast Cell Line Exosomes Reveals Disease Patterns and Potential Biomarkers. Sci. Rep. 2020, 10, 13572. [Google Scholar] [CrossRef]
- Joudaki, N.; Rashno, M.; Asadirad, A.; Khodadadi, A. Role of Breast Cancer-Derived Exosomes in Metabolism of Immune Cells through PD1-GLUT1-HK2 Metabolic Axis. Tissue Cell 2021, 71, 101576. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.; Chin, A.R.; et al. Breast-Cancer-Secreted MiR-122 Reprograms Glucose Metabolism in Premetastatic Niche to Promote Metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef]
- Kang, S.Y.; Lee, E.J.; Byun, J.W.; Han, D.; Choi, Y.; Hwang, D.W.; Lee, D.S. Extracellular Vesicles Induce an Aggressive Phenotype in Luminal Breast Cancer Cells Via PKM2 Phosphorylation. Front. Oncol. 2021, 11, 785450. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Wu, X.; Zhou, W.; Fong, M.Y.; Cao, M.; Liu, J.; Liu, X.; Chen, C.-H.; Fadare, O.; Pizzo, D.P.; et al. Cancer-Cell-Secreted Exosomal MiR-105 Promotes Tumour Growth through the MYC-Dependent Metabolic Reprogramming of Stromal Cells. Nat. Cell Biol. 2018, 20, 597–609. [Google Scholar] [CrossRef]
- Fong, M.Y.; Yan, W.; Ghassemian, M.; Wu, X.; Zhou, X.; Cao, M.; Jiang, L.; Wang, J.; Liu, X.; Zhang, J.; et al. Cancer-secreted MiRNAs Regulate Amino-acid-induced MTORC1 Signaling and Fibroblast Protein Synthesis. EMBO Rep. 2021, 22, e51239. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Sun, S.; Li, Z.; Yang, Q.; Li, B.; Zhu, S.; Wang, L.; Wu, J.; Yuan, J.; Yang, C.; et al. RETRACTED: Tumour-Originated Exosomal MiR-155 Triggers Cancer-Associated Cachexia to Promote Tumour Progression. Mol. Cancer 2018, 17, 155. [Google Scholar] [CrossRef]
- Balaban, S.; Shearer, R.F.; Lee, L.S.; van Geldermalsen, M.; Schreuder, M.; Shtein, H.C.; Cairns, R.; Thomas, K.C.; Fazakerley, D.J.; Grewal, T.; et al. Adipocyte Lipolysis Links Obesity to Breast Cancer Growth: Adipocyte-Derived Fatty Acids Drive Breast Cancer Cell Proliferation and Migration. Cancer Metab. 2017, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv. Clin. Chem. 2016, 74, 103–141. [Google Scholar] [CrossRef] [PubMed]
- Mineo, M.; Garfield, S.H.; Taverna, S.; Flugy, A.; Leo, G.D.; Alessandro, R.; Kohn, E.C. Exosomes Released by K562 Chronic Myeloid Leukemia Cells Promote Angiogenesis in a Src-Dependent Fashion. Angiogenesis 2012, 15, 33–45. [Google Scholar] [CrossRef]
- Madu, C.O.; Wang, S.; Madu, C.O.; Lu, Y. Angiogenesis in Breast Cancer Progression, Diagnosis, and Treatment. J. Cancer 2020, 11, 4474–4494. [Google Scholar] [CrossRef]
- Akil, A.; Gutiérrez-García, A.K.; Guenter, R.; Rose, J.B.; Beck, A.W.; Chen, H.; Ren, B. Notch Signaling in Vascular Endothelial Cells, Angiogenesis, and Tumor Progression: An Update and Prospective. Front. Cell Dev. Biol. 2021, 9, 642352. [Google Scholar] [CrossRef]
- Fagiani, E.; Christofori, G. Angiopoietins in Angiogenesis. Cancer Lett. 2013, 328, 18–26. [Google Scholar] [CrossRef]
- Claesson-Welsh, L.; Welsh, M. VEGFA and Tumour Angiogenesis. J. Intern. Med. 2013, 273, 114–127. [Google Scholar] [CrossRef]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic Enhancement of Exosome Release by Breast Cancer Cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef]
- Jung, K.O.; Youn, H.; Lee, C.-H.; Kang, K.W.; Chung, J.-K. Visualization of Exosome-Mediated MiR-210 Transfer from Hypoxic Tumor Cells. Oncotarget 2016, 8, 9899–9910. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral Sphingomyelinase 2 (NSMase2)-Dependent Exosomal Transfer of Angiogenic MicroRNAs Regulate Cancer Cell Metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef]
- Matesanz, N.; Park, G.; McAllister, H.; Leahey, W.; Devine, A.; McVeigh, G.E.; Gardiner, T.A.; McDonald, D.M. Docosahexaenoic Acid Improves the Nitroso-Redox Balance and Reduces VEGF-Mediated Angiogenic Signaling in Microvascular Endothelial Cells. Investig. Ophthalmol. Vis. Sci. 2010, 51, 6815–6825. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari-Makhmalbaf, P.; Sayyad, M.; Pakravan, K.; Razmara, E.; Bitaraf, A.; Bakhshinejad, B.; Goudarzi, P.; Yousefi, H.; Pournaghshband, M.; Nemati, F.; et al. Docosahexaenoic Acid Reverses the Promoting Effects of Breast Tumor Cell-Derived Exosomes on Endothelial Cell Migration and Angiogenesis. Life Sci. 2021, 264, 118719. [Google Scholar] [CrossRef]
- Shi, P.; Liu, Y.; Yang, H.; Hu, B. Breast Cancer Derived Exosomes Promoted Angiogenesis of Endothelial Cells in Microenvironment via CircHIPK3/MiR-124-3p/MTDH Axis. Cell Signal. 2022, 95, 110338. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wang, L.; Wang, T.; Li, Y.; Xun, Q.; Zhang, R.; Liu, L.; Li, L.; Wang, W.; Tian, Y.; et al. RETRACTED: Tumor Cell-Secreted Exosomal MiR-22-3p Inhibits Transgelin and Induces Vascular Abnormalization to Promote Tumor Budding. Mol. Ther. 2021, 29, 2151–2166. [Google Scholar] [CrossRef]
- Pan, S.; Zhao, X.; Shao, C.; Fu, B.; Huang, Y.; Zhang, N.; Dou, X.; Zhang, Z.; Qiu, Y.; Wang, R.; et al. STIM1 Promotes Angiogenesis by Reducing Exosomal MiR-145 in Breast Cancer MDA-MB-231 Cells. Cell Death Dis. 2021, 12, 38. [Google Scholar] [CrossRef]
- Chaudhary, P.; Gibbs, L.D.; Maji, S.; Lewis, C.M.; Suzuki, S.; Vishwanatha, J.K. Serum Exosomal-Annexin A2 Is Associated with African-American Triple-Negative Breast Cancer and Promotes Angiogenesis. Breast Cancer Res. 2020, 22, 11. [Google Scholar] [CrossRef]
- Han, B.; Zhang, H.; Tian, R.; Liu, H.; Wang, Z.; Wang, Z.; Tian, J.; Cui, Y.; Ren, S.; Zuo, X.; et al. Exosomal EPHA2 Derived from Highly Metastatic Breast Cancer Cells Promotes Angiogenesis by Activating the AMPK Signaling Pathway through Ephrin A1-EPHA2 Forward Signaling. Theranostics 2022, 12, 4127–4146. [Google Scholar] [CrossRef]
- Thompson, C.A.; Purushothaman, A.; Ramani, V.C.; Vlodavsky, I.; Sanderson, R.D. Heparanase Regulates Secretion, Composition, and Function of Tumor Cell-Derived Exosomes. J. Biol. Chem. 2013, 288, 10093–10099. [Google Scholar] [CrossRef]
- Lee, J.-K.; Park, S.-R.; Jung, B.-K.; Jeon, Y.-K.; Lee, Y.-S.; Kim, M.-K.; Kim, Y.-G.; Jang, J.-Y.; Kim, C.-W. Exosomes Derived from Mesenchymal Stem Cells Suppress Angiogenesis by Down-Regulating VEGF Expression in Breast Cancer Cells. PLoS ONE 2013, 8, e84256. [Google Scholar] [CrossRef]
- Pakravan, K.; Babashah, S.; Sadeghizadeh, M.; Mowla, S.J.; Mossahebi-Mohammadi, M.; Ataei, F.; Dana, N.; Javan, M. MicroRNA-100 Shuttled by Mesenchymal Stem Cell-Derived Exosomes Suppresses in Vitro Angiogenesis through Modulating the MTOR/HIF-1α/VEGF Signaling Axis in Breast Cancer Cells. Cell Oncol. 2017, 40, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Suppression and Promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef]
- Alcazar, C.R.G.D.; Alečković, M.; Polyak, K. Immune Escape during Breast Tumor Progression. Cancer Immunol. Res. 2020, 8, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Sivan, A.; Corrales, L.; Gajewski, T.F. Tumor and Host Factors Controlling Antitumor Immunity and Efficacy of Cancer Immunotherapy. Adv. Immunol. 2016, 130, 75–93. [Google Scholar] [CrossRef]
- Angelova, M.; Mlecnik, B.; Vasaturo, A.; Bindea, G.; Fredriksen, T.; Lafontaine, L.; Buttard, B.; Morgand, E.; Bruni, D.; Jouret-Mourin, A.; et al. Evolution of Metastases in Space and Time under Immune Selection. Cell 2018, 175, 751–765.e16. [Google Scholar] [CrossRef] [PubMed]
- Steven, A.; Seliger, B. The Role of Immune Escape and Immune Cell Infiltration in Breast Cancer. Breast Care 2018, 13, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Gatti-Mays, M.E.; Balko, J.M.; Gameiro, S.R.; Bear, H.D.; Prabhakaran, S.; Fukui, J.; Disis, M.L.; Nanda, R.; Gulley, J.L.; Kalinsky, K.; et al. If We Build It They Will Come: Targeting the Immune Response to Breast Cancer. NPJ Breast Cancer 2019, 5, 37. [Google Scholar] [CrossRef]
- Record, M.; Subra, C.; Silvente-Poirot, S.; Poirot, M. Exosomes as Intercellular Signalosomes and Pharmacological Effectors. Biochem. Pharmacol. 2011, 81, 1171–1182. [Google Scholar] [CrossRef]
- Whiteside, T.L. Exosomes in Cancer: Another Mechanism of Tumor-Induced Immune Suppression. Adv. Exp. Med. Biol. 2017, 1036, 81–89. [Google Scholar] [CrossRef]
- Whiteside, T.L. Exosomes Carrying Immunoinhibitory Proteins and Their Role in Cancer. Clin. Amp. Exp. Immunol. 2017, 189, 259–267. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Y.; Zhu, B. T-Cell Exhaustion in the Tumor Microenvironment. Cell Death Dis. 2015, 6, e1792. [Google Scholar] [CrossRef] [PubMed]
- RONG, L.; LI, R.; LI, S.; LUO, R. Immunosuppression of Breast Cancer Cells Mediated by Transforming Growth Factor-β in Exosomes from Cancer Cells. Oncol. Lett. 2016, 11, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Wieckowski, E.U.; Visus, C.; Szajnik, M.; Szczepanski, M.J.; Storkus, W.J.; Whiteside, T.L. Tumor-Derived Microvesicles Promote Regulatory T Cell Expansion and Induce Apoptosis in Tumor-Reactive Activated CD8+ T Lymphocytes. J. Immunol. 2009, 183, 3720–3730. [Google Scholar] [CrossRef]
- Ning, Y.; Shen, K.; Wu, Q.; Sun, X.; Bai, Y.; Xie, Y.; Pan, J.; Qi, C. Tumor Exosomes Block Dendritic Cells Maturation to Decrease the T Cell Immune Response. Immunol. Lett. 2018, 199, 36–43. [Google Scholar] [CrossRef]
- Ni, C.; Fang, Q.-Q.; Chen, W.-Z.; Jiang, J.-X.; Jiang, Z.; Ye, J.; Zhang, T.; Yang, L.; Meng, F.-B.; Xia, W.-J.; et al. Breast Cancer-Derived Exosomes Transmit LncRNA SNHG16 to Induce CD73+γδ1 Treg Cells. Signal. Transduct. Target. Ther. 2020, 5, 41. [Google Scholar] [CrossRef]
- Piao, Y.J.; Kim, H.S.; Hwang, E.H.; Woo, J.; Zhang, M.; Moon, W.K. Breast Cancer Cell-Derived Exosomes and Macrophage Polarization Are Associated with Lymph Node Metastasis. Oncotarget 2017, 9, 7398–7410. [Google Scholar] [CrossRef]
- Weng, Y.-S.; Tseng, H.-Y.; Chen, Y.-A.; Shen, P.-C.; Haq, A.T.A.; Chen, L.-M.; Tung, Y.-C.; Hsu, H.-L. MCT-1/MiR-34a/IL-6/IL-6R Signaling Axis Promotes EMT Progression, Cancer Stemness and M2 Macrophage Polarization in Triple-Negative Breast Cancer. Mol. Cancer 2019, 18, 42. [Google Scholar] [CrossRef]
- Xing, F.; Liu, Y.; Wu, S.-Y.; Wu, K.; Sharma, S.; Mo, Y.-Y.; Feng, J.; Sanders, S.; Jin, G.; Singh, R.; et al. Correction: Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal MiRNA to Promote Brain Metastasis. Cancer Res. 2021, 81, 5582. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.; Pioppini, C.; Ozpolat, B.; Calin, G.A. Non-Coding RNAs Regulation of Macrophage Polarization in Cancer. Mol. Cancer 2021, 20, 24. [Google Scholar] [CrossRef]
- Dou, D.; Ren, X.; Han, M.; Xu, X.; Ge, X.; Gu, Y.; Wang, X. Cancer-Associated Fibroblasts-Derived Exosomes Suppress Immune Cell Function in Breast Cancer via the MiR-92/PD-L1 Pathway. Front. Immunol. 2020, 11, 2026. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Xia, Y.; Wu, Y.; Zhang, Z.; Wang, X.; Lu, L.; Dai, C.; Song, Y.; Xu, K.; Ji, W.; et al. Lin28B-High Breast Cancer Cells Promote Immune Suppression in the Lung Pre-Metastatic Niche via Exosomes and Support Cancer Progression. Nat. Commun. 2022, 13, 897. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yu, S.; Zinn, K.; Wang, J.; Zhang, L.; Jia, Y.; Kappes, J.C.; Barnes, S.; Kimberly, R.P.; Grizzle, W.E.; et al. Murine Mammary Carcinoma Exosomes Promote Tumor Growth by Suppression of NK Cell Function. J. Immunol. 2006, 176, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Zhuyan, J.; Chen, M.; Zhu, T.; Bao, X.; Zhen, T.; Xing, K.; Wang, Q.; Zhu, S. Critical Steps to Tumor Metastasis: Alterations of Tumor Microenvironment and Extracellular Matrix in the Formation of Pre-Metastatic and Metastatic Niche. Cell Biosci. 2020, 10, 89. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.-J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating Breast Tumor Cells Exhibit Dynamic Changes in Epithelial and Mesenchymal Composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef]
- Vella, L. The Emerging Role of Exosomes in Epithelial–Mesenchymal-Transition in Cancer. Front. Oncol. 2014, 4, 361. [Google Scholar] [CrossRef]
- Kletukhina, S.; Neustroeva, O.; James, V.; Rizvanov, A.; Gomzikova, M. Role of Mesenchymal Stem Cell-Derived Extracellular Vesicles in Epithelial–Mesenchymal Transition. Int. J. Mol. Sci. 2019, 20, 4813. [Google Scholar] [CrossRef]
- Brena, D.; Huang, M.-B.; Bond, V. Extracellular Vesicle-Mediated Transport: Reprogramming a Tumor Microenvironment Conducive with Breast Cancer Progression and Metastasis. Transl. Oncol. 2022, 15, 101286. [Google Scholar] [CrossRef]
- Jiang, J.; Li, J.; Zhou, X.; Zhao, X.; Huang, B.; Qin, Y. Exosomes Regulate the Epithelial–Mesenchymal Transition in Cancer. Front. Oncol. 2022, 12, 864980. [Google Scholar] [CrossRef]
- Gwak, J.M.; Kim, H.J.; Kim, E.J.; Chung, Y.R.; Yun, S.; Seo, A.N.; Lee, H.J.; Park, S.Y. MicroRNA-9 Is Associated with Epithelial-Mesenchymal Transition, Breast Cancer Stem Cell Phenotype, and Tumor Progression in Breast Cancer. Breast Cancer Res. Treat. 2014, 147, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Drasin, D.J.; Guarnieri, A.L.; Neelakantan, D.; Kim, J.; Cabrera, J.H.; Wang, C.-A.; Zaberezhnyy, V.; Gasparini, P.; Cascione, L.; Huebner, K.; et al. TWIST1-Induced MiR-424 Reversibly Drives Mesenchymal Programming While Inhibiting Tumor Initiation. Cancer Res. 2015, 75, 1908–1921. [Google Scholar] [CrossRef] [PubMed]
- Green, T.M.; Alpaugh, M.L.; Barsky, S.H.; Rappa, G.; Lorico, A. Breast Cancer-Derived Extracellular Vesicles: Characterization and Contribution to the Metastatic Phenotype. Biomed. Res. Int. 2015, 2015, 634865. [Google Scholar] [CrossRef]
- Zhao, Y.; Zheng, X.; Zheng, Y.; Chen, Y.; Fei, W.; Wang, F.; Zheng, C. Extracellular Matrix: Emerging Roles and Potential Therapeutic Targets for Breast Cancer. Front. Oncol. 2021, 11, 650453. [Google Scholar] [CrossRef]
- Das, A.; Mohan, V.; Krishnaswamy, V.R.; Solomonov, I.; Sagi, I. Exosomes as a Storehouse of Tissue Remodeling Proteases and Mediators of Cancer Progression. Cancer Metastasis Rev. 2019, 38, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of Extracellular Matrix Remodelling in Tumour Progression and Metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- Han, W.; Chen, S.; Yuan, W.; Fan, Q.; Tian, J.; Wang, X.; Chen, L.; Zhang, X.; Wei, W.; Liu, R.; et al. Oriented Collagen Fibers Direct Tumor Cell Intravasation. Proc. Natl. Acad. Sci. USA 2016, 113, 11208–11213. [Google Scholar] [CrossRef]
- Patwardhan, S.; Mahadik, P.; Shetty, O.; Sen, S. ECM Stiffness-Tuned Exosomes Drive Breast Cancer Motility through Thrombospondin-1. Biomaterials 2021, 279, 121185. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Q.; Wang, D.; Yang, S.; Zhou, S.; Xu, H.; Zhang, H.; Zhong, S.; Feng, J. Microenvironment-induced TIMP2 Loss by Cancer-secreted Exosomal MiR-4443 Promotes Liver Metastasis of Breast Cancer. J. Cell Physiol. 2020, 235, 5722–5735. [Google Scholar] [CrossRef]
- Gupta, G.P.; Nguyen, D.X.; Chiang, A.C.; Bos, P.D.; Kim, J.Y.; Nadal, C.; Gomis, R.R.; Manova-Todorova, K.; Massagué, J. Mediators of Vascular Remodelling Co-Opted for Sequential Steps in Lung Metastasis. Nature 2007, 446, 765–770. [Google Scholar] [CrossRef]
- Ghoroghi, S.; Mary, B.; Larnicol, A.; Asokan, N.; Klein, A.; Osmani, N.; Busnelli, I.; Delalande, F.; Paul, N.; Halary, S.; et al. Ral GTPases Promote Breast Cancer Metastasis by Controlling Biogenesis and Organ Targeting of Exosomes. Elife 2021, 10, e61539. [Google Scholar] [CrossRef] [PubMed]
- Oskarsson, T.; Batlle, E.; Massagué, J. Metastatic Stem Cells: Sources, Niches, and Vital Pathways. Cell Stem Cell 2014, 14, 306–321. [Google Scholar] [CrossRef]
- Terceiro, L.E.L.; Edechi, C.A.; Ikeogu, N.M.; Nickel, B.E.; Hombach-Klonisch, S.; Sharif, T.; Leygue, E.; Myal, Y. The Breast Tumor Microenvironment: A Key Player in Metastatic Spread. Cancers 2021, 13, 4798. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, C.; Huang, X.; Li, J.; Fu, Z.; Li, W.; Yin, Y. The Advancing Roles of Exosomes in Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 731062. [Google Scholar] [CrossRef] [PubMed]
- Luga, V.; Zhang, L.; Viloria-Petit, A.M.; Ogunjimi, A.A.; Inanlou, M.R.; Chiu, E.; Buchanan, M.; Hosein, A.N.; Basik, M.; Wrana, J.L. Exosomes Mediate Stromal Mobilization of Autocrine Wnt-PCP Signaling in Breast Cancer Cell Migration. Cell 2012, 151, 1542–1556. [Google Scholar] [CrossRef]
- Shimoda, M.; Principe, S.; Jackson, H.W.; Luga, V.; Fang, H.; Molyneux, S.D.; Shao, Y.W.; Aiken, A.; Waterhouse, P.D.; Karamboulas, C.; et al. Loss of the Timp Gene Family Is Sufficient for the Acquisition of the CAF-like Cell State. Nat. Cell Biol. 2014, 16, 889–901. [Google Scholar] [CrossRef]
- Liu, Y.; Xiang, X.; Zhuang, X.; Zhang, S.; Liu, C.; Cheng, Z.; Michalek, S.; Grizzle, W.; Zhang, H.-G. Contribution of MyD88 to the Tumor Exosome-Mediated Induction of Myeloid Derived Suppressor Cells. Am. J. Pathol. 2010, 176, 2490–2499. [Google Scholar] [CrossRef]
- Chang, J.; Chaudhuri, O. Beyond Proteases: Basement Membrane Mechanics and Cancer Invasion. J. Cell Biol. 2019, 218, 2456–2469. [Google Scholar] [CrossRef]
- Condeelis, J.; Segall, J.E. Intravital Imaging of Cell Movement in Tumours. Nat. Rev. Cancer 2003, 3, 921–930. [Google Scholar] [CrossRef]
- Modica, M.D.; Regondi, V.; Sandri, M.; Iorio, M.V.; Zanetti, A.; Tagliabue, E.; Casalini, P.; Triulzi, T. Breast Cancer-Secreted MiR-939 Downregulates VE-Cadherin and Destroys the Barrier Function of Endothelial Monolayers. Cancer Lett. 2017, 384, 94–100. [Google Scholar] [CrossRef]
- Tominaga, N.; Kosaka, N.; Ono, M.; Katsuda, T.; Yoshioka, Y.; Tamura, K.; Lötvall, J.; Nakagama, H.; Ochiya, T. Brain Metastatic Cancer Cells Release MicroRNA-181c-Containing Extracellular Vesicles Capable of Destructing Blood–Brain Barrier. Nat. Commun. 2015, 6, 6716. [Google Scholar] [CrossRef]
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-Metastatic Niches: Organ-Specific Homes for Metastases. Nat. Rev. Cancer 2017, 17, 302–317. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-X.; Olivier, G. Tumor-Derived Extracellular Vesicles in Breast Cancer: From Bench to Bedside. Cancer Lett. 2019, 460, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Ochi, H.; Sunamura, S.; Kosaka, N.; Mabuchi, Y.; Fukuda, T.; Yao, K.; Kanda, H.; Ae, K.; Okawa, A.; et al. Cancer-Secreted Hsa-MiR-940 Induces an Osteoblastic Phenotype in the Bone Metastatic Microenvironment via Targeting ARHGAP1 and FAM134A. Proc. Natl. Acad. Sci. USA 2018, 115, 2204–2209. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.; Haider, A.; Rashid, S.; Al-Nabet, A.D.M.H. Paget’s “Seed and Soil” Theory of Cancer Metastasis: An Idea Whose Time Has Come. Adv. Anat. Pathol. 2019, 26, 69–74. [Google Scholar] [CrossRef]
- Chin, A.R.; Wang, S.E. Cancer-Derived Extracellular Vesicles: The ‘Soil Conditioner’ in Breast Cancer Metastasis? Cancer Metast. Rev 2016, 35, 669–676. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, H.; Jiang, X.; Qian, C.; Liu, Z.; Luo, D. Factors Involved in Cancer Metastasis: A Better Understanding to “Seed and Soil” Hypothesis. Mol. Cancer 2017, 16, 176. [Google Scholar] [CrossRef]
- Cox, T.R.; Rumney, R.M.H.; Schoof, E.M.; Perryman, L.; Høye, A.M.; Agrawal, A.; Bird, D.; Latif, N.A.; Forrest, H.; Evans, H.R.; et al. RETRACTED: The Hypoxic Cancer Secretome Induces Pre-Metastatic Bone Lesions through Lysyl Oxidase. Nature 2015, 522, 106–110. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Giannatale, A.D.; Ceder, S.; et al. Tumour Exosome Integrins Determine Organotropic Metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Loftus, A.; Cappariello, A.; George, C.; Ucci, A.; Shefferd, K.; Green, A.; Paone, R.; Ponzetti, M.; Monache, S.D.; Muraca, M.; et al. Extracellular Vesicles From Osteotropic Breast Cancer Cells Affect Bone Resident Cells. J. Bone Miner. Res. 2020, 35, 396–412. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, G.; Hoshino, A.; Kenific, C.M.; Matei, I.R.; Steiner, L.; Freitas, D.; Kim, H.S.; Oxley, P.R.; Scandariato, I.; Casanova-Salas, I.; et al. Tumour Exosomal CEMIP Protein Promotes Cancer Cell Colonization in Brain Metastasis. Nat. Cell Biol. 2019, 21, 1403–1412. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.-C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-Induced PTEN Loss by Exosomal MicroRNA Primes Brain Metastasis Outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Zhang, Y.; Zhang, S.; Qiu, L.; Zhuang, Z.; Wei, M.; Deng, X.; Wang, Z.; Han, J. The Key Role of Exosomes on the Pre-Metastatic Niche Formation in Tumors. Front. Mol. Biosci. 2021, 8, 703640. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.W.; Sceneay, J.; Lima, L.G.; Wong, C.S.F.; Becker, M.; Krumeich, S.; Lobb, R.J.; Castillo, V.; Wong, K.N.; Ellis, S.; et al. The Biodistribution and Immune Suppressive Effects of Breast Cancer–Derived Exosomes. Cancer Res. 2016, 76, 6816–6827. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, Y.; Zhou, L.; Ma, J.; Tang, K.; Xu, P.; Ji, T.; Liang, X.; Lv, J.; Dong, W.; et al. Circulating Tumor Microparticles Promote Lung Metastasis by Reprogramming Inflammatory and Mechanical Niches via a Macrophage-Dependent Pathway. Cancer Immunol. Res. 2018, 6, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wang, Z. Immune Modulation of Metastatic Niche Formation in the Bone. Front. Immunol. 2021, 12, 765994. [Google Scholar] [CrossRef] [PubMed]
- Keklikoglou, I.; Cianciaruso, C.; Güç, E.; Squadrito, M.L.; Spring, L.M.; Tazzyman, S.; Lambein, L.; Poissonnier, A.; Ferraro, G.B.; Baer, C.; et al. Chemotherapy Elicits Pro-Metastatic Extracellular Vesicles in Breast Cancer Models. Nat. Cell Biol. 2019, 21, 190–202. [Google Scholar] [CrossRef]
- Qian, B.; Deng, Y.; Im, J.H.; Muschel, R.J.; Zou, Y.; Li, J.; Lang, R.A.; Pollard, J.W. A Distinct Macrophage Population Mediates Metastatic Breast Cancer Cell Extravasation, Establishment and Growth. PLoS ONE 2009, 4, e6562. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.-Y.; Zhang, H.; Li, X.-F.; Zhang, C.-B.; Selli, C.; Tagliavini, G.; Lam, A.D.; Prost, S.; Sims, A.H.; Hu, H.-Y.; et al. Monocyte-Derived Macrophages Promote Breast Cancer Bone Metastasis Outgrowth. J. Exp. Med. 2020, 217, e20191820. [Google Scholar] [CrossRef]
- Qian, B.-Z.; Zhang, H.; Li, J.; He, T.; Yeo, E.-J.; Soong, D.Y.H.; Carragher, N.O.; Munro, A.; Chang, A.; Bresnick, A.R.; et al. FLT1 Signaling in Metastasis-Associated Macrophages Activates an Inflammatory Signature That Promotes Breast Cancer Metastasis. J. Cell Biol. 2015, 210, 2104OIA168. [Google Scholar] [CrossRef]
- Banys, M.; Hartkopf, A.D.; Krawczyk, N.; Kaiser, T.; Meier-Stiegen, F.; Fehm, T.; Neubauer, H. Dormancy in Breast Cancer. Breast Cancer Dove Med. Press 2012, 4, 183–191. [Google Scholar] [CrossRef]
- Klein, C.A. Framework Models of Tumor Dormancy from Patient-Derived Observations. Curr. Opin. Genet. Dev. 2011, 21, 42–49. [Google Scholar] [CrossRef]
- Bushnell, G.G.; Deshmukh, A.P.; den Hollander, P.; Luo, M.; Soundararajan, R.; Jia, D.; Levine, H.; Mani, S.A.; Wicha, M.S. Breast Cancer Dormancy: Need for Clinically Relevant Models to Address Current Gaps in Knowledge. NPJ Breast Cancer 2021, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Angelis, M.L.D.; Francescangeli, F.; Torre, F.L.; Zeuner, A. Stem Cell Plasticity and Dormancy in the Development of Cancer Therapy Resistance. Front. Oncol. 2019, 9, 626. [Google Scholar] [CrossRef]
- Angelis, M.L.D.; Francescangeli, F.; Zeuner, A. Breast Cancer Stem Cells as Drivers of Tumor Chemoresistance, Dormancy and Relapse: New Challenges and Therapeutic Opportunities. Cancers 2019, 11, 1569. [Google Scholar] [CrossRef]
- Ren, Q.; Khoo, W.H.; Corr, A.P.; Phan, T.G.; Croucher, P.I.; Stewart, S.A. Gene Expression Predicts Dormant Metastatic Breast Cancer Cell Phenotype. Breast Cancer Res. 2022, 24, 10. [Google Scholar] [CrossRef]
- Muscarella, A.M.; Aguirre, S.; Hao, X.; Waldvogel, S.M.; Zhang, X.H.-F. Exploiting Bone Niches: Progression of Disseminated Tumor Cells to Metastasis. J. Clin. Investig. 2021, 131, e143764. [Google Scholar] [CrossRef] [PubMed]
- Ghajar, C.M. Metastasis Prevention by Targeting the Dormant Niche. Nat. Rev. Cancer 2015, 15, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Jahangiri, L.; Ishola, T. Dormancy in Breast Cancer, the Role of Autophagy, LncRNAs, MiRNAs and Exosomes. Int. J. Mol. Sci. 2022, 23, 5271. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.K.; Bliss, S.A.; Patel, S.A.; Taborga, M.; Dave, M.A.; Gregory, L.A.; Greco, S.J.; Bryan, M.; Patel, P.S.; Rameshwar, P. Gap Junction–Mediated Import of MicroRNA from Bone Marrow Stromal Cells Can Elicit Cell Cycle Quiescence in Breast Cancer Cells. Cancer Res. 2011, 71, 1550–1560. [Google Scholar] [CrossRef]
- Lim, H.-Y.; Wang, W.; Wessells, R.J.; Ocorr, K.; Bodmer, R. Phospholipid Homeostasis Regulates Lipid Metabolism and Cardiac Function through SREBP Signaling in Drosophila. Genes Dev. 2011, 25, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Shupp, A.B.; Neupane, M.; Agostini, L.C.; Ning, G.; Brody, J.R.; Bussard, K.M. Stromal-Derived Extracellular Vesicles Suppress Proliferation of Bone Metastatic Cancer Cells Mediated By ERK2. Mol. Cancer Res. 2021, 19, 1763–1777. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Kosaka, N.; Tominaga, N.; Yoshioka, Y.; Takeshita, F.; Takahashi, R.; Yoshida, M.; Tsuda, H.; Tamura, K.; Ochiya, T. Exosomes from Bone Marrow Mesenchymal Stem Cells Contain a MicroRNA That Promotes Dormancy in Metastatic Breast Cancer Cells. Sci. Signal. 2014, 7, ra63. [Google Scholar] [CrossRef]
- Vallabhaneni, K.C.; Penfornis, P.; Xing, F.; Hassler, Y.; Adams, K.V.; Mo, Y.-Y.; Watabe, K.; Pochampally, R. Stromal Cell Extracellular Vesicular Cargo Mediated Regulation of Breast Cancer Cell Metastasis via Ubiquitin Conjugating Enzyme E2 N Pathway. Oncotarget 2017, 8, 109861–109876. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.M.; Yeap, S.K.; Ho, W.Y.; Boo, L.; Ky, H.; Satharasinghe, D.A.; Tan, S.W.; Cheong, S.K.; Huang, H.D.; Lan, K.C.; et al. Adipose MSCs Suppress MCF7 and MDA-MB-231 Breast Cancer Metastasis and EMT Pathways Leading to Dormancy via Exosomal-MiRNAs Following Co-Culture Interaction. Pharmaceuticals 2020, 14, 8. [Google Scholar] [CrossRef]
- Walker, N.D.; Elias, M.; Guiro, K.; Bhatia, R.; Greco, S.J.; Bryan, M.; Gergues, M.; Sandiford, O.A.; Ponzio, N.M.; Leibovich, S.J.; et al. Exosomes from Differentially Activated Macrophages Influence Dormancy or Resurgence of Breast Cancer Cells within Bone Marrow Stroma. Cell Death Dis. 2019, 10, 59. [Google Scholar] [CrossRef]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Sauer, A.G.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef]
- Nilsson, S.; Mäkelä, S.; Treuter, E.; Tujague, M.; Thomsen, J.; Andersson, G.; Enmark, E.; Pettersson, K.; Warner, M.; Gustafsson, J.-Å. Mechanisms of Estrogen Action. Physiol. Rev. 2001, 81, 1535–1565. [Google Scholar] [CrossRef]
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer Treatment and Survivorship Statistics, 2022. CA Cancer J. Clin. 2022, 72, 409–436. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.K.; Schiff, R. Mechanisms of Endocrine Resistance in Breast Cancer. Annu. Rev. Med. 2011, 62, 233–247. [Google Scholar] [CrossRef]
- Clarke, R.; Tyson, J.J.; Dixon, J.M. Endocrine Resistance in Breast Cancer—An Overview and Update. Mol. Cell Endocrinol. 2015, 418, 220–234. [Google Scholar] [CrossRef] [PubMed]
- AlFakeeh, A.; Brezden-Masley, C. Overcoming Endocrine Resistance in Hormone Receptor-Positive Breast Cancer. Curr. Oncol. 2018, 25, S18–S27. [Google Scholar] [CrossRef] [PubMed]
- Hanker, A.B.; Sudhan, D.R.; Arteaga, C.L. Overcoming Endocrine Resistance in Breast Cancer. Cancer Cell 2020, 37, 496–513. [Google Scholar] [CrossRef] [PubMed]
- Camera, G.L.; Gelsomino, L.; Caruso, A.; Panza, S.; Barone, I.; Bonofiglio, D.; Andò, S.; Giordano, C.; Catalano, S. The Emerging Role of Extracellular Vesicles in Endocrine Resistant Breast Cancer. Cancers 2021, 13, 1160. [Google Scholar] [CrossRef]
- Semina, S.E.; Scherbakov, A.M.; Vnukova, A.A.; Bagrov, D.V.; Evtushenko, E.G.; Safronova, V.M.; Golovina, D.A.; Lyubchenko, L.N.; Gudkova, M.V.; Krasil’nikov, M.A. Exosome-Mediated Transfer of Cancer Cell Resistance to Antiestrogen Drugs. Mol. J. Synth. Chem. Nat. Prod. Chem. 2018, 23, 829. [Google Scholar] [CrossRef]
- Wei, Y.; Lai, X.; Yu, S.; Chen, S.; Ma, Y.; Zhang, Y.; Li, H.; Zhu, X.; Yao, L.; Zhang, J. Exosomal MiR-221/222 Enhances Tamoxifen Resistance in Recipient ER-Positive Breast Cancer Cells. Breast Cancer Res. Treat. 2014, 147, 423–431. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, S.; Tang, W.; Huang, Q.; Mei, Y.; Yang, H. Exosomes from Tamoxifen-Resistant Breast Cancer Cells Transmit Drug Resistance Partly by Delivering MiR-9-5p. Cancer Cell Int. 2021, 21, 55. [Google Scholar] [CrossRef]
- Hu, K.; Liu, X.; Li, Y.; Li, Q.; Xu, Y.; Zeng, W.; Zhong, G.; Yu, C. Exosomes Mediated Transfer of Circ_UBE2D2 Enhances the Resistance of Breast Cancer to Tamoxifen by Binding to MiR-200a-3p. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e922253-1–e922253-12. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, X.; Zeng, C.; Liu, C.; Hao, Q.; Li, W.; Zhang, K.; Zhang, W.; Wang, S.; Zhao, H.; et al. CD63+ Cancer-Associated Fibroblasts Confer Tamoxifen Resistance to Breast Cancer Cells through Exosomal MiR-22. Adv. Sci. 2020, 7, 2002518. [Google Scholar] [CrossRef]
- Xu, C.-G.; Yang, M.-F.; Ren, Y.-Q.; Wu, C.-H.; Wang, L.-Q. Exosomes Mediated Transfer of LncRNA UCA1 Results in Increased Tamoxifen Resistance in Breast Cancer Cells. Eur. Rev. Med. Pharmacol. 2016, 20, 4362–4368. [Google Scholar]
- Sansone, P.; Berishaj, M.; Rajasekhar, V.K.; Ceccarelli, C.; Chang, Q.; Strillacci, A.; Savini, C.; Shapiro, L.; Bowman, R.L.; Mastroleo, C.; et al. Evolution of Cancer Stem-like Cells in Endocrine-Resistant Metastatic Breast Cancers Is Mediated by Stromal Microvesicles. Cancer Res. 2017, 77, 1927–1941. [Google Scholar] [CrossRef] [PubMed]
- Augimeri, G.; Camera, G.L.; Gelsomino, L.; Giordano, C.; Panza, S.; Sisci, D.; Morelli, C.; Győrffy, B.; Bonofiglio, D.; Andò, S.; et al. Evidence for Enhanced Exosome Production in Aromatase Inhibitor-Resistant Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 5841. [Google Scholar] [CrossRef]
- Gradishar, W.J.; Anderson, B.O.; Abraham, J.; Aft, R.; Agnese, D.; Allison, K.H.; Blair, S.L.; Burstein, H.J.; Dang, C.; Elias, A.D.; et al. Breast Cancer, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2020, 18, 452–478. [Google Scholar] [CrossRef]
- Shedden, K.; Xie, X.T.; Chandaroy, P.; Chang, Y.T.; Rosania, G.R. Expulsion of Small Molecules in Vesicles Shed by Cancer Cells: Association with Gene Expression and Chemosensitivity Profiles. Cancer Res. 2003, 63, 4331–4337. [Google Scholar] [PubMed]
- Ifergan, I.; Scheffer, G.L.; Assaraf, Y.G. Novel Extracellular Vesicles Mediate an ABCG2-Dependent Anticancer Drug Sequestration and Resistance. Cancer Res. 2005, 65, 10952–10958. [Google Scholar] [CrossRef]
- Lv, M.; Zhu, X.; Chen, W.; Zhong, S.; Hu, Q.; Ma, T.; Zhang, J.; Chen, L.; Tang, J.; Zhao, J. Exosomes Mediate Drug Resistance Transfer in MCF-7 Breast Cancer Cells and a Probable Mechanism Is Delivery of P-Glycoprotein. Tumor Biol. 2014, 35, 10773–10779. [Google Scholar] [CrossRef]
- Ma, X.; Cai, Y.; He, D.; Zou, C.; Zhang, P.; Lo, C.Y.; Xu, Z.; Chan, F.L.; Yu, S.; Chen, Y.; et al. Transient Receptor Potential Channel TRPC5 Is Essential for P-Glycoprotein Induction in Drug-Resistant Cancer Cells. Proc. Natl. Acad. Sci. USA 2012, 109, 16282–16287. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Pan, Q.; Jiang, L.; Chen, Z.; Zhang, F.; Liu, Y.; Xing, H.; Shi, M.; Li, J.; Li, X.; et al. Tumor Endothelial Expression of P-Glycoprotein upon Microvesicular Transfer of TrpC5 Derived from Adriamycin-Resistant Breast Cancer Cells. Biochem. Biophys. Res. Commun. 2014, 446, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ning, K.; Lu, T.; Sun, X.; Jin, L.; Qi, X.; Jin, J.; Hua, D. Increasing Circulating Exosomes-carrying TRPC5 Predicts Chemoresistance in Metastatic Breast Cancer Patients. Cancer Sci. 2017, 108, 448–454. [Google Scholar] [CrossRef]
- Ma, X.; Chen, Z.; Hua, D.; He, D.; Wang, L.; Zhang, P.; Wang, J.; Cai, Y.; Gao, C.; Zhang, X.; et al. Essential Role for TrpC5-Containing Extracellular Vesicles in Breast Cancer with Chemotherapeutic Resistance. Proc. Natl. Acad. Sci. USA 2014, 111, 6389–6394. [Google Scholar] [CrossRef]
- Ning, K.; Wang, T.; Sun, X.; Zhang, P.; Chen, Y.; Jin, J.; Hua, D. UCH-L1-containing Exosomes Mediate Chemotherapeutic Resistance Transfer in Breast Cancer. J. Surg. Oncol. 2017, 115, 932–940. [Google Scholar] [CrossRef]
- Kreger, B.T.; Johansen, E.R.; Cerione, R.A.; Antonyak, M.A. The Enrichment of Survivin in Exosomes from Breast Cancer Cells Treated with Paclitaxel Promotes Cell Survival and Chemoresistance. Cancers 2016, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Liu, X.; Lv, M.; Chen, L.; Zhao, J.; Zhong, S.; Ji, M.; Hu, Q.; Luo, Z.; Wu, J.; et al. Exosomes from Drug-Resistant Breast Cancer Cells Transmit Chemoresistance by a Horizontal Transfer of MicroRNAs. PLoS ONE 2014, 9, e95240. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.; Lima, N.d.S.; Sarian, L.O.; Matheu, A.; Ribeiro, M.L.; Derchain, S.F.M. Exosome-Mediated Breast Cancer Chemoresistance via MiR-155 Transfer. Sci. Rep. 2018, 8, 829. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wu, X.; Liang, Y.; Ye, G.; Che, Y.; Wu, X.; Zhu, X.; Fan, H.; Fan, X.; Xu, J. MiR-155 Increases Stemness and Decitabine Resistance in Triple-negative Breast Cancer Cells by Inhibiting TSPAN5. Mol. Carcinog. 2020, 59, 447–461. [Google Scholar] [CrossRef]
- Li, X.J.; Ren, Z.J.; Tang, J.H.; Yu, Q. Exosomal MicroRNA MiR-1246 Promotes Cell Proliferation, Invasion and Drug Resistance by Targeting CCNG2 in Breast Cancer. Cell Physiol. Biochem. 2018, 44, 1741–1748. [Google Scholar] [CrossRef]
- Wang, B.; Wang, Y.; Wang, X.; Gu, J.; Wu, W.; Wu, H.; Wang, Q.; Zhou, D. Extracellular Vesicles Carrying MiR-887-3p Promote Breast Cancer Cell Drug Resistance by Targeting BTBD7 and Activating the Notch1/Hes1 Signaling Pathway. Dis. Markers 2022, 2022, 5762686. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, Y.; Ye, M.; Wu, J.; Ma, L.; Chen, H. Cisplatin-Resistant MDA-MB-231 Cell-Derived Exosomes Increase the Resistance of Recipient Cells in an Exosomal MiR-423-5p-Dependent Manner. Curr. Drug Metab. 2019, 20, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pei, X.; Guo, G.; Qian, X.; Dou, D.; Zhang, Z.; Xu, X.; Duan, X. Exosome-mediated Transfer of Long Noncoding RNA H19 Induces Doxorubicin Resistance in Breast Cancer. J. Cell Physiol. 2020, 235, 6896–6904. [Google Scholar] [CrossRef]
- Zhong, S.; Chen, X.; Wang, D.; Zhang, X.; Shen, H.; Yang, S.; Lv, M.; Tang, J.; Zhao, J. MicroRNA Expression Profiles of Drug-Resistance Breast Cancer Cells and Their Exosomes. Oncotarget 2016, 7, 19601–19609. [Google Scholar] [CrossRef]
- Kavanagh, E.L.; Lindsay, S.; Halasz, M.; Gubbins, L.C.; Weiner-Gorzel, K.; Guang, M.H.Z.; McGoldrick, A.; Collins, E.; Henry, M.; Blanco-Fernández, A.; et al. Protein and Chemotherapy Profiling of Extracellular Vesicles Harvested from Therapeutic Induced Senescent Triple Negative Breast Cancer Cells. Oncogenesis 2017, 6, e388. [Google Scholar] [CrossRef]
- Chen, W.; Xu, L.; Qian, Q.; He, X.; Peng, W.; Zhu, Y.; Cheng, L. Analysis of MiRNA Signature Differentially Expressed in Exosomes from Adriamycin-Resistant and Parental Human Breast Cancer Cells. Biosci. Rep. 2018, 38, BSR20181090. [Google Scholar] [CrossRef]
- Chen, W.-X.; Xu, L.-Y.; Cheng, L.; Qian, Q.; He, X.; Peng, W.-T.; Zhu, Y.-L. Bioinformatics Analysis of Dysregulated MicroRNAs in Exosomes from Docetaxel-Resistant and Parental Human Breast Cancer Cells. Cancer Manag. Res. 2019, 11, 5425–5435. [Google Scholar] [CrossRef]
- Koh, M.Z.; Ho, W.Y.; Yeap, S.K.; Ali, N.M.; Yong, C.Y.; Boo, L.; Alitheen, N.B. Exosomal-MicroRNA Transcriptome Profiling of Parental and CSC-like MDA-MB-231 Cells in Response to Cisplatin Treatment. Pathol.-Res. Pract. 2022, 233, 153854. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Zhao, Z.; Fu, B.; Li, X.; Zhao, L.; Chen, Y.; Liu, L.; Liu, R.; Li, J. Exosomes Participate in the Radiotherapy Resistance of Cancers. Radiat. Res. 2022, 197, 559–565. [Google Scholar] [CrossRef]
- Payton, C.; Pang, L.Y.; Gray, M.; Argyle, D.J. Exosomes Derived from Radioresistant Breast Cancer Cells Promote Therapeutic Resistance in Naïve Recipient Cells. J. Pers. Med. 2021, 11, 1310. [Google Scholar] [CrossRef]
- Mutschelknaus, L.; Azimzadeh, O.; Heider, T.; Winkler, K.; Vetter, M.; Kell, R.; Tapio, S.; Merl-Pham, J.; Huber, S.M.; Edalat, L.; et al. Radiation Alters the Cargo of Exosomes Released from Squamous Head and Neck Cancer Cells to Promote Migration of Recipient Cells. Sci. Rep. 2017, 7, 12423. [Google Scholar] [CrossRef]
- Drucker, A.; Yoo, B.H.; Khan, I.A.; Choi, D.; Montermini, L.; Liu, X.; Jovanovic, S.; Younis, T.; Rosen, K.V. Trastuzumab-Induced Upregulation of a Protein Set in Extracellular Vesicles Emitted by ErbB2-Positive Breast Cancer Cells Correlates with Their Trastuzumab Sensitivity. Breast Cancer Res. 2020, 22, 105. [Google Scholar] [CrossRef]
- Jabbari, N.; Nawaz, M.; Rezaie, J. Ionizing Radiation Increases the Activity of Exosomal Secretory Pathway in MCF-7 Human Breast Cancer Cells: A Possible Way to Communicate Resistance against Radiotherapy. Int. J. Mol. Sci. 2019, 20, 3649. [Google Scholar] [CrossRef] [PubMed]
- Diamond, J.M.; Vanpouille-Box, C.; Spada, S.; Rudqvist, N.-P.; Chapman, J.R.; Ueberheide, B.M.; Pilones, K.A.; Sarfraz, Y.; Formenti, S.C.; Demaria, S. Exosomes Shuttle TREX1-Sensitive IFN-Stimulatory DsDNA from Irradiated Cancer Cells to DCs. Cancer Immunol. Res. 2018, 6, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Clark, G.C.; Hampton, J.D.; Koblinski, J.E.; Quinn, B.; Mahmoodi, S.; Metcalf, O.; Guo, C.; Peterson, E.; Fisher, P.B.; Farrell, N.P.; et al. Radiation Induces ESCRT Pathway Dependent CD44v3+ Extracellular Vesicle Production Stimulating Pro-Tumor Fibroblast Activity in Breast Cancer. Front. Oncol. 2022, 12, 913656. [Google Scholar] [CrossRef] [PubMed]
- Cortesi, L.; Toss, A.; Cirilli, C.; Marcheselli, L.; Braghiroli, B.; Sebastiani, F.; Federico, M. Twenty-years Experience with de Novo Metastatic Breast Cancer. Int. J. Cancer 2015, 137, 1417–1426. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, P.; Tang, H.; Xie, X.; Kong, Y.; Song, C.; Qiu, X.; Xiao, X. AFAP1-AS1 Promotes Epithelial-Mesenchymal Transition and Tumorigenesis Through Wnt/β-Catenin Signaling Pathway in Triple-Negative Breast Cancer. Front. Pharmacol. 2018, 9, 1248. [Google Scholar] [CrossRef]
- Zhang, X.; Li, F.; Zhou, Y.; Mao, F.; Lin, Y.; Shen, S.; Li, Y.; Zhang, S.; Sun, Q. Long noncoding RNA AFAP1-AS1 promotes tumor progression and invasion by regulating the miR-2110/Sp1 axis in triple-negative breast cancer. Cell Death Dis. 2020, 12, 627. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Chen, M.; Xing, P.; Yan, X.; Xie, B. Increased Expression of Exosomal AGAP2-AS1 (AGAP2 Antisense RNA 1) In Breast Cancer Cells Inhibits Trastuzumab-Induced Cell Cytotoxicity. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 2211–2220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yan, C.; Wang, Y. Exosome-Mediated Transfer of CircHIPK3 Promotes Trastuzumab Chemoresistance in Breast Cancer. J. Drug Target. 2021, 29, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, L.; Yu, G.; Sun, Z.; Wang, T.; Tian, X.; Duan, X.; Zhang, C. Exosomal MiR-1246 and MiR-155 as Predictive and Prognostic Biomarkers for Trastuzumab-Based Therapy Resistance in HER2-Positive Breast Cancer. Cancer Chemother. Pharmacol. 2020, 86, 761–772. [Google Scholar] [CrossRef]
- Martinez, V.G.; O’Neill, S.; Salimu, J.; Breslin, S.; Clayton, A.; Crown, J.; O’Driscoll, L. Resistance to HER2-Targeted Anti-Cancer Drugs Is Associated with Immune Evasion in Cancer Cells and Their Derived Extracellular Vesicles. Oncoimmunology 2017, 6, e1362530. [Google Scholar] [CrossRef]
- Cornell, L.; Wander, S.A.; Visal, T.; Wagle, N.; Shapiro, G.I. MicroRNA-Mediated Suppression of the TGF-β Pathway Confers Transmissible and Reversible CDK4/6 Inhibitor Resistance. Cell Rep. 2019, 26, 2667–2680.e7. [Google Scholar] [CrossRef] [PubMed]
- Re, M.D.; Bertolini, I.; Crucitta, S.; Fontanelli, L.; Rofi, E.; Angelis, C.D.; Diodati, L.; Cavallero, D.; Gianfilippo, G.; Salvadori, B.; et al. Overexpression of TK1 and CDK9 in Plasma-Derived Exosomes Is Associated with Clinical Resistance to CDK4/6 Inhibitors in Metastatic Breast Cancer Patients. Breast Cancer Res. Treat. 2019, 178, 57–62. [Google Scholar] [CrossRef]
- Zhang, Z.; Ma, X.; Ekiert, J.; Simons, Y.; Coleman, L.; Truica, C.I.; Blaes, A.H.; Rana, J.; Tandra, P.; Green, L.; et al. Identification of Exosome Protein Biomarkers in Patients with Advanced Hormone Receptor-Positive Breast Cancer Treated with Palbociclib and Tamoxifen. J. Clin. Oncol. 2022, 40, e13014. [Google Scholar] [CrossRef]
- Parise, C.A.; Caggiano, V. Risk of Mortality of Node-Negative, ER/PR/HER2 Breast Cancer Subtypes in T1, T2, and T3 Tumors. Breast Cancer Res. Treat. 2017, 165, 743–750. [Google Scholar] [CrossRef]
- Tirada, N.; Aujero, M.; Khorjekar, G.; Richards, S.; Chopra, J.; Dromi, S.; Ioffe, O. Breast Cancer Tissue Markers, Genomic Profiling, and Other Prognostic Factors: A Primer for Radiologists. Radiographics 2018, 38, 1902–1920. [Google Scholar] [CrossRef]
- Seale, K.N.; Tkaczuk, K.H.R. Circulating Biomarkers in Breast Cancer. Clin. Breast Cancer 2021, 22, e319–e331. [Google Scholar] [CrossRef]
- Veyssière, H.; Bidet, Y.; Penault-Llorca, F.; Radosevic-Robin, N.; Durando, X. Circulating Proteins as Predictive and Prognostic Biomarkers in Breast Cancer. Clin. Proteom. 2022, 19, 25. [Google Scholar] [CrossRef] [PubMed]
- Tay, T.K.Y.; Tan, P.H. Liquid Biopsy in Breast Cancer: A Focused Review. Arch. Pathol. Lab. Med. 2020, 145, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Nassar, F.J.; Chamandi, G.; Tfaily, M.A.; Zgheib, N.K.; Nasr, R. Peripheral Blood-Based Biopsy for Breast Cancer Risk Prediction and Early Detection. Front. Med. 2020, 7, 28. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, K.; Wang, Z.; Wang, Y.; Liu, J.; Lin, L.; Shao, Y.; Gao, L.; Yin, H.; Cui, C.; et al. DNA in Serum Extracellular Vesicles Is Stable under Different Storage Conditions. BMC Cancer 2016, 16, 753. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef]
- Ayala-Mar, S.; Donoso-Quezada, J.; Gallo-Villanueva, R.C.; Perez-Gonzalez, V.H.; González-Valdez, J. Recent Advances and Challenges in the Recovery and Purification of Cellular Exosomes. Electrophoresis 2019, 40, 3036–3049. [Google Scholar] [CrossRef]
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on Strategies and Technologies for Exosome Isolation and Purification. Front. Bioeng. Biotechnol. 2022, 9, 811971. [Google Scholar] [CrossRef]
- Kumar, D.N.; Chaudhuri, A.; Aqil, F.; Dehari, D.; Munagala, R.; Singh, S.; Gupta, R.C.; Agrawal, A.K. Exosomes as Emerging Drug Delivery and Diagnostic Modality for Breast Cancer: Recent Advances in Isolation and Application. Cancers 2022, 14, 1435. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of MRNAs and MicroRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Vila, M.; Yoshioka, Y.; Ochiya, T. Biological Functions Driven by MRNAs Carried by Extracellular Vesicles in Cancer. Front. Cell Dev. Biol. 2021, 9, 620498. [Google Scholar] [CrossRef] [PubMed]
- Zomer, A.; Maynard, C.; Verweij, F.J.; Kamermans, A.; Schäfer, R.; Beerling, E.; Schiffelers, R.M.; de Wit, E.; Berenguer, J.; Ellenbroek, S.I.J.; et al. In Vivo Imaging Reveals Extracellular Vesicle-Mediated Phenocopying of Metastatic Behavior. Cell 2015, 161, 1046–1057. [Google Scholar] [CrossRef]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in Extracellular Vesicles: RNA in Extracellular Vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, e1413. [Google Scholar] [CrossRef]
- Jenjaroenpun, P.; Kremenska, Y.; Nair, V.M.; Kremenskoy, M.; Joseph, B.; Kurochkin, I.V. Characterization of RNA in Exosomes Secreted by Human Breast Cancer Cell Lines Using Next-Generation Sequencing. PeerJ 2013, 1, e201. [Google Scholar] [CrossRef]
- Conley, A.; Minciacchi, V.R.; Lee, D.H.; Knudsen, B.S.; Karlan, B.Y.; Citrigno, L.; Viglietto, G.; Tewari, M.; Freeman, M.R.; Demichelis, F.; et al. High-Throughput Sequencing of Two Populations of Extracellular Vesicles Provides an MRNA Signature That Can Be Detected in the Circulation of Breast Cancer Patients. RNA Biol. 2016, 14, 305–316. [Google Scholar] [CrossRef]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef]
- Zhao, F.; Cheng, L.; Shao, Q.; Chen, Z.; Lv, X.; Li, J.; He, L.; Sun, Y.; Ji, Q.; Lu, P.; et al. Characterization of Serum Small Extracellular Vesicles and Their Small RNA Contents across Humans, Rats, and Mice. Sci. Rep. 2020, 10, 4197. [Google Scholar] [CrossRef]
- Ozawa, P.M.M.; Jucoski, T.S.; Vieira, E.; Carvalho, T.M.; Malheiros, D.; Ribeiro, E.M.d.S.F. Liquid Biopsy for Breast Cancer Using Extracellular Vesicles and Cell-Free MicroRNAs as Biomarkers. Transl. Res. 2020, 223, 40–60. [Google Scholar] [CrossRef]
- Sempere, L.F.; Keto, J.; Fabbri, M. Exosomal MicroRNAs in Breast Cancer towards Diagnostic and Therapeutic Applications. Cancers 2017, 9, 71. [Google Scholar] [CrossRef]
- Aggarwal, T.; Wadhwa, R.; Gupta, R.; Paudel, K.R.; Collet, T.; Chellappan, D.K.; Gupta, G.; Perumalsamy, H.; Mehta, M.; Satija, S.; et al. MicroRNAs as Biomarker for Breast Cancer. Endocr. Metab. Immune Disord.-Drug Targets 2019, 20, 1597–1610. [Google Scholar] [CrossRef]
- Khalife, H.; Skafi, N.; Fayyad-Kazan, M.; Badran, B. MicroRNAs in Breast Cancer: New Maestros Defining the Melody. Cancer Genet. 2020, 246–247, 18–40. [Google Scholar] [CrossRef] [PubMed]
- Richard, V.; Davey, M.G.; Annuk, H.; Miller, N.; Dwyer, R.M.; Lowery, A.; Kerin, M.J. MicroRNAs in Molecular Classification and Pathogenesis of Breast Tumors. Cancers 2021, 13, 5332. [Google Scholar] [CrossRef] [PubMed]
- Shi, J. Considering Exosomal MiR-21 as a Biomarker for Cancer. J. Clin. Med. 2016, 5, 42. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Wang, J.; Wang, X.; Shi, S.; Wang, W.; Chen, Z. Appraising MicroRNA-155 as a Noninvasive Diagnostic Biomarker for Cancer Detection. Medicine 2016, 95, e2450. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Iinuma, H.; Umemoto, Y.; Yanagisawa, T.; Matsumoto, A.; Jinno, H. Exosome-Encapsulated MicroRNA-223-3p as a Minimally Invasive Biomarker for the Early Detection of Invasive Breast Cancer. Oncol. Lett. 2018, 15, 9584–9592. [Google Scholar] [CrossRef]
- Eichelser, C.; Stückrath, I.; Müller, V.; Milde-Langosch, K.; Wikman, H.; Pantel, K.; Schwarzenbach, H. Increased Serum Levels of Circulating Exosomal MicroRNA-373 in Receptor-Negative Breast Cancer Patients. Oncotarget 2014, 5, 9650–9663. [Google Scholar] [CrossRef]
- Shen, S.; Song, Y.; Zhao, B.; Xu, Y.; Ren, X.; Zhou, Y.; Sun, Q. Cancer-Derived Exosomal MiR-7641 Promotes Breast Cancer Progression and Metastasis. Cell Commun. Signal. 2021, 19, 20. [Google Scholar] [CrossRef]
- Hannafon, B.N.; Trigoso, Y.D.; Calloway, C.L.; Zhao, Y.D.; Lum, D.H.; Welm, A.L.; Zhao, Z.J.; Blick, K.E.; Dooley, W.C.; Ding, W.Q. Plasma Exosome MicroRNAs Are Indicative of Breast Cancer. Breast Cancer Res. 2016, 18, 90. [Google Scholar] [CrossRef] [PubMed]
- Motawi, T.M.K.; Sadik, N.A.H.; Shaker, O.G.; Masry, M.R.E.; Mohareb, F. Study of MicroRNAs-21/221 as Potential Breast Cancer Biomarkers in Egyptian Women. Gene 2016, 590, 210–219. [Google Scholar] [CrossRef]
- Ali, S.A.; Abdulrahman, Z.F.A.; Faraidun, H.N. Circulatory MiRNA-155, MiRNA-21 Target PTEN Expression and Activity as a Factor in Breast Cancer Development. Cell Mol. Biol. 2020, 66, 44–50. [Google Scholar] [CrossRef]
- Konoshenko, M.; Sagaradze, G.; Orlova, E.; Shtam, T.; Proskura, K.; Kamyshinsky, R.; Yunusova, N.; Alexandrova, A.; Efimenko, A.; Tamkovich, S. Total Blood Exosomes in Breast Cancer: Potential Role in Crucial Steps of Tumorigenesis. Int. J. Mol. Sci. 2020, 21, 7341. [Google Scholar] [CrossRef] [PubMed]
- Ni, Q.; Stevic, I.; Pan, C.; Müller, V.; Oliveira-Ferrer, L.; Pantel, K.; Schwarzenbach, H. Different Signatures of MiR-16, MiR-30b and MiR-93 in Exosomes from Breast Cancer and DCIS Patients. Sci. Rep. 2018, 8, 12974. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, A.; de Miguel-Pérez, D.; Ortega, F.G.; García-Puche, J.L.; Robles-Fernández, I.; Exposito, J.; Martorell-Marugan, J.; Carmona-Sáez, P.; Garrido-Navas, M.d.C.; Rolfo, C.; et al. Exosomal MiRNA Profile as Complementary Tool in the Diagnostic and Prediction of Treatment Response in Localized Breast Cancer under Neoadjuvant Chemotherapy. Breast Cancer Res. 2019, 21, 21. [Google Scholar] [CrossRef] [PubMed]
- Han, J.-G.; Jiang, Y.-D.; Zhang, C.-H.; Yang, Y.-M.; Pang, D.; Song, Y.-N.; Zhang, G.-Q. A Novel Panel of Serum MiR-21/MiR-155/MiR-365 as a Potential Diagnostic Biomarker for Breast Cancer. Ann. Surg. Treat. Res. 2017, 92, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Villasana, V.; Rashed, M.H.; Gonzalez-Cantú, Y.; Bayraktar, R.; Menchaca-Arredondo, J.L.; Vazquez-Guillen, J.M.; Rodriguez-Padilla, C.; Lopez-Berestein, G.; Resendez-Perez, D. Presence of Circulating MiR-145, MiR-155, and MiR-382 in Exosomes Isolated from Serum of Breast Cancer Patients and Healthy Donors. Dis. Markers 2019, 2019, 6852917. [Google Scholar] [CrossRef] [PubMed]
- Khalighfard, S.; Alizadeh, A.M.; Irani, S.; Omranipour, R. Plasma MiR-21, MiR-155, MiR-10b, and Let-7a as the Potential Biomarkers for the Monitoring of Breast Cancer Patients. Sci. Rep. 2018, 8, 17981. [Google Scholar] [CrossRef]
- Fang, R.; Zhu, Y.; Hu, L.; Khadka, V.S.; Ai, J.; Zou, H.; Ju, D.; Jiang, B.; Deng, Y.; Hu, X. Plasma MicroRNA Pair Panels as Novel Biomarkers for Detection of Early Stage Breast Cancer. Front. Physiol. 2019, 9, 1879. [Google Scholar] [CrossRef]
- Shamsizadeh, S.; Goliaei, S.; Moghadam, Z.R. CAMIRADA: Cancer MicroRNA Association Discovery Algorithm, a Case Study on Breast Cancer. J. Biomed. Inform. 2019, 94, 103180. [Google Scholar] [CrossRef]
- Lv, S.; Wang, Y.; Xu, W.; Dong, X. Serum Exosomal MiR-17-5p as a Promising Biomarker Diagnostic Biomarker for Breast Cancer. Clin. Lab. 2020, 66. [Google Scholar] [CrossRef]
- Shimomura, A.; Shiino, S.; Kawauchi, J.; Takizawa, S.; Sakamoto, H.; Matsuzaki, J.; Ono, M.; Takeshita, F.; Niida, S.; Shimizu, C.; et al. Novel Combination of Serum MicroRNA for Detecting Breast Cancer in the Early Stage. Cancer Sci. 2016, 107, 326–334. [Google Scholar] [CrossRef]
- Asadirad, A.; Khodadadi, A.; Talaiezadeh, A.; Shohan, M.; Rashno, M.; Joudaki, N. Evaluation of MiRNA-21-5p and MiRNA-10b-5p Levels in Serum-Derived Exosomes of Breast Cancer Patients in Different Grades. Mol. Cell Probes 2022, 64, 101831. [Google Scholar] [CrossRef] [PubMed]
- Hamam, R.; Ali, A.M.; Alsaleh, K.A.; Kassem, M.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. MicroRNA Expression Profiling on Individual Breast Cancer Patients Identifies Novel Panel of Circulating MicroRNA for Early Detection. Sci. Rep. 2016, 6, 25997. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, B.; Shi, X.; Zhang, J.; Chen, A.M.; Xu, J.; Wang, W.; Huang, K.; Gao, J.; Zheng, Z.; et al. Cross-Platform Genomic Identification and Clinical Validation of Breast Cancer Diagnostic Biomarkers. Aging 2021, 13, 4258–4273. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Hu, J.; Yang, Y.; Hu, H.; Zhou, D.; Ma, M.; Xu, N. Plasma Extracellular Vesicle-Packaged MicroRNAs as Candidate Diagnostic Biomarkers for Early-Stage Breast Cancer. Mol. Med. Rep. 2019, 20, 3991–4002. [Google Scholar] [CrossRef] [PubMed]
- Adam-Artigues, A.; Garrido-Cano, I.; Carbonell-Asins, J.A.; Lameirinhas, A.; Simón, S.; Ortega-Morillo, B.; Martínez, M.T.; Hernando, C.; Constâncio, V.; Burgues, O.; et al. Identification of a Two-MicroRNA Signature in Plasma as a Novel Biomarker for Very Early Diagnosis of Breast Cancer. Cancers 2021, 13, 2848. [Google Scholar] [CrossRef]
- Jang, J.Y.; Kim, Y.S.; Kang, K.N.; Kim, K.H.; Park, Y.J.; Kim, C.W. Multiple MicroRNAs as Biomarkers for Early Breast Cancer Diagnosis. Mol. Clin. Oncol. 2021, 14, 31. [Google Scholar] [CrossRef] [PubMed]
- Borsos, B.N.; Páhi, Z.G.; Ujfaludi, Z.; Sükösd, F.; Nikolényi, A.; Bankó, S.; Pankotai-Bodó, G.; Oláh-Németh, O.; Pankotai, T. BC-MiR: Monitoring Breast Cancer-Related MiRNA Profile in Blood Sera—A Prosperous Approach for Tumor Detection. Cells 2022, 11, 2721. [Google Scholar] [CrossRef] [PubMed]
- Zografos, E.; Zagouri, F.; Kalapanida, D.; Zakopoulou, R.; Kyriazoglou, A.; Apostolidou, K.; Gazouli, M.; Dimopoulos, M.-A. Prognostic Role of MicroRNAs in Breast Cancer: A Systematic Review. Oncotarget 2019, 10, 7156–7178. [Google Scholar] [CrossRef] [PubMed]
- Pourteimoor, V.; Paryan, M.; Mohammadi-Yeganeh, S. MicroRNA as a Systemic Intervention in the Specific Breast Cancer Subtypes with C-MYC Impacts; Introducing Subtype-based Appraisal Tool. J. Cell Physiol. 2018, 233, 5655–5669. [Google Scholar] [CrossRef] [PubMed]
- Stevic, I.; Müller, V.; Weber, K.; Fasching, P.A.; Karn, T.; Marmé, F.; Schem, C.; Stickeler, E.; Denkert, C.; van Mackelenbergh, M.; et al. Specific MicroRNA Signatures in Exosomes of Triple-Negative and HER2-Positive Breast Cancer Patients Undergoing Neoadjuvant Therapy within the GeparSixto Trial. BMC Med. 2018, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- Huo, D.; Clayton, W.M.; Yoshimatsu, T.F.; Chen, J.; Olopade, O.I. Identification of a Circulating MicroRNA Signature to Distinguish Recurrence in Breast Cancer Patients. Oncotarget 2016, 7, 55231–55248. [Google Scholar] [CrossRef]
- Papadaki, C.; Thomopoulou, K.; Koronakis, G.; Monastirioti, A.A.; Papadaki, M.A.; Kalbakis, K.; Agelaki, S.; Mavroudis, D. MicroRNAs Involved in Immune Response as Prognostic Markers in Early and Metastatic Breast Cancer. J. Clin. Oncol. 2020, 38, e15528. [Google Scholar] [CrossRef]
- Thomopoulou, K.; Papadaki, C.; Monastirioti, A.; Koronakis, G.; Mala, A.; Kalapanida, D.; Mavroudis, D.; Agelaki, S. MicroRNAs Regulating Tumor Immune Response in the Prediction of the Outcome in Patients With Breast Cancer. Front. Mol. Biosci. 2021, 8, 668534. [Google Scholar] [CrossRef]
- Joshi, S.; Garlapati, C.; Bhattarai, S.; Su, Y.; Rios-Colon, L.; Deep, G.; Torres, M.A.; Aneja, R. Exosomal Metabolic Signatures Are Associated with Differential Response to Neoadjuvant Chemotherapy in Patients with Breast Cancer. Int. J. Mol. Sci. 2022, 23, 5324. [Google Scholar] [CrossRef]
- Todorova, V.K.; Byrum, S.D.; Gies, A.J.; Haynie, C.; Smith, H.; Reyna, N.S.; Makhoul, I. Circulating Exosomal MicroRNAs as Predictive Biomarkers of Neoadjuvant Chemotherapy Response in Breast Cancer. Curr. Oncol. 2022, 29, 55. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, H.; Yu, J.; Xu, L.; Pang, X.; Xiang, Q.; Liu, Q.; Cui, Y. MiRNAs as Therapeutic Predictors and Prognostic Biomarkers of Neoadjuvant Chemotherapy in Breast Cancer: A Systematic Review and Meta-Analysis. Breast Cancer Res. Treat. 2022, 194, 483–505. [Google Scholar] [CrossRef]
- Zhu, W.; Liu, M.; Fan, Y.; Ma, F.; Xu, N.; Xu, B. Dynamics of Circulating MicroRNAs as a Novel Indicator of Clinical Response to Neoadjuvant Chemotherapy in Breast Cancer. Cancer Med. 2018, 7, 4420–4433. [Google Scholar] [CrossRef]
- Sueta, A.; Fujiki, Y.; Goto-Yamaguchi, L.; Tomiguchi, M.; Yamamoto-Ibusuki, M.; Iwase, H.; Yamamoto, Y. Exosomal MiRNA Profiles of Triple-Negative Breast Cancer in Neoadjuvant Treatment. Oncol. Lett. 2021, 22, 819. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, X.; Yang, J.; Zhen, J.; Zhang, D. Serum MicroRNA-21 as a Potential Diagnostic Biomarker for Breast Cancer: A Systematic Review and Meta-Analysis. Clin. Exp. Med. 2016, 16, 29–35. [Google Scholar] [CrossRef]
- Salim, B.; Athira, M.V.; Kandaswamy, A.; Vijayakumar, M. Investigation on Staging of Breast Cancer Using MiR-21 as a Biomarker in the Serum. Int. J. Biomed. Eng. Technol. 2020, 33, 211–221. [Google Scholar] [CrossRef]
- Allah, A.W.; Yahya, M.; Elsaeidy, K.S.; Alkanj, S.; Hamam, K.; El-Saady, M.; Ebada, M.A. Clinical Assessment of MiRNA-23b as a Prognostic Factor for Various Carcinomas: A Systematic Review and Meta-Analysis. Meta Gene 2020, 24, 100651. [Google Scholar] [CrossRef]
- Li, X.; Zeng, Z.; Wang, J.; Wu, Y.; Chen, W.; Zheng, L.; Xi, T.; Wang, A.; Lu, Y. MicroRNA-9 and Breast Cancer. Biomed. Pharmacother. 2020, 122, 109687. [Google Scholar] [CrossRef] [PubMed]
- Sehovic, E.; Urru, S.; Chiorino, G.; Doebler, P. Meta-Analysis of Diagnostic Cell-Free Circulating MicroRNAs for Breast Cancer Detection. BMC Cancer 2022, 22, 634. [Google Scholar] [CrossRef]
- Hamam, R.; Hamam, D.; Alsaleh, K.A.; Kassem, M.; Zaher, W.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. Circulating MicroRNAs in Breast Cancer: Novel Diagnostic and Prognostic Biomarkers. Cell Death Dis. 2017, 8, e3045. [Google Scholar] [CrossRef]
- Li, J.; Guan, X.; Fan, Z.; Ching, L.-M.; Li, Y.; Wang, X.; Cao, W.-M.; Liu, D.-X. Non-Invasive Biomarkers for Early Detection of Breast Cancer. Cancers 2020, 12, 2767. [Google Scholar] [CrossRef]
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular Functions of Long Noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Qian, Y.; Shi, L.; Luo, Z. Long Non-Coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 2020, 7, 612393. [Google Scholar] [CrossRef]
- Liu, S.J.; Dang, H.X.; Lim, D.A.; Feng, F.Y.; Maher, C.A. Long Noncoding RNAs in Cancer Metastasis. Nat. Rev. Cancer 2021, 21, 446–460. [Google Scholar] [CrossRef] [PubMed]
- Ming, H.; Li, B.; Zhou, L.; Goel, A.; Huang, C. Long Non-Coding RNAs and Cancer Metastasis: Molecular Basis and Therapeutic Implications. Biochim. Biophys. Acta BBA-Rev. Cancer 2021, 1875, 188519. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Jin, X.; Wang, Y.; Zhang, Q.; Yang, L. High Serum Exosomal Long Non-coding RNA DANCR Expression Confers Poor Prognosis in Patients with Breast Cancer. J. Clin. Lab. Anal. 2022, 36, e24186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhou, H.; Lu, K.; Lu, Y.; Wang, Y.; Feng, T. Exosome-Mediated Delivery of MALAT1 Induces Cell Proliferation in Breast Cancer. Oncotargets Ther. 2018, 11, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Zhang, X.; Li, H.; Yue, X.; Sun, Q. Serum Exosomal LncRNA XIST Is a Potential Non-invasive Biomarker to Diagnose Recurrence of Triple-negative Breast Cancer. J. Cell Mol. Med. 2021, 25, 7602–7607. [Google Scholar] [CrossRef]
- Qiu, P.; Guo, Q.; Lin, J.; Pan, K.; Chen, J.; Ding, M. An Exosome-Related Long Non-Coding RNAs Risk Model Could Predict Survival Outcomes in Patients with Breast Cancer. Sci. Rep. 2022, 12, 22322. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Peng, S.; Li, J.; Tan, X. Exosomal-mediated transfer of OIP5-AS1 enhanced cell chemoresistance to trastuzumab in breast cancer via up-regulating HMGB3 by sponging miR-381-3p. Open Med (Wars) 2021, 16, 512–525. [Google Scholar] [CrossRef]
- Shi, S.-J.; Wang, L.-J.; Yu, B.; Li, Y.-H.; Jin, Y.; Bai, X.-Z. LncRNA-ATB Promotes Trastuzumab Resistance and Invasion-Metastasis Cascade in Breast Cancer. Oncotarget 2015, 6, 11652–11663. [Google Scholar] [CrossRef]
- Qian, X.; Qu, H.; Zhang, F.; Peng, S.; Dou, D.; Yang, Y.; Ding, Y.; Xie, M.; Dong, H.; Liao, Y.; et al. Exosomal Long Noncoding RNA AGAP2-AS1 Regulates Trastuzumab Resistance via Inducing Autophagy in Breast Cancer. Am. J. Cancer Res. 2020, 11, 1962–1981. [Google Scholar]
- Dong, H.; Wang, W.; Chen, R.; Zhang, Y.; Zou, K.; Ye, M.; He, X.; Zhang, F.; Han, J. Exosome-Mediated Transfer of LncRNA-SNHG14 Promotes Trastuzumab Chemoresistance in Breast Cancer. Int. J. Oncol. 2022, 61, 92. [Google Scholar] [CrossRef]
- Wu, Z.; Xu, Z.; Yu, B.; Zhang, J.; Yu, B. The Potential Diagnostic Value of Exosomal Long Noncoding RNAs in Solid Tumors: A Meta-Analysis and Systematic Review. Biomed. Res. Int. 2020, 2020, 6786875. [Google Scholar] [CrossRef] [PubMed]
- Møller, H.D.; Mohiyuddin, M.; Prada-Luengo, I.; Sailani, M.R.; Halling, J.F.; Plomgaard, P.; Maretty, L.; Hansen, A.J.; Snyder, M.P.; Pilegaard, H.; et al. Circular DNA Elements of Chromosomal Origin Are Common in Healthy Human Somatic Tissue. Nat. Commun. 2018, 9, 1069. [Google Scholar] [CrossRef]
- HOLDENRIEDER, S.; STIEBER, P. Therapy Control in Oncology by Circulating Nucleosomes. Ann. N. Y. Acad. Sci. 2004, 1022, 211–216. [Google Scholar] [CrossRef]
- Li, Z.; Chen, Z.; Hu, G.; Jiang, Y. Roles of Circular RNA in Breast Cancer: Present and Future. Am. J. Transl. Res. 2019, 11, 3945–3954. [Google Scholar]
- Suzuki, H.; Tsukahara, T. A View of Pre-MRNA Splicing from RNase R Resistant RNAs. Int. J. Mol. Sci. 2014, 15, 9331. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Zheng, J.; Cao, M. Circ_0004771 Accelerates Cell Carcinogenic Phenotypes via Suppressing MiR-1253-Mediated DDAH1 Inhibition in Breast Cancer. Cancer Manag. Res. 2021, 13, 1–11. [Google Scholar] [CrossRef]
- Liu, J.; Peng, X.; Liu, Y.; Hao, R.; Zhao, R.; Zhang, L.; Zhao, F.; Liu, Q.; Liu, Y.; Qi, Y. The Diagnostic Value of Serum Exosomal Has_circ_0000615 for Breast Cancer Patients. Int. J. Gen. Med. 2021, 14, 4545–4554. [Google Scholar] [CrossRef]
- Turco, C.; Esposito, G.; Iaiza, A.; Goeman, F.; Benedetti, A.; Gallo, E.; Daralioti, T.; Perracchio, L.; Sacconi, A.; Pasanisi, P.; et al. MALAT1-Dependent Hsa_circ_0076611 Regulates Translation Rate in Triple-Negative Breast Cancer. Commun. Biol. 2022, 5, 598. [Google Scholar] [CrossRef]
- Yang, S.; Tang, J. Exosomal Circular RNAs Derived from Serum: Promising Biomarkers for Therapeutic Targets and Prognosis of Triple-Negative Breast Cancer (TNBC). J. Clin. Oncol. 2020, 38, 3528. [Google Scholar] [CrossRef]
- Yang, S.; Wang, D.; Zhong, S.; Chen, W.; Wang, F.; Zhang, J.; Xu, W.; Xu, D.; Zhang, Q.; Li, J.; et al. Tumor-Derived Exosomal CircPSMA1 Facilitates the Tumorigenesis, Metastasis, and Migration in Triple-Negative Breast Cancer (TNBC) through MiR-637/Akt1/β-Catenin (Cyclin D1) Axis. Cell. Death Dis. 2021, 12, 420. [Google Scholar] [CrossRef]
- Halvaei, S.; Daryani, S.; Eslami-S, Z.; Samadi, T.; Jafarbeik-Iravani, N.; Bakhshayesh, T.O.; Majidzadeh-A., K.; Esmaeili, R. Exosomes in Cancer Liquid Biopsy: A Focus on Breast Cancer. Mol. Ther.-Nucleic Acids 2018, 10, 131–141. [Google Scholar] [CrossRef]
- Tutanov, O.; Proskura, K.; Kamyshinsky, R.; Shtam, T.; Tsentalovich, Y.; Tamkovich, S. Proteomic Profiling of Plasma and Total Blood Exosomes in Breast Cancer: A Potential Role in Tumor Progression, Diagnosis, and Prognosis. Front. Oncol. 2020, 10, 580891. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, W.; Bu, J.; Li, Y.; Li, R.; Nie, R.; Xiao, C.; Ma, K.; Huang, X.; Li, Y. Exosomal Protein CD82 as a Diagnostic Biomarker for Precision Medicine for Breast Cancer. Mol. Carcinog. 2019, 58, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Yang, S.; Pi, H.; Li, Z.; Yao, P.; Zhang, Q.; Wang, Q.; Shen, P.; Li, X.; et al. Proteomic Landscape of Exosomes Reveals the Functional Contributions of CD151 in Triple-Negative Breast Cancer. Mol. Cell Proteom. 2021, 20, 100121. [Google Scholar] [CrossRef]
- Rupp, A.-K.; Rupp, C.; Keller, S.; Brase, J.C.; Ehehalt, R.; Fogel, M.; Moldenhauer, G.; Marmé, F.; Sültmann, H.; Altevogt, P. Loss of EpCAM Expression in Breast Cancer Derived Serum Exosomes: Role of Proteolytic Cleavage. Gynecol. Oncol. 2011, 122, 437–446. [Google Scholar] [CrossRef]
- Galindo-Hernandez, O.; Villegas-Comonfort, S.; Candanedo, F.; González-Vázquez, M.-C.; Chavez-Ocaña, S.; Jimenez-Villanueva, X.; Sierra-Martinez, M.; Salazar, E.P. Elevated Concentration of Microvesicles Isolated from Peripheral Blood in Breast Cancer Patients. Arch. Med. Res. 2013, 44, 208–214. [Google Scholar] [CrossRef]
- Jordan, K.R.; Hall, J.K.; Schedin, T.; Borakove, M.; Xian, J.J.; Dzieciatkowska, M.; Lyons, T.R.; Schedin, P.; Hansen, K.C.; Borges, V.F. Extracellular Vesicles from Young Women’s Breast Cancer Patients Drive Increased Invasion of Non-Malignant Cells via the Focal Adhesion Kinase Pathway: A Proteomic Approach. Breast Cancer Res. 2020, 22, 128. [Google Scholar] [CrossRef]
- Higginbotham, J.N.; Demory Beckler, M.; Gephart, J.D.; Franklin, J.L.; Bogatcheva, G.; Kremers, G.-J.; Piston, D.W.; Ayers, G.D.; McConnell, R.E.; Tyska, M.J.; et al. Amphiregulin Exosomes Increase Cancer Cell Invasion. Curr. Biol. 2011, 21, 779–786. [Google Scholar] [CrossRef]
- Bi, T.-L.; Sun, J.-J.; Tian, Y.-Z.; Zhou, Y.-F. Research Progress of Relationship between Exosomes and Breast Cancer. Sheng Li Xue Bao Acta Physiol. Sin. 2016, 68, 352–358. [Google Scholar]
- Moon, P.-G.; Lee, J.-E.; Cho, Y.-E.; Lee, S.J.; Chae, Y.S.; Jung, J.H.; Kim, I.-S.; Park, H.Y.; Baek, M.-C. Fibronectin on Circulating Extracellular Vesicles as a Liquid Biopsy to Detect Breast Cancer. Oncotarget 2016, 7, 40189–40199. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Lee, J.; Jung, J.H.; Park, H.Y.; Moon, P.-G.; Chae, Y.S.; Baek, M.-C. Exosomal Del-1 as a Potent Diagnostic Marker for Breast Cancer: Prospective Cohort Study. Clin. Breast Cancer 2021, 21, e748–e756. [Google Scholar] [CrossRef]
- Li, K.; Liu, T.; Chen, J.; Ni, H.; Li, W. Survivin in Breast Cancer–Derived Exosomes Activates Fibroblasts by up-Regulating SOD1, Whose Feedback Promotes Cancer Proliferation and Metastasis. J. Biol. Chem. 2020, 295, 13737–13752. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cheng, K.; Zhang, G.; Jia, Z.; Yu, Y.; Guo, J.; Hua, Y.; Guo, F.; Li, X.; Zou, W.; et al. Enrichment of CD44 in Exosomes From Breast Cancer Cells Treated With Doxorubicin Promotes Chemoresistance. Front. Oncol. 2020, 10, 960. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, Y.; Yu, X.; Jiang, X.; Li, G.; Zhao, J. Identification of Programmed Death Ligand-1 Positive Exosomes in Breast Cancer Based on DNA Amplification-Responsive Metal-Organic Frameworks. Biosens. Bioelectron. 2020, 166, 112452. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.H.; Kim, J.-Y.; Cho, E.Y.; Oh, J.M.; Lee, J.E.; Kim, S.W.; Nam, S.J.; Park, Y.H.; Ahn, J.S.; Im, Y.-H. Elevated Level of Nerve Growth Factor (NGF) in Serum-Derived Exosomes Predicts Poor Survival in Patients with Breast Cancer Undergoing Neoadjuvant Chemotherapy. Cancers 2021, 13, 5260. [Google Scholar] [CrossRef] [PubMed]
- Bastos, N.; Ruivo, C.F.; da Silva, S.; Melo, S.A. Exosomes in Cancer: Use Them or Target Them? Semin. Cell Dev. Biol. 2018, 78, 13–21. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and Mechanisms of Extracellular Vesicle Uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef]
- O’Donoghue, E.J.; Krachler, A.M. Mechanisms of Outer Membrane Vesicle Entry into Host Cells. Cell Microbiol. 2016, 18, 1508–1517. [Google Scholar] [CrossRef]
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.-P.; Belting, M. Cancer Cell Exosomes Depend on Cell-Surface Heparan Sulfate Proteoglycans for Their Internalization and Functional Activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385. [Google Scholar] [CrossRef]
- Escrevente, C.; Keller, S.; Altevogt, P.; Costa, J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Svensson, K.J.; Christianson, H.C.; Wittrup, A.; Bourseau-Guilmain, E.; Lindqvist, E.; Svensson, L.M.; Mörgelin, M.; Belting, M. Exosome Uptake Depends on ERK1/2-Heat Shock Protein 27 Signaling and Lipid Raft-Mediated Endocytosis Negatively Regulated by Caveolin-1. J. Biol. Chem. 2013, 288, 17713–17724. [Google Scholar] [CrossRef]
- Wang, L.H.; Rothberg, K.G.; Anderson, R.G. Mis-Assembly of Clathrin Lattices on Endosomes Reveals a Regulatory Switch for Coated Pit Formation. J. Cell Biol. 1993, 123, 1107–1117. [Google Scholar] [CrossRef]
- Ehrlich, M.; Boll, W.; van Oijen, A.; Hariharan, R.; Chandran, K.; Nibert, M.L.; Kirchhausen, T. Endocytosis by Random Initiation and Stabilization of Clathrin-Coated Pits. Cell 2004, 118, 591–605. [Google Scholar] [CrossRef]
- Koivusalo, M.; Welch, C.; Hayashi, H.; Scott, C.C.; Kim, M.; Alexander, T.; Touret, N.; Hahn, K.M.; Grinstein, S. Amiloride Inhibits Macropinocytosis by Lowering Submembranous PH and Preventing Rac1 and Cdc42 Signaling. J. Cell Biol. 2010, 188, 547–563. [Google Scholar] [CrossRef]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; Milito, A.D.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental PH Is a Key Factor for Exosome Traffic in Tumor Cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [PubMed]
- Hait, N.C.; Maiti, A. The Role of Sphingosine-1-Phosphate and Ceramide-1-Phosphate in Inflammation and Cancer. Mediat. Inflamm. 2017, 2017, 4806541. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Menck, K.; Sönmezer, C.; Worst, T.S.; Schulz, M.; Dihazi, G.H.; Streit, F.; Erdmann, G.; Kling, S.; Boutros, M.; Binder, C.; et al. Neutral Sphingomyelinases Control Extracellular Vesicles Budding from the Plasma Membrane. J. Extracell. Vesicles 2017, 6, 1378056. [Google Scholar] [CrossRef] [PubMed]
- Catalano, M.; O’Driscoll, L. Inhibiting Extracellular Vesicles Formation and Release: A Review of EV Inhibitors. J. Extracell. Vesicles 2019, 9, 1703244. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory Mechanisms and Intercellular Transfer of MicroRNAs in Living Cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef]
- Wang, J.; Yeung, B.Z.; Wientjes, M.G.; Cui, M.; Peer, C.J.; Lu, Z.; Figg, W.D.; Woo, S.; Au, J.L.-S. A Quantitative Pharmacology Model of Exosome-Mediated Drug Efflux and Perturbation-Induced Synergy. Pharmaceutics 2021, 13, 997. [Google Scholar] [CrossRef]
- Bianco, F.; Perrotta, C.; Novellino, L.; Francolini, M.; Riganti, L.; Menna, E.; Saglietti, L.; Schuchman, E.H.; Furlan, R.; Clementi, E.; et al. Acid Sphingomyelinase Activity Triggers Microparticle Release from Glial Cells. EMBO J. 2009, 28, 1043–1054. [Google Scholar] [CrossRef]
- Kosgodage, U.; Trindade, R.; Thompson, P.; Inal, J.; Lange, S. Chloramidine/Bisindolylmaleimide-I-Mediated Inhibition of Exosome and Microvesicle Release and Enhanced Efficacy of Cancer Chemotherapy. Int. J. Mol. Sci. 2017, 18, 1007. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan–Syntenin–ALIX Regulates the Biogenesis of Exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Song, M.; Cui, Z.S.; Li, C.Y.; Xue, X.X.; Yu, M.; Lu, Y.; Zhang, S.Y.; Wang, E.H.; Wen, Y.Y. Down-Regulation of TSG101 by Small Interfering RNA Inhibits the Proliferation of Breast Cancer Cells through the MAPK/ERK Signal Pathway. Histol. Histopathol. 2010, 26, 87–94. [Google Scholar] [CrossRef]
- Li, Z.; Fang, R.; Fang, J.; He, S.; Liu, T. Functional Implications of Rab27 GTPases in Cancer. Cell Commun. Signal. 2018, 16, 44. [Google Scholar] [CrossRef]
- Li, W.; Hu, Y.; Jiang, T.; Han, Y.; Han, G.; Chen, J.; Li, X. Rab27A Regulates Exosome Secretion from Lung Adenocarcinoma Cells A549: Involvement of EPI64. Apmis 2014, 122, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Pereira, F.; de Matos, A.P.A.; Fernandes, M.; Baptista, P.V.; Fernandes, A.R. Smuggling Gold Nanoparticles across Cell Types—A New Role for Exosomes in Gene Silencing. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1389–1398. [Google Scholar] [CrossRef]
- Hoshino, D.; Kirkbride, K.C.; Costello, K.; Clark, E.S.; Sinha, S.; Grega-Larson, N.; Tyska, M.J.; Weaver, A.M. Exosome Secretion Is Enhanced by Invadopodia and Drives Invasive Behavior. Cell Rep. 2013, 5, 1159–1168. [Google Scholar] [CrossRef]
- Sinha, S.; Hoshino, D.; Hong, N.H.; Kirkbride, K.C.; Grega-Larson, N.E.; Seiki, M.; Tyska, M.J.; Weaver, A.M. Cortactin Promotes Exosome Secretion by Controlling Branched Actin Dynamics. J. Cell Biol. 2016, 214, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Twafra, S.; Sokolik, C.G.; Sneh, T.; Srikanth, K.D.; Meirson, T.; Genna, A.; Chill, J.H.; Gil-Henn, H. A Novel Pyk2-Derived Peptide Inhibits Invadopodia-Mediated Breast Cancer Metastasis. Oncogene 2023, 42, 278–292. [Google Scholar] [CrossRef]
- Huang, M.-B.; Brena, D.; Wu, J.Y.; Roth, W.W.; Owusu, S.; Bond, V.C. Novel Secretion Modification Region (SMR) Peptide Exhibits Anti-Metastatic Properties in Human Breast Cancer Cells. Sci. Rep. 2022, 12, 13204. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-X.; Cheng, L.; Pan, M.; Qian, Q.; Zhu, Y.-L.; Xu, L.-Y.; Ding, Q. D Rhamnose β-Hederin against Human Breast Cancer by Reducing Tumor-Derived Exosomes. Oncol. Lett. 2018, 16, 5172–5178. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Li, M.; Cui, S.; Wang, D.; Zhang, C.-Y.; Zen, K.; Li, L. Shikonin Inhibits the Proliferation of Human Breast Cancer Cells by Reducing Tumor-Derived Exosomes. Molecules 2016, 21, 777. [Google Scholar] [CrossRef] [PubMed]
- Kosgodage, U.S.; Mould, R.; Henley, A.B.; Nunn, A.V.; Guy, G.W.; Thomas, E.L.; Inal, J.M.; Bell, J.D.; Lange, S. Cannabidiol (CBD) Is a Novel Inhibitor for Exosome and Microvesicle (EMV) Release in Cancer. Front. Pharmacol. 2018, 9, 889. [Google Scholar] [CrossRef]
- Huang, M.-B.; Gonzalez, R.R.; Lillard, J.; Bond, V.C. Secretion Modification Region-Derived Peptide Blocks Exosome Release and Mediates Cell Cycle Arrest in Breast Cancer Cells. Oncotarget 2017, 8, 11302–11315. [Google Scholar] [CrossRef]
- Conlan, R.S.; Pisano, S.; Oliveira, M.I.; Ferrari, M.; Pinto, I.M. Exosomes as Reconfigurable Therapeutic Systems. Trends Mol. Med. 2017, 23, 636–650. [Google Scholar] [CrossRef]
- Pullan, J.E.; Confeld, M.I.; Osborn, J.K.; Kim, J.; Sarkar, K.; Mallik, S. Exosomes as Drug Carriers for Cancer Therapy. Mol. Pharm. 2019, 16, 1789–1798. [Google Scholar] [CrossRef]
- Kim, H.; Jang, H.; Cho, H.; Choi, J.; Hwang, K.Y.; Choi, Y.; Kim, S.H.; Yang, Y. Recent Advances in Exosome-Based Drug Delivery for Cancer Therapy. Cancers 2021, 13, 4435. [Google Scholar] [CrossRef]
- Chen, L.; Wang, L.; Zhu, L.; Xu, Z.; Liu, Y.; Li, Z.; Zhou, J.; Luo, F. Exosomes as Drug Carriers in Anti-Cancer Therapy. Front. Cell Dev. Biol. 2022, 10, 728616. [Google Scholar] [CrossRef]
- Gorshkov, A.; Purvinsh, L.; Brodskaia, A.; Vasin, A. Exosomes as Natural Nanocarriers for RNA-Based Therapy and Prophylaxis. Nanomaterials 2022, 12, 524. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Stephens, E.; Zhang, Y. Chapter Nine—Extracellular Vesicles in Tumor Immunotherapy. In Systemic Drug Delivery Strategies; Amiji, M.M., Milane, L.S., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 231–256. [Google Scholar] [CrossRef]
- Muhammad, S.A.; Mohammed, J.S.; Rabiu, S. Exosomes as Delivery Systems for Targeted Tumour Therapy: A Systematic Review and Meta-Analysis of In Vitro Studies. Pharm. Nanotechnol. 2023, 11, 93–104. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Du, S.-L.; Zhang, J.; Liang, A.-L.; Liu, Y.-J. Exosomes and Breast Cancer: A Comprehensive Review of Novel Therapeutic Strategies from Diagnosis to Treatment. Cancer Gene Ther. 2017, 24, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Allahverdiyev, A.M.; Parlar, E.; Dinparvar, S.; Bagirova, M.; Abamor, E.Ş. Current Aspects in Treatment of Breast Cancer Based of Nanodrug Delivery Systems and Future Prospects. Artif. Cells Nanomed. Biotechnol. 2018, 46, S755–S762. [Google Scholar] [CrossRef]
- Mughees, M.; Kumar, K.; Wajid, S. Exosome Vesicle as a Nano-Therapeutic Carrier for Breast Cancer. J. Drug Target. 2021, 29, 121–130. [Google Scholar] [CrossRef]
- Al-Humaidi, R.B.; Fayed, B.; Sharif, S.I.; Noreddin, A.; Soliman, S.S.M. Role of Exosomes in Breast Cancer Management: Evidence-Based Review. Curr. Cancer Drug Targets 2021, 21, 666–675. [Google Scholar] [CrossRef]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as Therapeutic Drug Carriers and Delivery Vehicles across Biological Membranes: Current Perspectives and Future Challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef]
- Yang, B.; Chen, Y.; Shi, J. Exosome Biochemistry and Advanced Nanotechnology for Next-Generation Theranostic Platforms. Adv. Mater. 2019, 31, e1802896. [Google Scholar] [CrossRef] [PubMed]
- Rufino-Ramos, D.; Albuquerque, P.R.; Carmona, V.; Perfeito, R.; Nobre, R.J.; de Almeida, L.P. Extracellular Vesicles: Novel Promising Delivery Systems for Therapy of Brain Diseases. J. Control Release 2017, 262, 247–258. [Google Scholar] [CrossRef]
- Kutova, O.M.; Guryev, E.L.; Sokolova, E.A.; Alzeibak, R.; Balalaeva, I.V. Targeted Delivery to Tumors: Multidirectional Strategies to Improve Treatment Efficiency. Cancers 2019, 11, 68. [Google Scholar] [CrossRef]
- Smyth, T.J.; Redzic, J.S.; Graner, M.W.; Anchordoquy, T.J. Examination of the Specificity of Tumor Cell Derived Exosomes with Tumor Cells in Vitro. Biochim. Biophys. Acta BBA-Biomembr. 2014, 1838, 2954–2965. [Google Scholar] [CrossRef]
- Agrawal, A.K.; Aqil, F.; Jeyabalan, J.; Spencer, W.A.; Beck, J.; Gachuki, B.W.; Alhakeem, S.S.; Oben, K.; Munagala, R.; Bondada, S.; et al. Milk-Derived Exosomes for Oral Delivery of Paclitaxel. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1627–1636. [Google Scholar] [CrossRef]
- Kandimalla, R.; Aqil, F.; Alhakeem, S.S.; Jeyabalan, J.; Tyagi, N.; Agrawal, A.; Yan, J.; Spencer, W.; Bondada, S.; Gupta, R.C. Targeted Oral Delivery of Paclitaxel Using Colostrum-Derived Exosomes. Cancers 2021, 13, 3700. [Google Scholar] [CrossRef]
- Kumar, D.N.; Chaudhuri, A.; Dehari, D.; Shekher, A.; Gupta, S.C.; Majumdar, S.; Krishnamurthy, S.; Singh, S.; Kumar, D.; Agrawal, A.K. Combination Therapy Comprising Paclitaxel and 5-Fluorouracil by Using Folic Acid Functionalized Bovine Milk Exosomes Improves the Therapeutic Efficacy against Breast Cancer. Life 2022, 12, 1143. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, L.; Qin, Z.; Hua, S.; Guo, Z.; Chu, C.; Lin, H.; Zhang, Y.; Li, W.; Zhang, X.; et al. Genetically Engineered Liposome-like Nanovesicles as Active Targeted Transport Platform. Adv. Mater. 2018, 30, 1705350. [Google Scholar] [CrossRef]
- Gomari, H.; Moghadam, M.F.; Soleimani, M.; Ghavami, M.; Khodashenas, S. Targeted Delivery of Doxorubicin to HER2 Positive Tumor Models. Int. J. Nanomed. 2019, 14, 5679–5690. [Google Scholar] [CrossRef]
- Pullan, J.; Dailey, K.; Bhallamudi, S.; Feng, L.; Alhalhooly, L.; Froberg, J.; Osborn, J.; Sarkar, K.; Molden, T.; Sathish, V.; et al. Modified Bovine Milk Exosomes for Doxorubicin Delivery to Triple-Negative Breast Cancer Cells. Acs. Appl. Biol. Mater. 2022, 5, 2163–2175. [Google Scholar] [CrossRef]
- Schindler, C.; Collinson, A.; Matthews, C.; Pointon, A.; Jenkinson, L.; Minter, R.R.; Vaughan, T.J.; Tigue, N.J. Exosomal Delivery of Doxorubicin Enables Rapid Cell Entry and Enhanced in Vitro Potency. PLoS ONE 2019, 14, e0214545. [Google Scholar] [CrossRef]
- Zhang, G.; Yang, X.; Su, X.; An, N.; Yang, F.; Li, X.; Jiang, Y.; Xing, Y. Understanding the Protective Role of Exosomes in Doxorubicin-Induced Cardiotoxicity. Oxidative Med. Cell Longev. 2022, 2022, 2852251. [Google Scholar] [CrossRef]
- Hadla, M.; Palazzolo, S.; Corona, G.; Caligiuri, I.; Canzonieri, V.; Toffoli, G.; Rizzolio, F. Exosomes Increase the Therapeutic Index of Doxorubicin in Breast and Ovarian Cancer Mouse Models. Nanomedicine 2016, 11, 2431–2441. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Mi, Y.; Wang, B.; Zhu, Y.; Ding, Y.; Ding, Y.; Li, X. Naturally Equipped Urinary Exosomes Coated Poly (2−ethyl−2−oxazoline)−Poly (D, L−lactide) Nanocarriers for the Pre−Clinical Translation of Breast Cancer. Bioengineering 2022, 9, 363. [Google Scholar] [CrossRef] [PubMed]
- Kanchanapally, R.; Brown, K. Cancer Cell-Derived Exosomes as the Delivery Vehicle of Paclitaxel to Inhibit Cancer Cell Growth. J. Cancer Discov. 2022, 1, 49–58. [Google Scholar] [CrossRef]
- Liu, J.; Tang, Y.; Li, Y.; Hu, X.; Huang, S.; Xu, W.; Hao, X.; Zhou, M.; Wu, J.; Xiang, D. Paclitaxel-Loaded Hybrid Exosome for Targeted Chemotherapy of Triple-Negative Breast Cancer. Res. Sq. 2022. preprint. [Google Scholar] [CrossRef]
- Wang, K.; Ye, H.; Zhang, X.; Wang, X.; Yang, B.; Luo, C.; Zhao, Z.; Zhao, J.; Lu, Q.; Zhang, H.; et al. An Exosome-like Programmable-Bioactivating Paclitaxel Prodrug Nanoplatform for Enhanced Breast Cancer Metastasis Inhibition. Biomaterials 2020, 257, 120224. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wu, J.; Gu, W.; Huang, Y.; Tong, Z.; Huang, L.; Tan, J. Exosome–Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs. Adv. Sci. 2018, 5, 1700611. [Google Scholar] [CrossRef]
- Naseri, Z.; Oskuee, R.K.; Jaafari, M.R.; Forouzandeh, M. Exosome-Mediated Delivery of Functionally Active MiRNA-142-3p Inhibitor Reduces Tumorigenicity of Breast Cancer in Vitro and in Vivo. Int. J. Nanomed. 2018, 13, 7727–7747. [Google Scholar] [CrossRef]
- Kim, H.; Rhee, W.J. Exosome-Mediated Let7c-5p Delivery for Breast Cancer Therapeutic Development. Biotechnol. Bioproc. E 2020, 25, 513–520. [Google Scholar] [CrossRef]
- Ohno, S.; Takanashi, M.; Sudo, K.; Ueda, S.; Ishikawa, A.; Matsuyama, N.; Fujita, K.; Mizutani, T.; Ohgi, T.; Ochiya, T.; et al. Systemically Injected Exosomes Targeted to EGFR Deliver Antitumor MicroRNA to Breast Cancer Cells. Mol. Ther. 2013, 21, 185–191. [Google Scholar] [CrossRef]
- O’Brien, K.; Lowry, M.C.; Corcoran, C.; Martinez, V.G.; Daly, M.; Rani, S.; Gallagher, W.M.; Radomski, M.W.; MacLeod, R.A.F.; O’Driscoll, L. MiR-134 in Extracellular Vesicles Reduces Triple-Negative Breast Cancer Aggression and Increases Drug Sensitivity. Oncotarget 2015, 6, 32774–32789. [Google Scholar] [CrossRef]
- Shojaei, S.; Hashemi, S.M.; Ghanbarian, H.; Sharifi, K.; Salehi, M.; Mohammadi-Yeganeh, S. Delivery of MiR-381-3p Mimic by Mesenchymal Stem Cell-Derived Exosomes Inhibits Triple Negative Breast Cancer Aggressiveness; an In Vitro Study. Stem Cell Rev. Rep. 2021, 17, 1027–1038. [Google Scholar] [CrossRef]
- Sheykhhasan, M.; Kalhor, N.; Sheikholeslami, A.; Dolati, M.; Amini, E.; Fazaeli, H. Exosomes of Mesenchymal Stem Cells as a Proper Vehicle for Transfecting MiR-145 into the Breast Cancer Cell Line and Its Effect on Metastasis. Biomed. Res. Int. 2021, 2021, 5516078. [Google Scholar] [CrossRef]
- Gong, C.; Tian, J.; Wang, Z.; Gao, Y.; Wu, X.; Ding, X.; Qiang, L.; Li, G.; Han, Z.; Yuan, Y.; et al. Functional Exosome-Mediated Co-Delivery of Doxorubicin and Hydrophobically Modified MicroRNA 159 for Triple-Negative Breast Cancer Therapy. J. Nanobiotechnol. 2019, 17, 93. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, G.; Yu, T.; He, J.; Liu, J.; Chai, X.; Zhao, G.; Yin, D.; Zhang, C. Exosomes Deliver LncRNA DARS-AS1 SiRNA to Inhibit Chronic Unpredictable Mild Stress-Induced TNBC Metastasis. Cancer Lett. 2022, 543, 215781. [Google Scholar] [CrossRef]
- Markov, O.; Oshchepkova, A.; Mironova, N. Immunotherapy Based on Dendritic Cell-Targeted/-Derived Extracellular Vesicles—A Novel Strategy for Enhancement of the Anti-Tumor Immune Response. Front. Pharmacol. 2019, 10, 1152. [Google Scholar] [CrossRef]
- Kitai, Y.; Kawasaki, T.; Sueyoshi, T.; Kobiyama, K.; Ishii, K.J.; Zou, J.; Akira, S.; Matsuda, T.; Kawai, T. DNA-Containing Exosomes Derived from Cancer Cells Treated with Topotecan Activate a STING-Dependent Pathway and Reinforce Antitumor Immunity. J. Immunol. 2017, 198, 1649–1659. [Google Scholar] [CrossRef]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of Established Murine Tumors Using a Novel Cell-Free Vaccine: Dendritic Cell Derived Exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef]
- Pitt, J.M.; Charrier, M.; Viaud, S.; André, F.; Besse, B.; Chaput, N.; Zitvogel, L. Dendritic Cell–Derived Exosomes as Immunotherapies in the Fight against Cancer. J. Immunol. 2014, 193, 1006–1011. [Google Scholar] [CrossRef]
- Xie, Y.; Wu, J.; Xu, A.; Ahmeqd, S.; Sami, A.; Chibbar, R.; Freywald, A.; Zheng, C.; Xiang, J. Heterologous Human/Rat HER2-Specific Exosome-Targeted T Cell Vaccine Stimulates Potent Humoral and CTL Responses Leading to Enhanced Circumvention of HER2 Tolerance in Double Transgenic HLA-A2/HER2 Mice. Vaccine 2018, 36, 1414–1422. [Google Scholar] [CrossRef]
- Liu, H.; Chen, L.; Peng, Y.; Yu, S.; Liu, J.; Wu, L.; Zhang, L.; Wu, Q.; Chang, X.; Yu, X.; et al. Dendritic Cells Loaded with Tumor Derived Exosomes for Cancer Immunotherapy. Oncotarget 2017, 9, 2887–2894. [Google Scholar] [CrossRef]
- Huang, L.; Rong, Y.; Tang, X.; Yi, K.; Qi, P.; Hou, J.; Liu, W.; He, Y.; Gao, X.; Yuan, C.; et al. Engineered Exosomes as an in Situ DC-Primed Vaccine to Boost Antitumor Immunity in Breast Cancer. Mol. Cancer 2022, 21, 45. [Google Scholar] [CrossRef]
- Taghikhani, A.; Hassan, Z.M.; Ebrahimi, M.; Moazzeni, S. MicroRNA Modified Tumor-derived Exosomes as Novel Tools for Maturation of Dendritic Cells. J. Cell Physiol. 2019, 234, 9417–9427. [Google Scholar] [CrossRef]
- Jung, K.O.; Jo, H.; Yu, J.H.; Gambhir, S.S.; Pratx, G. Development and MPI Tracking of Novel Hypoxia-Targeted Theranostic Exosomes. Biomaterials 2018, 177, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Limoni, S.K.; Moghadam, M.F.; Moazzeni, S.M.; Gomari, H.; Salimi, F. Engineered Exosomes for Targeted Transfer of SiRNA to HER2 Positive Breast Cancer Cells. Appl. Biochem. Biotech. 2019, 187, 352–364. [Google Scholar] [CrossRef]
- Cheng, Q.; Shi, X.; Han, M.; Smbatyan, G.; Lenz, H.-J.; Zhang, Y. Reprogramming Exosomes as Nanoscale Controllers of Cellular Immunity. J. Am. Chem. Soc. 2018, 140, 16413–16417. [Google Scholar] [CrossRef]
- Shi, X.; Cheng, Q.; Hou, T.; Han, M.; Smbatyan, G.; Lang, J.E.; Epstein, A.L.; Lenz, H.-J.; Zhang, Y. Genetically Engineered Cell-Derived Nanoparticles for Targeted Breast Cancer Immunotherapy. Mol. Ther. 2020, 28, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The Biology and Role of CD44 in Cancer Progression: Therapeutic Implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef]
- Hong, Y.; Nam, G.; Koh, E.; Jeon, S.; Kim, G.B.; Jeong, C.; Kim, D.; Yang, Y.; Kim, I. Exosome as a Vehicle for Delivery of Membrane Protein Therapeutics, PH20, for Enhanced Tumor Penetration and Antitumor Efficacy. Adv. Funct. Mater. 2018, 28, 1703074. [Google Scholar] [CrossRef]
- Feng, C.; Xiong, Z.; Wang, C.; Xiao, W.; Xiao, H.; Xie, K.; Chen, K.; Liang, H.; Zhang, X.; Yang, H. Folic Acid-Modified Exosome-PH20 Enhances the Efficiency of Therapy via Modulation of the Tumor Microenvironment and Directly Inhibits Tumor Cell Metastasis. Bioact. Mater. 2021, 6, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yao, S.; Zhou, Z.; Shi, J.; Huang, Z.; Wu, Z. Hyaluronan Decoration of Milk Exosomes Directs Tumor-Specific Delivery of Doxorubicin. Carbohydr. Res. 2020, 493, 108032. [Google Scholar] [CrossRef] [PubMed]
- Piffoux, M.; Nicolás-Boluda, A.; Mulens-Arias, V.; Richard, S.; Rahmi, G.; Gazeau, F.; Wilhelm, C.; Silva, A.K.A. Extracellular Vesicles for Personalized Medicine: The Input of Physically Triggered Production, Loading and Theranostic Properties. Adv. Adv. Drug Deliv. Rev. 2018, 138, 247–258. [Google Scholar] [CrossRef] [PubMed]
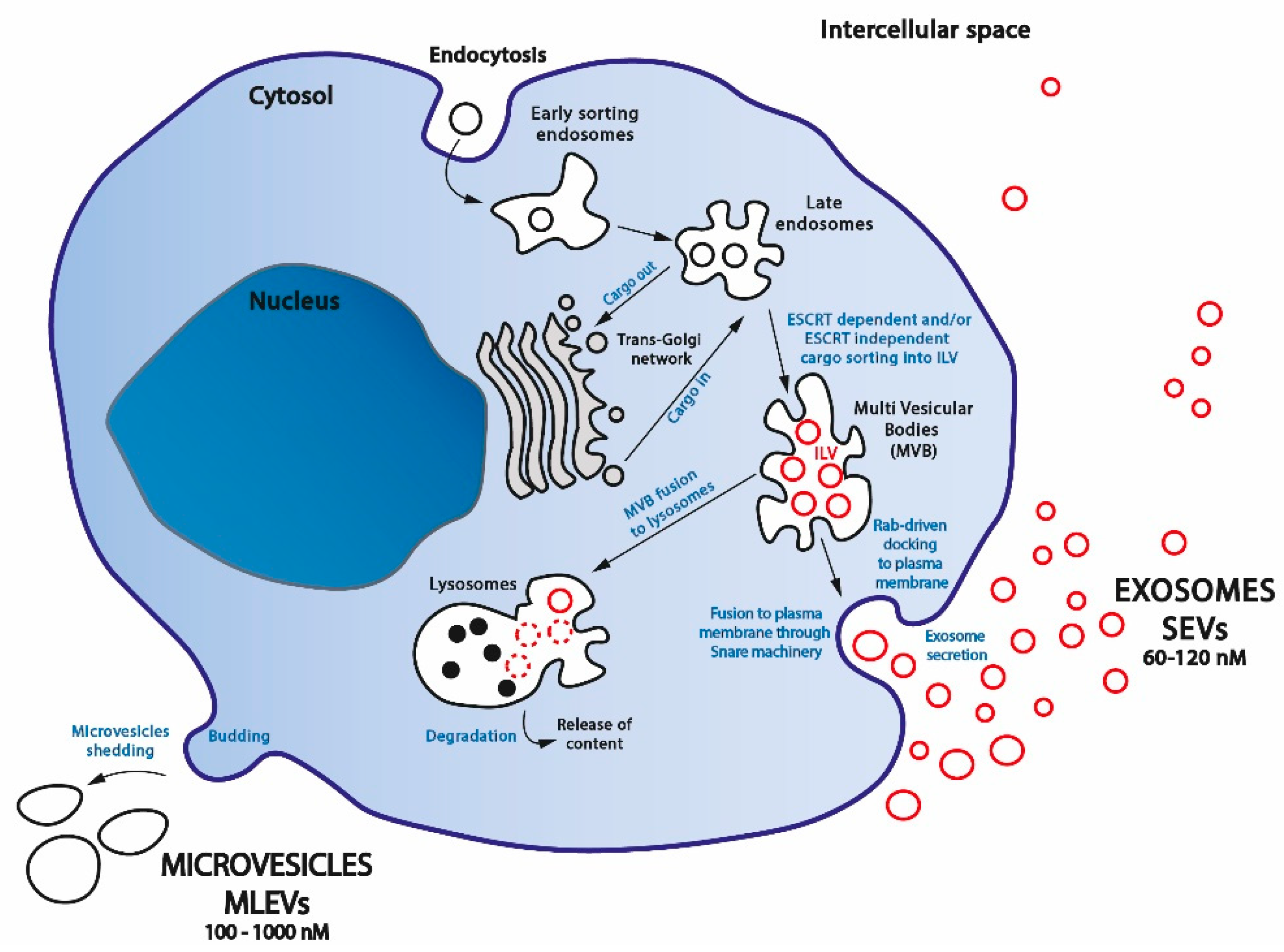
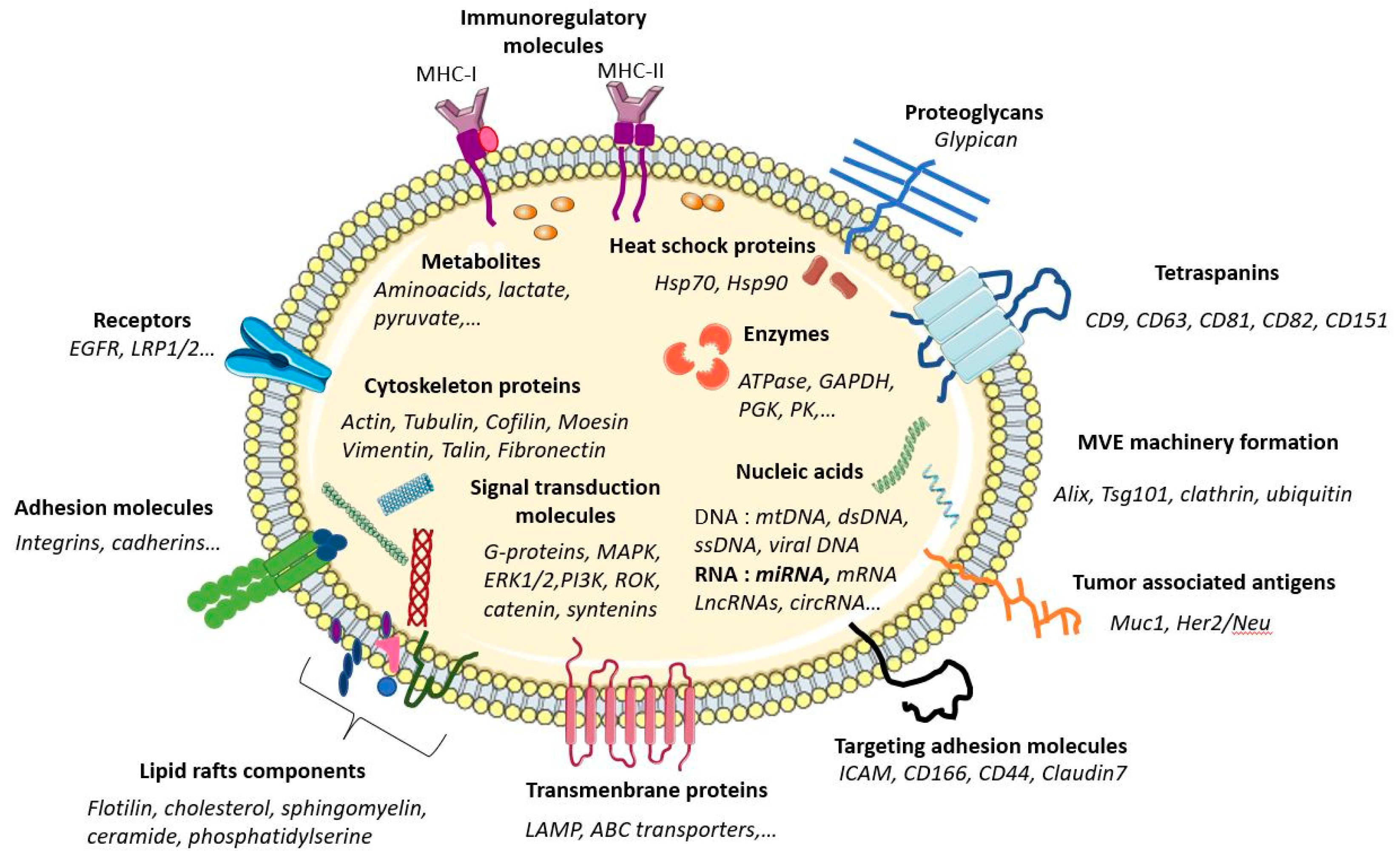
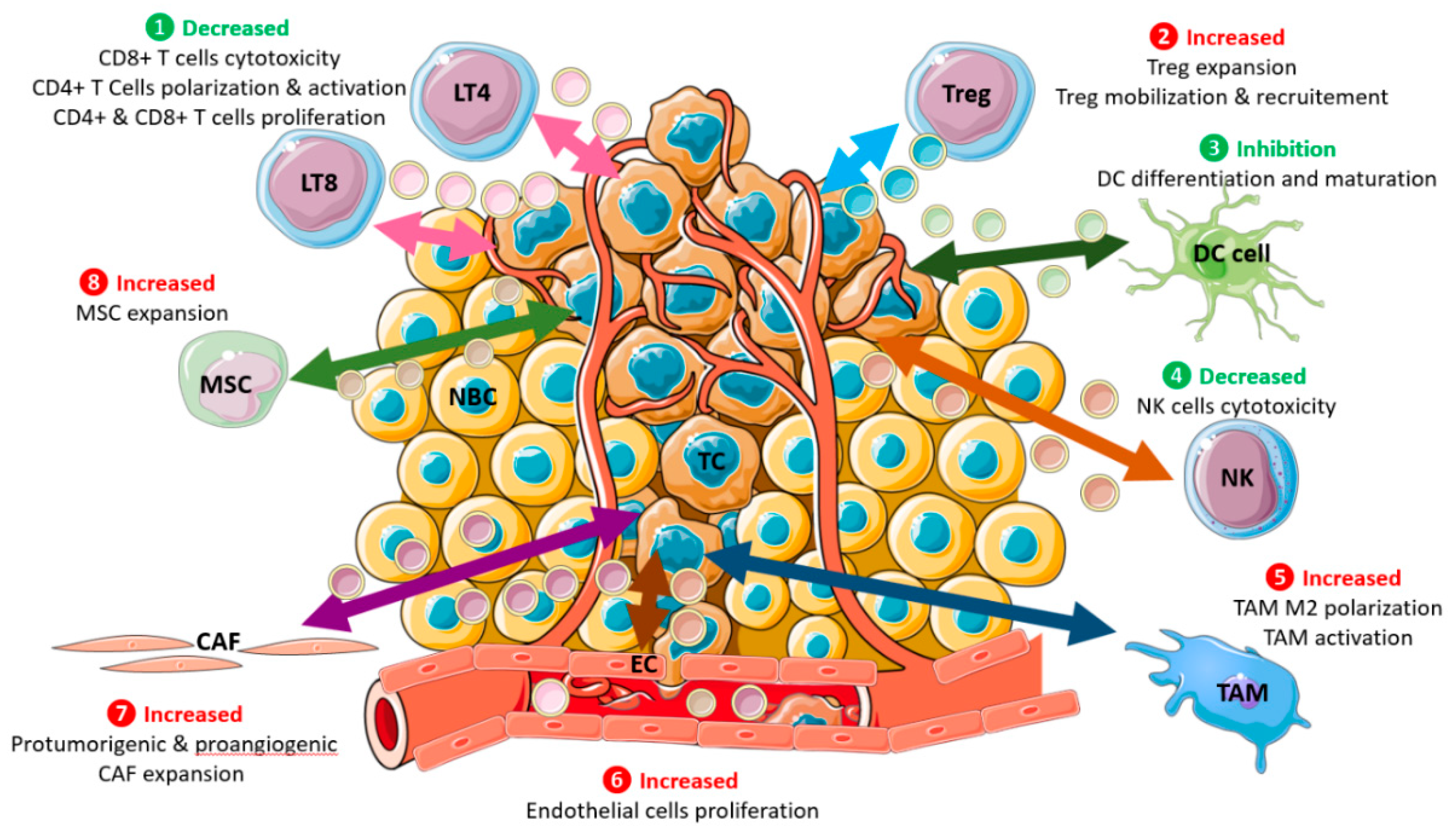
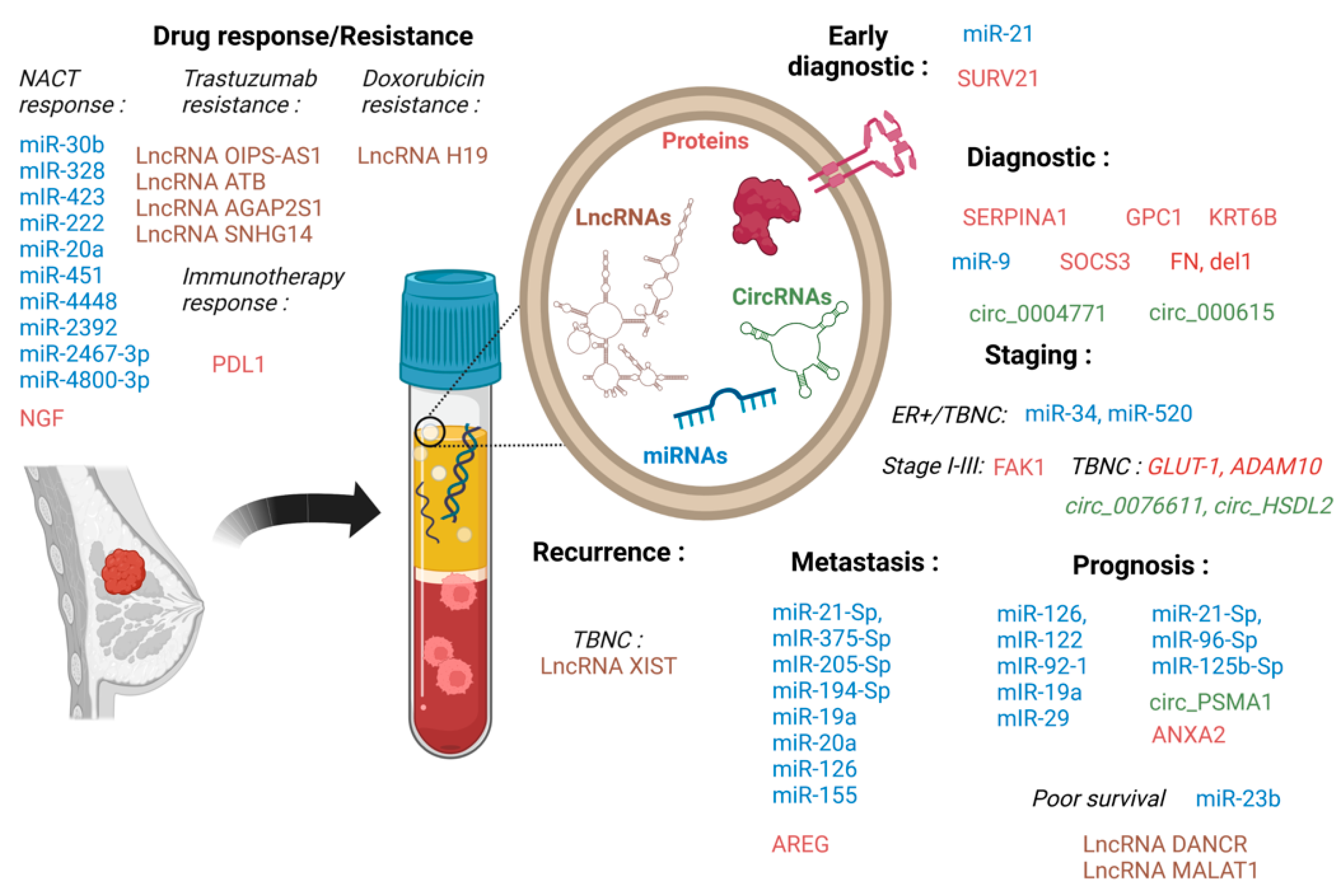
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loric, S.; Denis, J.A.; Desbene, C.; Sabbah, M.; Conti, M. Extracellular Vesicles in Breast Cancer: From Biology and Function to Clinical Diagnosis and Therapeutic Management. Int. J. Mol. Sci. 2023, 24, 7208. https://doi.org/10.3390/ijms24087208
Loric S, Denis JA, Desbene C, Sabbah M, Conti M. Extracellular Vesicles in Breast Cancer: From Biology and Function to Clinical Diagnosis and Therapeutic Management. International Journal of Molecular Sciences. 2023; 24(8):7208. https://doi.org/10.3390/ijms24087208
Chicago/Turabian StyleLoric, Sylvain, Jérôme Alexandre Denis, Cédric Desbene, Michèle Sabbah, and Marc Conti. 2023. "Extracellular Vesicles in Breast Cancer: From Biology and Function to Clinical Diagnosis and Therapeutic Management" International Journal of Molecular Sciences 24, no. 8: 7208. https://doi.org/10.3390/ijms24087208
APA StyleLoric, S., Denis, J. A., Desbene, C., Sabbah, M., & Conti, M. (2023). Extracellular Vesicles in Breast Cancer: From Biology and Function to Clinical Diagnosis and Therapeutic Management. International Journal of Molecular Sciences, 24(8), 7208. https://doi.org/10.3390/ijms24087208









