Conventional and State-of-the-Art Detection Methods of Bovine Spongiform Encephalopathy (BSE)
Abstract
1. Introduction
2. Sampling
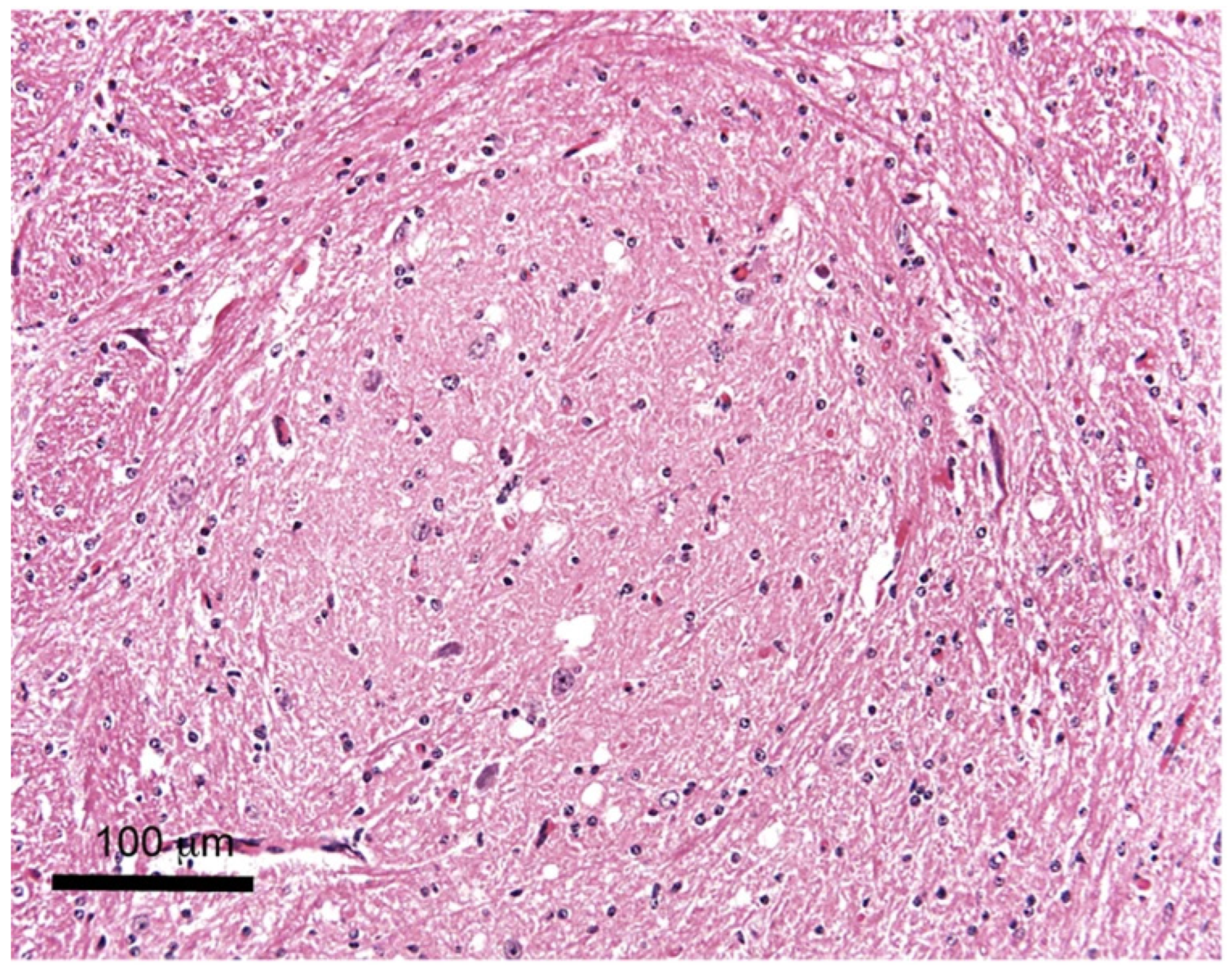
3. Histopathological Examination
4. Immunohistochemistry (IHC)
5. The Paraffin-Embedded Tissue (PET) Blot and Histoblot
6. Western Blot Methods
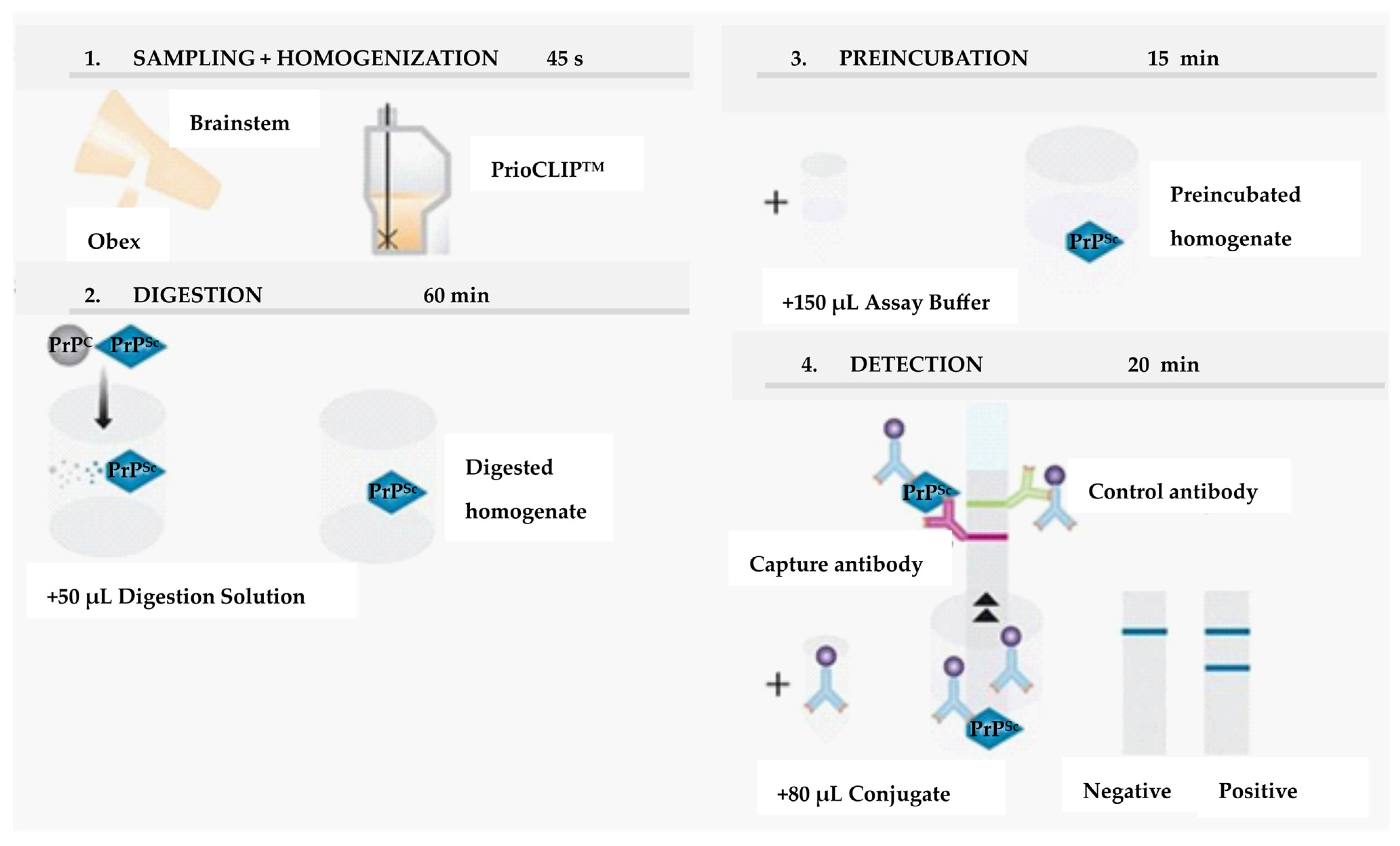
7. EU-Approved Rapid Tests
- Prionics®-Check Western test,
- Prionics®-Check LIA test,
- Bio-Rad TeSeE SAP rapid test,
- Roboscreen Beta Prion BSE EIA Test Kit,
- IDEXX HerdChek BSE-Scrapie Antigen Test Kit, EIA,
- Prionics®-Check PrioSTRIP.
- Bio-Rad TeSeE SAP rapid test,
- Bio-Rad TeSeE Sheep/Goat rapid test,
- IDEXX HerdChek BSE-Scrapie Antigen Test Kit, EIA.
8. The Immunofluorescence Assay (IFA), Flow Cytometry, and Immunofluorometric Assay (IFMA)
9. Scrapie-Associated Fibrils (SAFs)
10. In Vivo Bioassay
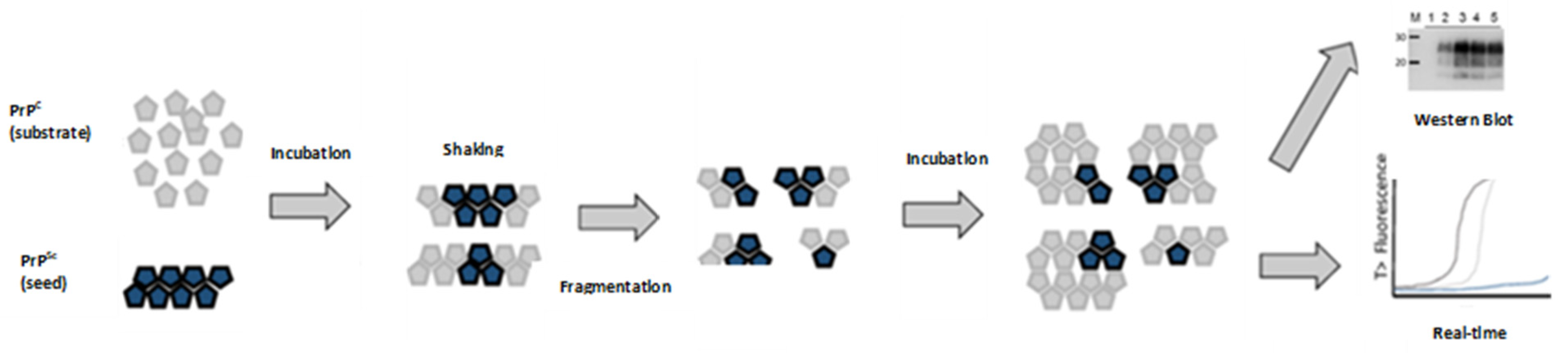
11. Protein Misfolding Cyclic Amplification (PMCA)
12. Real-Time Quaking-Induced Conversion (RT-QuIC)
13. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Konold, T.; Bone, G.; Ryder, S.; Hawkins, S.A.; Courtin, F.; Berthelin-Baker, C. Clinical findings in 78 suspected cases of bovine spongiform encephalopathy in Great Britain. Vet. Rec. 2004, 155, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Saegerman, C.; Speybroeck, N.; Roels, S.; Vanopdenbosch, E.; Thiry, E.; Berkvens, D. Decision support tools for clinical diagnosis of disease in cows with suspected bovine spongiform encephalopathy. J. Clin. Microbiol. 2004, 42, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, S.B.; Scott, M.R.; DeArmond, S.J.; Cohen, F.E. Prion protein biology. Cell 1998, 93, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.R. What is the basis of transmissible spongiform encephalopathy induced neurodegeneration and can it be repaired? Neuropathol. Appl. Neurobiol. 2002, 28, 1–11. [Google Scholar] [CrossRef]
- Casalone, C.; Zanusso, G.; Acutis, P.; Ferrari, S.; Capucci, L.; Tagliavini, F.; Monaco, S.; Caramelli, M. Identification of a second bovine amyloidotic spongiform encephalopathy: Molecular similarities with sporadic Creutzfeldt-Jakob disease. Proc. Natl. Acad. Sci. USA 2004, 101, 3065–3070. [Google Scholar] [CrossRef]
- Biacabe, A.G.; Laplanche, J.L.; Ryder, S.; Baron, T. Distinct molecular phenotypes in bovine prion diseases. EMBO Rep. 2004, 5, 110–115. [Google Scholar] [CrossRef]
- Hagiwara, K.; Iwamaru, Y.; Tabeta, N.; Yokoyama, T.; Tobiume, M. Evaluation of rapid post-mortem test kits for bovine spongiform encephalopathy (BSE) screening in Japan: Their analytical sensitivity to atypical BSE prions. Prion 2017, 11, 113–127. [Google Scholar] [CrossRef]
- Meloni, D.; Davidse, A.; Langeveld, J.P.; Varello, K.; Casalone, C.; Corona, C.; Balkema-Buschmann, A.; Groschup, M.H.; Ingravalle, F.; Bozzetta, E. EU-approved rapid tests for bovine spongiform encephalopathy detect atypical forms: A study for their sensitivities. PLoS ONE 2012, 7, e43133. [Google Scholar] [CrossRef]
- Béringue, V.; Vilotte, J.L.; Laude, H. Prion agent diversity and species barrier. Vet. Res. 2008, 39, 47. [Google Scholar] [CrossRef]
- Comoy, E.E.; Casalone, C.; Lescoutra-Etchegaray, N.; Zanusso, G.; Freire, S.; Marcé, D.; Auvré, F.; Ruchoux, M.M.; Ferrari, S.; Monaco, S.; et al. Atypical BSE (BASE) transmitted from asymptomatic aging cattle to a primate. PLoS ONE 2008, 3, e3017. [Google Scholar] [CrossRef]
- Kong, Q.; Zheng, M.; Casalone, C.; Qing, L.; Huang, S.; Chakraborty, B.; Wang, P.; Chen, F.; Cali, I.; Corona, C.; et al. Evaluation of the human transmission risk of an atypical bovine spongiform encephalopathy prion strain. J. Virol. 2008, 82, 3697–3701. [Google Scholar] [CrossRef] [PubMed]
- Mestre-Francés, N.; Nicot, S.; Rouland, S.; Biacabe, A.G.; Quadrio, I.; Perret-Liaudet, A.; Baron, T.; Verdier, J.M. Oral transmission of L-type bovine spongiform encephalopathy in primate model. Emerg. Infect. Dis. 2012, 18, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Ono, F.; Tase, N.; Kurosawa, A.; Hiyaoka, A.; Ohyama, A.; Tezuka, Y.; Wada, N.; Sato, Y.; Tobiume, M.; Hagiwara, K.; et al. Atypical L-type bovine spongiform encephalopathy (L-BSE) transmission to cynomolgus macaques, a non-human primate. Jpn. J. Infect. Dis. 2011, 64, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Eloit, M.; Adjou, K.; Coulpier, M.; Fontaine, J.J.; Hamel, R.; Lilin, T.; Messiaen, S.; Andreoletti, O.; Baron, T.; Bencsik, A.; et al. BSE agent signatures in a goat. Vet. Rec. 2005, 156, 523–524. [Google Scholar] [CrossRef]
- Foster, J.D.; Hope, J.; Fraser, H. Transmission of bovine spongiform encephalopathy to sheep and goats. Vet. Rec. 1993, 133, 339–341. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, M.; González, L.; Chong, A.; Foster, J.; Goldmann, W.; Hunter, N.; Martin, S. Ovine infection with the agents of scrapie (CH1641 isolate) and bovine spongiform encephalopathy: Immunochemical similarities can be resolved by immunohistochemistry. J. Comp. Pathol. 2006, 134, 17–29. [Google Scholar] [CrossRef]
- Manual for Diagnostic Tests and Vaccines for Terrestrial Animals, chapter 3.4.5.: Bovine spongiform encephalopathy. OIE, 2021.
- Masujin, K.; Okada, H.; Miyazawa, K.; Matsuura, Y.; Imamura, M.; Iwamaru, Y.; Murayama, Y.; Yokoyama, T. Emergence of a novel bovine spongiform encepha-lopathy (BSE) prion from an atypical H-type BSE. Sci. Rep. 2016, 6, 22753. [Google Scholar] [CrossRef]
- Fix, A.S.; Garman, R.H. Practical aspects of neuropathology: A technical guide for working with the nervous system. Toxicol. Pathol. 2000, 28, 122–131. [Google Scholar] [CrossRef]
- Gavier-Widén, D.; Stack, M.J.; Baron, T.; Balachandran, A.; Simmons, M. Diagnosis of transmissible spongiform encephalopathies in animals: A review. J. Vet. Diagn. Investig. 2005, 17, 509–527. [Google Scholar] [CrossRef]
- Konold, T.; Bone, G.E.; Clifford, D.; Chaplin, M.J.; Cawthraw, S.; Stack, M.J.; Simmons, M.M. Experimental H-type and L-type bovine spongiform encephalopathy in cattle: Observation of two clinical syndromes and diagnostic challenges. BMC Vet. Res. 2012, 8, 22. [Google Scholar] [CrossRef]
- Fast, C.; Graham, C.; Kaatz, M.; Santiago-Mateo, K.; Kaatz, T.; MacPherson, K.; Balkema-Buschmann, A.; Ziegler, U.; Groschup, M.H.; Czub, S. Discrimination of Classical and Atypical BSE by a Distinct Immunohistochemical PrPSc Profile. Pathogens 2023, 12, 353. [Google Scholar] [CrossRef] [PubMed]
- Casalone, C.; Hope, J. Atypical and classic bovine spongiform encephalopathy. Handb. Clin. Neurol. 2018, 153, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Wells, G.A.; McGill, I.S. Recently described scrapie-like encephalopathies of animals: Case definitions. Res. Vet. Sci. 1992, 53, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Debeer, S.O.; Baron, T.G.; Bencsik, A.A. Immunohistochemistry of PrPsc within bovine spongiform encephalopathy brain samples with graded autolysis. J. Histochem. Cytochem. 2001, 49, 1519–1524. [Google Scholar] [CrossRef] [PubMed]
- Bencsik, A.A.; Debeer, S.O.; Baron, T.G. An alternative pretreatment procedure in animal transmissible spongiform encephalopathies diagnosis using PrPsc immunohistochemistry. J. Histochem. Cytochem. 2005, 53, 1199–1202. [Google Scholar] [CrossRef]
- Okada, H.; Iwamaru, Y.; Imamura, M.; Masujin, K.; Yokoyama, T.; Mohri, S. Immunohistochemical detection of disease-associated prion protein in the intestine of cattle naturally affected with bovine spongiform encephalopathy by using an alkaline-based chemical antigen retrieval method. J. Vet. Med. Sci. 2010, 72, 1423–1429. [Google Scholar] [CrossRef]
- Shi, S.R.; Cote, R.J.; Taylor, C.R. Antigen retrieval techniques: Current perspectives. J. Histochem. Cytochem. 2001, 49, 931–937. [Google Scholar] [CrossRef]
- Okada, H.; Iwamaru, Y.; Yokoyama, T.; Mohri, S. Immunohistochemical detection of disease-associated prion protein in the peripheral nervous system in experimental H-type bovine spongiform encephalopathy. Vet. Pathol. 2013, 50, 659–663. [Google Scholar] [CrossRef]
- González, L.; Martin, S.; Jeffrey, M. Distinct profiles of PrP(d) immunoreactivity in the brain of scrapie- and BSE-infected sheep: Implications for differential cell targeting and PrP processing. J. Gen. Virol. 2003, 84 Pt 5, 1339–1350. [Google Scholar] [CrossRef]
- González, L.; Martin, S.; Houston, F.E.; Hunter, N.; Reid, H.W.; Bellworthy, S.J.; Jeffrey, M. Phenotype of disease-associated PrP accumulation in the brain of bovine spongiform encephalopathy experimentally infected sheep. J. Gen. Virol. 2005, 86 Pt 3, 827–838. [Google Scholar] [CrossRef]
- Dustan, B.H.; Spencer, Y.I.; Casalone, C.; Brownlie, J.; Simmons, M.M. A histopathologic and immunohistochemical review of archived UK caprine scrapie cases. Vet. Pathol. 2008, 45, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Orge, L.; Lima, C.; Machado, C.; Tavares, P.; Mendonça, P.; Carvalho, P.; Silva, J.; Pinto, M.L.; Bastos, E.; Pereira, J.C.; et al. Neuropathology of Animal Prion Diseases. Biomolecules 2021, 11, 466. [Google Scholar] [CrossRef] [PubMed]
- Stack, M.J.; Moore, S.J.; Davis, A.; Webb, P.R.; Bradshaw, J.M.; Lee, Y.H.; Chaplin, M.; Focosi-Snyman, R.; Thurston, L.; Spencer, Y.I.; et al. Bovine spongiform encephalopathy: Investigation of phenotypic variation among passive surveillance cases. J. Comp. Pathol. 2011, 144, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Porcario, C.; Hall, S.M.; Martucci, F.; Corona, C.; Iulini, B.; Perazzini, A.Z.; Acutis, P.; Hamir, A.N.; Loiacono, C.M.; Greenlee, J.J.; et al. Evaluation of two sets of immunohistochemical and Western blot confirmatory methods in the detection of typical and atypical BSE cases. BMC Res. Notes 2011, 4, 376. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, M.; González, L. Classical sheep transmissible spongiform encephalopathies: Pathogenesis, pathological phenotypes and clinical disease. Neuropathol. Appl. Neurobiol. 2007, 33, 373–394. [Google Scholar] [CrossRef]
- Jeffrey, M.; Martin, S.; González, L.; Ryder, S.J.; Bellworthy, S.J.; Jackman, R. Differential diagnosis of infections with the bovine spongiform encephalopathy (BSE) and scrapie agents in sheep. J. Comp. Pathol. 2001, 125, 271–284. [Google Scholar] [CrossRef]
- Jeffrey, M.; Martin, S.; González, L. Cell-associated variants of disease-specific prion protein immunolabelling are found in different sources of sheep transmissible spongiform encephalopathy. J. Gen. Virol. 2003, 84 Pt 4, 1033–1046. [Google Scholar] [CrossRef]
- Matsuura, Y.; Iwamaru, Y.; Masujin, K.; Imamura, M.; Mohri, S.; Yokoyama, T.; Okada, H. Distribution of abnormal prion protein in a sheep affected with L-type bovine spongiform encephalopathy. J. Comp. Pathol. 2013, 149, 113–118. [Google Scholar] [CrossRef]
- Iwamaru, Y.; Imamura, M.; Matsuura, Y.; Masujin, K.; Shimizu, Y.; Shu, Y.; Kurachi, M.; Kasai, K.; Murayama, Y.; Fukuda, S.; et al. Accumulation of L-type bovine prions in peripheral nerve tissues. Emerg. Infect. Dis. 2010, 16, 1151–1154. [Google Scholar] [CrossRef]
- Iwata, N.; Sato, Y.; Higuchi, Y.; Nohtomi, K.; Nagata, N.; Hasegawa, H.; Tobiume, M.; Nakamura, Y.; Hagiwara, K.; Furuoka, H.; et al. Distribution of PrP(Sc) in cattle with bovine spongiform encephalopathy slaughtered at abattoirs in Japan. Jpn. J. Infect. Dis. 2006, 59, 100–107. [Google Scholar]
- Zhao, L.; Hou, X.; Ji, R.; Han, C.; Yu, X.; Hong, T. Establishment of bovine prion peptide-based monoclonal antibodies for identifying bovine prion. Sci. China C Life Sci. 2009, 52, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Féraudet, C.; Morel, N.; Simon, S.; Volland, H.; Frobert, Y.; Créminon, C.; Vilette, D.; Lehmann, S.; Grassi, J. Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J. Biol. Chem. 2005, 280, 11247–11258. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, M.; Yamazaki, N.; Ikeda, T.; Ishiguro, N.; Shinagawa, M. A cellular form of prion protein (PrPC) exists in many non-neuronal tissues of sheep. J. Gen. Virol. 1995, 76 Pt 10, 2583–2587. [Google Scholar] [CrossRef] [PubMed]
- Harmeyer, S.; Pfaff, E.; Groschup, M.H. Synthetic peptide vaccines yield monoclonal antibodies to cellular and pathological prion proteins of ruminants. J. Gen. Virol. 1998, 79 Pt 4, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Krasemann, S.; Groschup, M.H.; Harmeyer, S.; Hunsmann, G.; Bodemer, W. Generation of monoclonal antibodies against human prion proteins in PrP0/0 mice. Mol. Med. 1996, 2, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Garssen, G.J.; Van Keulen, L.J.; Farquhar, C.F.; Smits, M.A.; Jacobs, J.G.; Bossers, A.; Meloen, R.H.; Langeveld, J.P. Applicability of three anti-PrP peptide sera including staining of tonsils and brainstem of sheep with scrapie. Microsc. Res. Tech. 2000, 50, 32–39. [Google Scholar] [CrossRef]
- Langeveld, J.P.; Jacobs, J.G.; Erkens, J.H.; Bossers, A.; van Zijderveld, F.G.; van Keulen, L.J. Rapid and discriminatory diagnosis of scrapie and BSE in retro-pharyngeal lymph nodes of sheep. BMC Vet. Res. 2006, 2, 19. [Google Scholar] [CrossRef]
- O’Rourke, K.I.; Baszler, T.V.; Miller, J.M.; Spraker, T.R.; Sadler-Riggleman, I.; Knowles, D.P. Monoclonal antibody F89/160.1.5 defines a conserved epitope on the ruminant prion protein. J. Clin. Microbiol. 1998, 36, 1750–1755. [Google Scholar] [CrossRef]
- Schaller, O.; Fatzer, R.; Stack, M.; Clark, J.; Cooley, W.; Biffiger, K.; Egli, S.; Doherr, M.; Vandevelde, M.; Heim, D.; et al. Validation of a western immunoblotting procedure for bovine PrP(Sc) detection and its use as a rapid surveillance method for the diagnosis of bovine spongiform encephalopathy (BSE). Acta Neuropathol. 1999, 98, 437–443. [Google Scholar] [CrossRef]
- Korth, C.; Stierli, B.; Streit, P.; Moser, M.; Schaller, O.; Fischer, R.; Schulz-Schaeffer, W.; Kretzschmar, H.; Raeber, A.; Braun, U.; et al. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature 1997, 390, 74–77. [Google Scholar] [CrossRef]
- Shimizu, Y.; Kaku-Ushiki, Y.; Iwamaru, Y.; Muramoto, T.; Kitamoto, T.; Yokoyama, T.; Mohri, S.; Tagawa, Y. A novel anti-prion protein monoclonal antibody and its single-chain fragment variable derivative with ability to inhibit abnormal prion protein accumulation in cultured cells. Microbiol. Immunol. 2010, 54, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Thuring, C.M.; Erkens, J.H.; Jacobs, J.G.; Bossers, A.; Van Keulen, L.J.; Garssen, G.J.; Van Zijderveld, F.G.; Ryder, S.J.; Groschup, M.H.; Sweeney, T.; et al. Discrimination between scrapie and bovine spongiform encephalopathy in sheep by molecular size, immunoreactivity, and glycoprofile of prion protein. J. Clin. Microbiol. 2004, 42, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.L.; Umetani, A.; Matsui, T.; Ishiguro, N.; Shinagawa, M.; Horiuchi, M. Antigenic characterization of an abnormal isoform of prion protein using a new diverse panel of monoclonal antibodies. Virology 2004, 320, 40–51. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, K.I.; Baszler, T.V.; Besser, T.E.; Miller, J.M.; Cutlip, R.C.; Wells, G.A.; Ryder, S.J.; Parish, S.M.; Hamir, A.N.; Cockett, N.E.; et al. Preclinical diagnosis of scrapie by immunohistochemistry of third eyelid lymphoid tissue. J. Clin. Microbiol. 2000, 38, 3254–3259. [Google Scholar] [CrossRef] [PubMed]
- Demart, S.; Fournier, J.G.; Creminon, C.; Frobert, Y.; Lamoury, F.; Marce, D.; Lasmézas, C.; Dormont, D.; Grassi, J.; Deslys, J.P. New insight into abnormal prion protein using monoclonal antibodies. Biochem. Biophys. Res. Commun. 1999, 265, 652–657. [Google Scholar] [CrossRef]
- Thuring, C.M.; van Keulen, L.J.; Langeveld, J.P.; Vromans, M.E.; van Zijderveld, F.G.; Sweeney, T. Immunohistochemical distinction between preclinical bovine spongiform encephalopathy and scrapie infection in sheep. J. Comp. Pathol. 2005, 132, 59–69. [Google Scholar] [CrossRef]
- Horiuchi, M.; Karino, A.; Furuoka, H.; Ishiguro, N.; Kimura, K.; Shinagawa, M. Generation of monoclonal antibody that distinguishes PrPSc from PrPC and neutralizes prion infectivity. Virology 2009, 394, 200–207. [Google Scholar] [CrossRef]
- Chaplin, M.J.; Barlow, N.; Ryder, S.; Simmons, M.M.; Spencer, Y.; Hughes, R.; Stack, M.J. Evaluation of the effects of controlled autolysis on the immunodetection of PrP(Sc) by immunoblotting and immunohistochemistry from natural cases of scrapie and BSE. Res. Vet. Sci. 2002, 72, 37–43. [Google Scholar] [CrossRef]
- Debeer, S.O.; Baron, T.G.; Bencsik, A.A. Transmissible spongiform encephalopathy diagnosis using PrPsc immunohistochemistry on fixed but previously frozen brain samples. J. Histochem. Cytochem. 2002, 50, 611–616. [Google Scholar] [CrossRef]
- Monleón, E.; Monzón, M.; Hortells, P.; Vargas, A.; Badiola, J.J. Detection of PrP(sc) in samples presenting a very advanced degree of autolysis (BSE liquid state) by immunocytochemistry. J. Histochem. Cytochem. 2003, 51, 15–18. [Google Scholar] [CrossRef]
- Sarasa, R.; Becher, D.; Badiola, J.J.; Monzón, M. A comparative study of modified confirmatory techniques and additional immuno-based methods for non-conclusive autolytic bovine spongiform encephalopathy cases. BMC Vet. Res. 2013, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- Schulz-Schaeffer, W.J.; Fatzer, R.; Vandevelde, M.; Kretzschmar, H.A. Detection of PrP(Sc) in subclinical BSE with the paraffin-embedded tissue (PET) blot. Arch. Virol. Suppl. 2000, 16, 173–180. [Google Scholar] [CrossRef]
- Lezmi, S.; Bencsik, A.; Baron, T. PET-blot analysis contributes to BSE strain recognition in C57Bl/6 mice. J. Histochem. Cytochem. 2006, 54, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Webb, P.R.; Denyer, M.; Gough, J.; Spiropoulos, J.; Simmons, M.M.; Spencer, Y.I. Paraffin-embedded tissue blot as a sensitive method for discrimination between classical scrapie and experimental bovine spongiform encephalopathy in sheep. J. Vet. Diagn. Investig. 2011, 23, 492–498. [Google Scholar] [CrossRef]
- Polak, M.P.; Rożek, W.; Żmudziński, J.F. Implementation of dot-blot in rapid diagnosis of BSE. Bull. Vet. Inst. Pulawy. 2005, 49, 263–266. [Google Scholar]
- Stack, M.J.; Balachandran, A.; Chaplin, M.; Davis, L.; Czub, S.; Miller, B. The first Canadian indigenous case of bovine spongiform encephalopathy (BSE) has molecular characteristics for prion protein that are similar to those of BSE in the United Kingdom but differ from those of chronic wasting disease in captive elk and deer. Can Vet. J. 2004, 45, 825–830. [Google Scholar]
- Method for the Provisional Discrimination of Bovine TSE Isolates (C-TYPE, L-TYPE & H-TYPE BSE). Available online: https://science.vla.gov.uk/tseglobalnet/documents/tse-rl-blot.pdf (accessed on 15 March 2023).
- Jacobs, J.G.; Langeveld, J.P.; Biacabe, A.G.; Acutis, P.L.; Polak, M.P.; Gavier-Widen, D.; Buschmann, A.; Caramelli, M.; Casalone, C.; Mazza, M.; et al. Molecular discrimination of atypical bovine spongiform encephalopathy strains from a geographical region spanning a wide area in Europe. J. Clin. Microbiol. 2007, 45, 1821–1829. [Google Scholar] [CrossRef]
- Priemer, G.; Balkema-Buschmann, A.; Hills, B.; Groschup, M.H. Biochemical Characteristics and PrP(Sc) Distribution Pattern in the Brains of Cattle Experimentally Challenged with H-type and L-type Atypical BSE. PLoS ONE 2013, 8, e67599. [Google Scholar] [CrossRef]
- Langeveld, J.P.; Erkens, J.H.; Rammel, I.; Jacobs, J.G.; Davidse, A.; van Zijderveld, F.G.; Bossers, A.; Schildorfer, H. Four independent molecular prion protein parameters for discriminating new cases of C, L, and h bovine spongiform encephalopathy in cattle. J. Clin. Microbiol. 2011, 49, 3026–3028. [Google Scholar] [CrossRef]
- Biacabe, A.G.; Jacobs, J.G.; Bencsik, A.; Langeveld, J.P.; Baron, T.G. H-type bovine spongiform encephalopathy: Complex molecular features and similarities with human prion diseases. Prion 2007, 1, 61–68. [Google Scholar] [CrossRef]
- Dudas, S.; Czub, S. Atypical BSE: Current Knowledge and Knowledge Gaps. Food Saf. 2017, 5, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Polak, M.P.; Zmudzinski, J.F.; Jacobs, J.G.; Langeveld, J.P. Atypical status of bovine spongiform encephalopathy in Poland: A molecular typing study. Arch. Virol. 2008, 153, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Stack, M.; Jeffrey, M.; Gubbins, S.; Grimmer, S.; González, L.; Martin, S.; Chaplin, M.; Webb, P.; Simmons, M.; Spencer, Y.; et al. Monitoring for bovine spongiform encephalopathy in sheep in Great Britain, 1998-2004. J Gen Virol. 2006, 87 Pt 7, 2099–2107. [Google Scholar] [CrossRef] [PubMed]
- Stack, M.J.; Chaplin, M.J.; Davis, L.A.; Everitt, S.; Simmons, M.M.; Windl, O.; Hope, J.; Burke, P. Four BSE cases with an L-BSE molecular profile in cattle from Great Britain. Vet. Rec. 2013, 172, 70. [Google Scholar] [CrossRef] [PubMed]
- Baron, T.G.; Madec, J.Y.; Calavas, D.; Richard, Y.; Barillet, F. Comparison of French natural scrapie isolates with bovine spongiform encephalopathy and experimental scrapie infected sheep. Neurosci. Lett. 2000, 284, 175–178. [Google Scholar] [CrossRef]
- Stack, M.J.; Chaplin, M.J.; Clark, J. Differentiation of prion protein glycoforms from naturally occurring sheep scrapie, sheep-passaged scrapie strains (CH1641 and SSBP1), bovine spongiform encephalopathy (BSE) cases and Romney and Cheviot breed sheep experimentally inoculated with BSE using two monoclonal antibodies. Acta Neuropathol. 2002, 104, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Gough, K.C.; Rees, H.C.; Ives, S.E.; Maddison, B.C. Methods for Differentiating Prion Types in Food-Producing Animals. Biology 2015, 4, 785–813. [Google Scholar] [CrossRef] [PubMed]
- Vulin, J.; Biacabe, A.G.; Cazeau, G.; Calavas, D.; Baron, T. Molecular typing of protease-resistant prion protein in transmissible spongiform encephalopathies of small ruminants, France, 2002–2009. Emerg. Infect. Dis. 2011, 17, 55–63. [Google Scholar] [CrossRef]
- Lezmi, S.; Martin, S.; Simon, S.; Comoy, E.; Bencsik, A.; Deslys, J.P.; Grassi, J.; Jeffrey, M.; Baron, T. Comparative molecular analysis of the abnormal prion protein in field scrapie cases and experimental bovine spongiform encephalopathy in sheep by use of Western blotting and immunohistochemical methods. J. Virol. 2004, 78, 3654–3662. [Google Scholar] [CrossRef]
- Acutis, P.L.; Martucci, F.; Mazza, M.; Nodari, S.; Maurella, C.; Ru, G.; Casalone, C.; Caramelli, M. Molecular typing of transmissible spongiform encephalopathy from Italian disease outbreaks in small ruminants. Vet Rec. 2006, 159, 746–747. [Google Scholar] [CrossRef]
- Baron, T.; Bencsik, A.; Vulin, J.; Biacabe, A.G.; Morignat, E.; Verchere, J.; Betemps, D. A C-terminal protease-resistant prion fragment distinguishes ovine "CH1641-like" scrapie from bovine classical and L-Type BSE in ovine transgenic mice. PLoS Pathog. 2008, 4, e1000137. [Google Scholar] [CrossRef] [PubMed]
- Owen, J.P.; Rees, H.C.; Maddison, B.C.; Terry, L.A.; Thorne, L.; Jackman, R.; Whitelam, G.C.; Gough, K.C. Molecular profiling of ovine prion diseases by using thermolysin-resistant PrPSc and endogenous C2 PrP fragments. J. Virol. 2007, 81, 10532–10539. [Google Scholar] [CrossRef] [PubMed]
- Pirisinu, L.; Migliore, S.; Di Bari, M.A.; Esposito, E.; Baron, T.; D’Agostino, C.; De Grossi, L.; Vaccari, G.; Agrimi, U.; Nonno, R. Molecular discrimination of sheep bovine spongiform encephalopathy from scrapie. Emerg. Infect. Dis. 2011, 17, 695–698. [Google Scholar] [CrossRef] [PubMed]
- Gretzschel, A.; Buschmann, A.; Langeveld, J.; Groschup, M.H. Immunological characterization of abnormal prion protein from atypical scrapie cases in sheep using a panel of monoclonal antibodies. J. Gen. Virol. 2006, 87 Pt 12, 3715–3722. [Google Scholar] [CrossRef] [PubMed]
- Klingeborn, M.; Wik, L.; Simonsson, M.; Renström, L.H.M.; Ottinger, T.; Linné, T. Characterization of proteinase K-resistant N- and C-terminally truncated PrP in Nor98 atypical scrapie. J. Gen. Virol. 2006, 87 Pt 6, 1751–1760. [Google Scholar] [CrossRef] [PubMed]
- Dudas, S.; James, J.; Anderson, R.; Czub, S. Exploring the cause of initially reactive bovine brains on rapid tests for BSE. Prion 2015, 9, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Grassi, J. Pre-clinical diagnosis of transmissible spongiform encephalopathies using rapid tests. Transfus. Clin. Biol. 2003, 10, 19–22. [Google Scholar] [CrossRef]
- Wells, G.A.; Hawkins, S.A.; Green, R.B.; Austin, A.R.; Dexter, I.; Spencer, Y.I.; Chaplin, M.J.; Stack, M.J.; Dawson, M. Preliminary observations on the pathogenesis of experimental bovine spongiform encephalopathy (BSE): An update. Vet. Rec. 1998, 142, 103–106. [Google Scholar] [CrossRef]
- Gray, J.G.; Dudas, S.; Czub, S. A study on the analytical sensitivity of 6 BSE tests used by the Canadian BSE reference laboratory. PLoS ONE 2011, 6, e17633. [Google Scholar] [CrossRef]
- Prionics®—Check PrioSTRIP; Prionics AG: Schlieren-Zurich, Switzerland, 2023.
- Webster, K.; Flowers, M.; Cassar, C.; Bayliss, D. Determination of Analytical Sensitivity (Detection Limit) for Currently Approved TSE Rapid Tests. 2009. Available online: http://www.efsa.europa.eu/de/scdocs/doc/1436.pdf (accessed on 15 March 2023).
- Prionics®—Check LIA BSE Antigen Test Kit; Prionics AG: Schlieren-Zurich, Switzerland, 2023.
- IDEXX® HerdCheck Bovine Spongiform Encephalopathy Antigen Test Kit, EIA. IDEXX Laboratories, Westbrook, ME, USA.
- Simon, S.; Nugier, J.; Morel, N.; Boutal, H.; Créminon, C.; Benestad, S.L.; Andréoletti, O.; Lantier, F.; Bilheude, J.M.; Feyssaguet, M.; et al. Rapid typing of transmissible spongiform encephalopathy strains with differential ELISA. Emerg. Infect. Dis. 2008, 14, 608–616. [Google Scholar] [CrossRef]
- Meloni, D.; Bozzetta, E.; Langeveld, J.P.; Groschup, M.H.; Goldmann, W.; Andrèoletti, O.; Lantier, I.; Van Keulen, L.; Bossers, A.; Pitardi, D.; et al. EU-approved rapid tests might underestimate bovine spongiform encephalopathy infection in goats. J. Vet. Diagn. Investig. 2017, 29, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.D.; Greenlee, J.J. Detection of misfolded prion protein in retina samples of sheep and cattle by use of a commercially available enzyme immunoassay. Am J Vet Res. 2014, 75, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Wear, A.; Henderson, K.; Webster, K.; Patel, I. A comparison of rapid bovine spongiform encephalopathy testing methods on autolyzed bovine brain tissue. J. Vet. Diagn. Investig. 2005, 17, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Meloni, D.; Varello, K.; Pezzolato, M.; Manzardo, E.; Cavarretta, M.C.; Ingravalle, F.; Caramelli, M.; Bozzetta, E. Effect of autolysis on the specificity of bovine spongiform encephalopathy rapid tests. BMC Res. Notes. 2010, 3, 193. [Google Scholar] [CrossRef]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Stöckel, J.; Hartl, F.U. Chaperonin-mediated de novo generation of prion protein aggregates. J. Mol. Biol. 2001, 313, 861–872. [Google Scholar] [CrossRef]
- Saleem, F.; Bjorndahl, T.C.; Ladner, C.L.; Perez-Pineiro, R.; Ametaj, B.N.; Wishart, D.S. Lipopolysaccharide induced conversion of recombinant prion protein. Prion 2014, 8, 221–232. [Google Scholar] [CrossRef]
- Gray, J.G.; Dudas, S.; Graham, C.; Czub, S. Performance analysis of rapid diagnostic tests on atypical bovine spongiform encephalopathy. J. Vet. Diagn. Investig. 2012, 24, 976–980. [Google Scholar] [CrossRef]
- Taraboulos, A.; Serban, D.; Prusiner, S.B. Scrapie prion proteins accumulate in the cytoplasm of persistently infected cultured cells. J. Cell Biol. 1990, 110, 2117–2132. [Google Scholar] [CrossRef]
- Yamasaki, T.; Suzuki, A.; Shimizu, T.; Watarai, M.; Hasebe, R.; Horiuchi, M. Characterization of intracellular localization of PrP(Sc) in prion-infected cells using a mAb that recognizes the region consisting of aa 119-127 of mouse PrP. J. Gen. Virol. 2012, 93 Pt 3, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Pimpinelli, F.; Lehmann, S.; Maridonneau-Parini, I. The scrapie prion protein is present in flotillin-1-positive vesicles in central- but not peripheral-derived neuronal cell lines. Eur. J. Neurosci. 2005, 21, 2063–2072. [Google Scholar] [CrossRef] [PubMed]
- Veith, N.M.; Plattner, H.; Stuermer, C.A.; Schulz-Schaeffer, W.J.; Bürkle, A. Immunolocalisation of PrPSc in scrapie-infected N2a mouse neuroblastoma cells by light and electron microscopy. Eur. J. Cell Biol. 2009, 88, 45–63. [Google Scholar] [CrossRef] [PubMed]
- Marijanovic, Z.; Caputo, A.; Campana, V.; Zurzolo, C. Identification of an intracellular site of prion conversion. PLoS Pathog. 2009, 5, e1000426. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Shi, Q.; Wang, J.; Xiao, K.; Sun, J.; Lv, Y.; Guo, M.; Zhou, W.; Chen, C.; Gao, C.; et al. Reduction of NF-κB (p65) in Scrapie-Infected Cultured Cells and in the Brains of Scrapie-Infected Rodents. ACS Chem. Neurosci. 2017, 8, 2535–2548. [Google Scholar] [CrossRef]
- Shan, Z.; Yamasaki, T.; Suzuki, A.; Hasebe, R.; Horiuchi, M. Establishment of a simple cell-based ELISA for the direct detection of abnormal isoform of prion protein from prion-infected cells without cell lysis and proteinase K treatment. Prion 2016, 10, 305–318. [Google Scholar] [CrossRef]
- Suzuki, A.; Yamasaki, T.; Hasebe, R.; Horiuchi, M. Enhancement of binding avidity by bivalent binding enables PrPSc-specific detection by anti-PrP monoclonal antibody 132. PLoS ONE 2019, 14, e0217944. [Google Scholar] [CrossRef]
- Dunbar, S.A. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta. 2006, 363, 71–82. [Google Scholar] [CrossRef]
- Vignali, D.A. Multiplexed particle-based flow cytometric assays. J. Immunol. Methods. 2000, 243, 243–255. [Google Scholar] [CrossRef]
- Tang, Y.; Thorne, J.; Whatling, K.; Jacobs, J.G.; Langeveld, J.; Sauer, M.J. A single step multiplex immunofluorometric assay for differential diagnosis of BSE and scrapie. J. Immunol. Methods. 2010, 356, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Gielbert, A.; Jacobs, J.G.; Baron, T.; Andreoletti, O.; Yokoyama, T.; Langeveld, J.P.; Sauer, M.J. All major prion types recognised by a multiplex immunofluorometric assay for disease screening and confirmation in sheep. J. Immunol. Methods. 2012, 380, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Merz, P.A.; Somerville, R.A.; Wisniewski, H.M.; Iqbal, K. Abnormal fibrils from scrapie-infected brain. Acta Neuropathol. 1981, 54, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Corona, C.; Vallino Costassa, E.; Iulini, B.; Caramelli, M.; Bozzetta, E.; Mazza, M.; Desiato, R.; Ru, G.; Casalone, C. Phenotypical Variability in Bovine Spongiform Encephalopathy: Epidemiology, Pathogenesis, and Diagnosis of Classical and Atypical Forms. Prog. Mol. Biol. Transl. Sci. 2017, 150, 241–265. [Google Scholar] [CrossRef] [PubMed]
- Hope, J.; Multhaup, G.; Reekie, L.J.; Kimberlin, R.H.; Beyreuther, K. Molecular pathology of scrapie-associated fibril protein (PrP) in mouse brain affected by the ME7 strain of scrapie. Eur. J. Biochem. 1988, 172, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.C.; Wells, G.A.; Stack, M.J.; White, H.; Dawson, M. Bovine spongiform encephalopathy: Detection and quantitation of fibrils, fibril protein (PrP) and vacuolation in brain. Vet. Microbiol. 1990, 23, 295–304. [Google Scholar] [CrossRef]
- Wells, G.A.; Scott, A.C.; Wilesmith, J.W.; Simmons, M.M.; Matthews, D. Correlation between the results of a histopathological examination and the detection of abnormal brain fibrils in the diagnosis of bovine spongiform encephalopathy. Res. Vet. Sci. 1994, 56, 346–351. [Google Scholar] [CrossRef]
- Cooley, W.A.; Clark, J.K.; Stack, M.J. Comparison of scrapie-associated fibril detection and Western immunoblotting for the diagnosis of natural ovine scrapie. J. Comp. Pathol. 1998, 118, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Stack, M.J.; Aldrich, A.M.; Kitching, A.D.; Scott, A.C. Comparative study of electron microscopical techniques for the detection of scrapie-associated fibrils. Res. Vet. Sci. 1995, 59, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Cooley, W.A.; Clark, J.K.; Ryder, S.J.; Davis, L.A.; Farrelly, S.S.; Stack, M.J. Evaluation of a rapid western immunoblotting procedure for the diagnosis of bovine spongiform encephalopathy (BSE) in the UK. J. Comp. Pathol. 2001, 125, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, A.; Pfaff, E.; Reifenberg, K.; Müller, H.M.; Groschup, M.H. Detection of cattle-derived BSE prions using transgenic mice overexpressing bovine PrP(C). Arch. Virol. Suppl. 2000, 16, 75–86. [Google Scholar] [CrossRef]
- Buschmann, A.; Groschup, M.H. Highly bovine spongiform encephalopathy-sensitive transgenic mice confirm the essential restriction of infectivity to the nervous system in clinically diseased cattle. J. Infect. Dis. 2005, 192, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.; Hart, P.; Piccardo, P.; Hunter, N.; Casalone, C.; Baron, T.; Barron, R.M. Bovine PrP expression levels in transgenic mice influence transmission characteristics of atypical bovine spongiform encephalopathy. J. Gen. Virol. 2012, 93 Pt 5, 1132–1140. [Google Scholar] [CrossRef] [PubMed]
- Thackray, A.M.; Andréoletti, O.; Spiropoulos, J.; Bujdoso, R. A new model for sensitive detection of zoonotic prions by PrP transgenic Drosophila. J. Biol. Chem. 2021, 297, 100878. [Google Scholar] [CrossRef] [PubMed]
- Beck, K.E.; Vickery, C.M.; Lockey, R.; Holder, T.; Thorne, L.; Terry, L.A.; Denyer, M.; Webb, P.; Simmons, M.M.; Spiropoulos, J. The interpretation of disease phenotypes to identify TSE strains following murine bioassay: Characterisation of classical scrapie. Vet. Res. 2012, 43, 77. [Google Scholar] [CrossRef] [PubMed]
- Saborio, G.P.; Permanne, B.; Soto, C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 2001, 411, 810–813. [Google Scholar] [CrossRef]
- Castilla, J.; Saá, P.; Morales, R.; Abid, K.; Maundrell, K.; Soto, C. Protein misfolding cyclic amplification for diagnosis and prion propagation studies. Methods Enzymol. 2006, 412, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Nishina, K.; Deleault, N.R.; Lucassen, R.W.; Supattapone, S. In vitro prion protein conversion in detergent-solubilized membranes. Biochemistry 2004, 43, 2613–2621. [Google Scholar] [CrossRef]
- Colby, D.W.; Zhang, Q.; Wang, S.; Groth, D.; Legname, G.; Riesner, D.; Prusiner, S.B. Prion detection by an amyloid seeding assay. Proc. Natl. Acad. Sci. USA 2007, 104, 20914–20919. [Google Scholar] [CrossRef]
- Atarashi, R.; Wilham, J.M.; Christensen, L.; Hughson, A.G.; Moore, R.A.; Johnson, L.M.; Onwubiko, H.A.; Priola, S.A.; Caughey, B. Simplified ultrasensitive prion detection by recombinant PrP conversion with shaking. Nat. Methods. 2008, 5, 211–212. [Google Scholar] [CrossRef]
- Panza, G.; Stöhr, J.; Dumpitak, C.; Papathanassiou, D.; Weiss, J.; Riesner, D.; Willbold, D.; Birkmann, E. Spontaneous and BSE-prion-seeded amyloid formation of full length recombinant bovine prion protein. Biochem. Biophys. Res. Commun. 2008, 373, 493–497. [Google Scholar] [CrossRef]
- Wang, F.; Wang, X.; Yuan, C.G.; Ma, J. Generating a prion with bacterially expressed recombinant prion protein. Science 2010, 327, 1132–1135. [Google Scholar] [CrossRef]
- Kim, J.I.; Cali, I.; Surewicz, K.; Kong, Q.; Raymond, G.J.; Atarashi, R.; Race, B.; Qing, L.; Gambetti, P.; Caughey, B.; et al. Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J. Biol. Chem. 2010, 285, 14083–14087. [Google Scholar] [CrossRef] [PubMed]
- Mays, C.E.; Yeom, J.; Kang, H.E.; Bian, J.; Khaychuk, V.; Kim, Y.; Bartz, J.C.; Telling, G.C.; Ryou, C. In vitro amplification of misfolded prion protein using lysate of cultured cells. PLoS ONE 2011, 6, e18047. [Google Scholar] [CrossRef] [PubMed]
- Deleault, N.R.; Harris, B.T.; Rees, J.R.; Supattapone, S. Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. USA 2007, 104, 9741–9746. [Google Scholar] [CrossRef] [PubMed]
- Thorne, L.; Terry, L.A. In vitro amplification of PrPSc derived from the brain and blood of sheep infected with scrapie. J. Gen. Virol. 2008, 89 Pt 12, 3177–3184. [Google Scholar] [CrossRef] [PubMed]
- Murayama, Y.; Yoshioka, M.; Masujin, K.; Okada, H.; Iwamaru, Y.; Imamura, M.; Matsuura, Y.; Fukuda, S.; Onoe, S.; Yokoyama, T.; et al. Sulfated dextrans enhance in vitro amplification of bovine spongiform encephalopathy PrP(Sc) and enable ultrasensitive detection of bovine PrP(Sc). PLoS ONE 2010, 5, e13152. [Google Scholar] [CrossRef]
- Moudjou, M.; Sibille, P.; Fichet, G.; Reine, F.; Chapuis, J.; Herzog, L.; Jaumain, E.; Laferrière, F.; Richard, C.A.; Laude, H.; et al. Highly infectious prions generated by a single round of microplate-based protein misfolding cyclic amplification. mBio 2013, 5, e00829-13. [Google Scholar] [CrossRef]
- Gonzalez-Montalban, N.; Makarava, N.; Ostapchenko, V.G.; Savtchenk, R.; Alexeeva, I.; Rohwer, R.G.; Baskakov, I.V. Highly efficient protein misfolding cyclic amplification. PLoS Pathog 2011, 7, e1001277. [Google Scholar] [CrossRef]
- Park, J.H.; Choi, Y.G.; Park, S.J.; Choi, H.S.; Choi, E.K.; Kim, Y.S. Ultra-efficient Amplification of Abnormal Prion Protein by Modified Protein Misfolding Cyclic Amplification with Electric Current. Mol. Neurobiol. 2018, 55, 1630–1638. [Google Scholar] [CrossRef]
- Gough, K.C.; Bishop, K.; Maddison, B.C. Highly sensitive detection of small ruminant bovine spongiform encephalopathy within transmissible spongiform encephalopathy mixes by serial protein misfolding cyclic amplification. J. Clin. Microbiol. 2014, 52, 3863–3868. [Google Scholar] [CrossRef]
- Simmons, M.M.; Chaplin, M.J.; Vickery, C.M.; Simon, S.; Davis, L.; Denyer, M.; Lockey, R.; Stack, M.J.; O’Connor, M.J.; Bishop, K.; et al. Does the Presence of Scrapie Affect the Ability of Current Statutory Discriminatory Tests to Detect the Presence of Bovine Spongiform Encephalopathy? J. Clin. Microbiol. 2015, 53, 2593–2604. [Google Scholar] [CrossRef]
- Chen, B.; Morales, R.; Barria, M.A.; Soto, C. Estimating prion concentration in fluids and tissues by quantitative PMCA. Nat. Methods. 2010, 7, 519–520. [Google Scholar] [CrossRef] [PubMed]
- Orem, N.R.; Geoghegan, J.C.; Deleault, N.R.; Kascsak, R.; Supattapone, S. Copper (II) ions potently inhibit purified PrPres amplification. J. Neurochem. 2006, 96, 1409–1415. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, A.; Atarashi, R.; Fuse, T.; Ubagai, K.; Nakagaki, T.; Yamaguchi, N.; Ishibashi, D.; Katamine, S.; Nishida, N. Hyperefficient PrP Sc amplification of mouse-adapted BSE and scrapie strain by protein misfolding cyclic amplification technique. FEBS J. 2009, 276, 2841–2848. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, M.J.; Bishop, K.; Workman, R.G.; Maddison, B.C.; Gough, K.C. In vitro amplification of H-type atypical bovine spongiform encephalopathy by protein misfolding cyclic amplification. Prion 2017, 11, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Murayama, Y.; Ono, F.; Shimozaki, N.; Shibata, H. L-Arginine ethylester enhances in vitro amplification of PrP(Sc) in macaques with atypical L-type bovine spongiform encephalopathy and enables presymptomatic detection of PrP(Sc) in the bodily fluids. Biochem. Biophys. Res. Commun. 2016, 470, 563–568. [Google Scholar] [CrossRef]
- Castilla, J.; Gonzalez-Romero, D.; Saá, P.; Morales, R.; De Castro, J.; Soto, C. Crossing the species barrier by PrP(Sc) replication in vitro generates unique infectious prions. Cell 2008, 134, 757–768. [Google Scholar] [CrossRef]
- Peden, A.H.; Suleiman, S.; Barria, M.A. Understanding Intra-Species and Inter-Species Prion Conversion and Zoonotic Potential Using Protein Misfolding Cyclic Amplification. Front. Aging Neurosci. 2021, 13, 716452. [Google Scholar] [CrossRef]
- Barria, M.A.; Balachandran, A.; Morita, M.; Kitamoto, T.; Barron, R.; Manson, J.; Knight, R.; Ironside, J.W.; Head, M.W. Molecular barriers to zoonotic transmission of prions. Emerg. Infect. Dis. 2014, 20, 88–97. [Google Scholar] [CrossRef]
- Béringue, V.; Herzog, L.; Reine, F.; Le Dur, A.; Casalone, C.; Vilotte, J.L.; Laude, H. Transmission of atypical bovine prions to mice transgenic for human prion protein. Emerg. Infect. Dis. 2008, 14, 1898–1901. [Google Scholar] [CrossRef]
- Cosseddu, G.M.; Nonno, R.; Vaccari, G.; Bucalossi, C.; Fernandez-Borges, N.; Di Bari, M.A.; Castilla, J.; Agrimi, U. Ultra-efficient PrP(Sc) amplification highlights potentialities and pitfalls of PMCA technology. PLoS Pathog. 2011, 7, e1002370. [Google Scholar] [CrossRef]
- Chang, B.; Cheng, X.; Yin, S.; Pan, T.; Zhang, H.; Wong, P.; Kang, S.C.; Xiao, F.; Yan, H.; Li, C.; et al. Test for detection of disease-associated prion aggregate in the blood of infected but asymptomatic animals. Clin. Vaccine Immunol. 2007, 14, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, R.; Sano, K.; Satoh, K.; Nishida, N. Real-time quaking-induced conversion: A highly sensitive assay for prion detection. Prion 2011, 5, 150–153. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, M.; Cramm, M.; Llorens, F.; Müller-Cramm, D.; Collins, S.; Atarashi, R.; Satoh, K.; Orrù, C.D.; Groveman, B.R.; Zafar, S.; et al. The real-time quaking-induced conversion assay for detection of human prion disease and study of other protein misfolding diseases. Nat. Protoc. 2016, 11, 2233–2242. [Google Scholar] [CrossRef]
- Wilham, J.M.; Orrú, C.D.; Bessen, R.A.; Atarashi, R.; Sano, K.; Race, B.; Meade-White, K.D.; Taubner, L.M.; Timmes, A.; Caughey, B. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010, 6, e1001217. [Google Scholar] [CrossRef] [PubMed]
- Masujin, K.; Orrú, C.D.; Miyazawa, K.; Groveman, B.R.; Raymond, L.D.; Hughson, A.G.; Caughey, B. Detection of Atypical H-Type Bovine Spongiform Encephalopathy and Discrimination of Bovine Prion Strains by Real-Time Quaking-Induced Conversion. J. Clin. Microbiol. 2016, 54, 676–686. [Google Scholar] [CrossRef]
- Peden, A.H.; McGuire, L.I.; Appleford, N.E.J.; Mallinson, G.; Wilham, J.M.; Orrú, C.D.; Caughey, B.; Ironside, J.W.; Knight, R.S.; Will, R.G.; et al. Sensitive and specific detection of sporadic Creutzfeldt-Jakob disease brain prion protein using real-time quaking-induced conversion. J. Gen. Virol. 2012, 93 Pt 2, 438–449. [Google Scholar] [CrossRef]
- Dassanayake, R.P.; Orrú, C.D.; Hughson, A.G.; Caughey, B.; Graça, T.; Zhuang, D.; Madsen-Bouterse, S.A.; Knowles, D.P.; Schneider, D.A. Sensitive and specific detection of classical scrapie prions in the brains of goats by real-time quaking-induced conversion. J. Gen. Virol. 2016, 97, 803–812. [Google Scholar] [CrossRef]
- Samorodnitsky, D.; Nicholson, E.M. Differential effects of divalent cations on elk prion protein fibril formation and stability. Prion 2018, 12, 63–71. [Google Scholar] [CrossRef]
- Moore, S.J.; Vrentas, C.E.; Hwang, S.; West Greenlee, M.H.; Nicholson, E.M.; Greenlee, J.J. Pathologic and biochemical characterization of PrPSc from elk with PRNP polymorphisms at codon 132 after experimental infection with the chronic wasting disease agent. BMC Vet. Res. 2018, 14, 80. [Google Scholar] [CrossRef]
- Di Bari, M.A.; Nonno, R.; Castilla, J.; D’Agostino, C.; Pirisinu, L.; Riccardi, G.; Conte, M.; Richt, J.; Kunkle, R.; Langeveld, J.; et al. Chronic wasting disease in bank voles: Characterisation of the shortest incubation time model for prion diseases. PLoS Pathog. 2013, 9, e1003219. [Google Scholar] [CrossRef]
- Nonno, R.; Di Bari, M.A.; Cardone, F.; Vaccari, G.; Fazzi, P.; Dell’Omo, G.; Cartoni, C.; Ingrosso, L.; Boyle, A.; Galeno, R.; et al. Efficient transmission and characterization of Creutzfeldt-Jakob disease strains in bank voles. PLoS Pathog. 2006, 2, e12. [Google Scholar] [CrossRef] [PubMed]
- Agrimi, U.; Nonno, R.; Dell’Omo, G.; Di Bari, M.A.; Conte, M.; Chiappini, B.; Esposito, E.; Di Guardo, G.; Windl, O.; Vaccari, G.; et al. Prion protein amino acid determinants of differential susceptibility and molecular feature of prion strains in mice and voles. PLoS Pathog. 2008, 4, e1000113. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Tatum, T.; Lebepe-Mazur, S.; Nicholson, E.M. Preparation of lyophilized recombinant prion protein for TSE diagnosis by RT-QuIC. BMC Res. Notes 2018, 11, 895. [Google Scholar] [CrossRef]
- Orrú, C.D.; Hughson, A.G.; Groveman, B.R.; Campbell, K.J.; Anson, K.J.; Manca, M.; Kraus, A.; Caughey, B. Factors That Improve RT-QuIC Detection of Prion Seeding Activity. Viruses 2016, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.E.; Mo, Y.; Abd Rahim, R.; Lee, H.M.; Ryou, C. Prion Diagnosis: Application of Real-Time Quaking-Induced Conversion. Biomed. Res. Int. 2017, 2017, 5413936. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Mitteregger-Kretzschmar, G.; Giese, A.; Kretzschmar, H.A. Establishing quantitative real-time quaking-induced conversion (qRT-QuIC) for highly sensitive detection and quantification of PrPSc in prion-infected tissues. Acta Neuropathol. Commun. 2013, 1, 44. [Google Scholar] [CrossRef]
- Henderson, D.M.; Davenport, K.A.; Haley, N.J.; Denkers, N.D.; Mathiason, C.K.; Hoover, E.A. Quantitative assessment of prion infectivity in tissues and body fluids by real-time quaking-induced conversion. J. Gen. Virol. 2015, 96 Pt 1, 210–219. [Google Scholar] [CrossRef]
- Orrú, C.D.; Wilham, J.M.; Raymond, L.D.; Kuhn, F.; Schroeder, B.; Raeber, A.J.; Caughey, B. Prion disease blood test using immunoprecipitation and improved quaking-induced conversion. mBio. 2011, 2, e00078-11. [Google Scholar] [CrossRef]
- Elder, A.M.; Henderson, D.M.; Nalls, A.V.; Wilham, J.M.; Caughey, B.W.; Hoover, E.A.; Kincaid, A.E.; Bartz, J.C.; Mathiason, C.K. In vitro detection of prionemia in TSE-infected cervids and hamsters. PLoS ONE 2013, 8, e80203. [Google Scholar] [CrossRef]
- John, T.R.; Schätzl, H.M.; Gilch, S. Early detection of chronic wasting disease prions in urine of pre-symptomatic deer by real-time quaking-induced conversion assay. Prion. 2013, 7, 253–258. [Google Scholar] [CrossRef]
- Henderson, D.M.; Manca, M.; Haley, N.J.; Denkers, N.D.; Nalls, A.V.; Mathiason, C.K.; Caughey, B.; Hoover, E.A. Rapid antemortem detection of CWD prions in deer saliva. PLoS ONE 2013, 8, e74377. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Greenlee, J.J.; Nicholson, E.M. Real-Time Quaking-Induced Conversion Detection of PrPSc in Fecal Samples From Chronic Wasting Disease Infected White-Tailed Deer Using Bank Vole Substrate. Front. Vet. Sci. 2021, 8, 643754. [Google Scholar] [CrossRef] [PubMed]
- Favole, A.; Mazza, M.; D’Angelo, A.; Lombardi, G.; Palmitessa, C.; Dell’Atti, L.; Cagnotti, G.; Berrone, E.; Gallo, M.; Avanzato, T.; et al. RT-QuIC detection of pathological prion protein in subclinical goats following experimental oral transmission of L-type BSE. BMC Res. Notes. 2021, 14, 442. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Manca, M.; Foutz, A.; Camacho, M.V.; Raymond, G.J.; Race, B.; Orru, C.D.; Yuan, J.; Shen, P.; Li, B.; et al. Early preclinical detection of prions in the skin of prion-infected animals. Nat Commun. 2019, 10, 247. [Google Scholar] [CrossRef]
- Orrú, C.D.; Favole, A.; Corona, C.; Mazza, M.; Manca, M.; Groveman, B.R.; Hughson, A.G.; Acutis, P.L.; Caramelli, M.; Zanusso, G.; et al. Detection and discrimination of classical and atypical L-type bovine spongiform encephalopathy by real-time quaking-induced conversion. J. Clin. Microbiol. 2015, 53, 1115–1120. [Google Scholar] [CrossRef]
- Levavasseur, E.; Biacabe, A.G.; Comoy, E.; Culeux, A.; Grznarova, K.; Privat, N.; Simoneau, S.; Flan, B.; Sazdovitch, V.; Seilhean, D.; et al. Detection and partial discrimination of atypical and classical bovine spongiform encephalopathies in cattle and primates using real-time quaking-induced conversion assay. PLoS ONE 2017, 12, e0172428. [Google Scholar] [CrossRef]
- Sawada, K.; Suzuki, A.; Yamasaki, T.; Iwamaru, Y.; Matsuura, Y.; Miyazawa, K.; Masujin, K.; Atarashi, R.; Horiuchi, M. Estimation of prion infectivity in tissues of cattle infected with atypical BSE by real time-quaking induced conversion assay. J. Vet. Med. Sci. 2019, 81, 846–850. [Google Scholar] [CrossRef]
- Favole, A.; Mazza, M.; Vallino Costassa, E.; D’Angelo, A.; Lombardi, G.; Marconi, P.; Crociara, P.; Berrone, E.; Gallo, M.; Palmitessa, C.; et al. Early and Pre-Clinical Detection of Prion Seeding Activity in Cerebrospinal Fluid of Goats using Real-Time Quaking-Induced Conversion Assay. Sci. Rep. 2019, 9, 6173. [Google Scholar] [CrossRef]
- Trieschmann, L.; Navarrete Santos, A.; Kaschig, K.; Torkler, S.; Maas, E.; Schätzl, H.; Böhm, G. Ultra-sensitive detection of prion protein fibrils by flow cytometry in blood from cattle affected with bovine spongiform encephalopathy. BMC Biotechnol. 2005, 5, 26. [Google Scholar] [CrossRef]
- World Organisation for Animal Health. Available online: https://www.oie.int/en/what-we-offer/veterinary-products/diagnostic-kits/ (accessed on 15 March 2023).
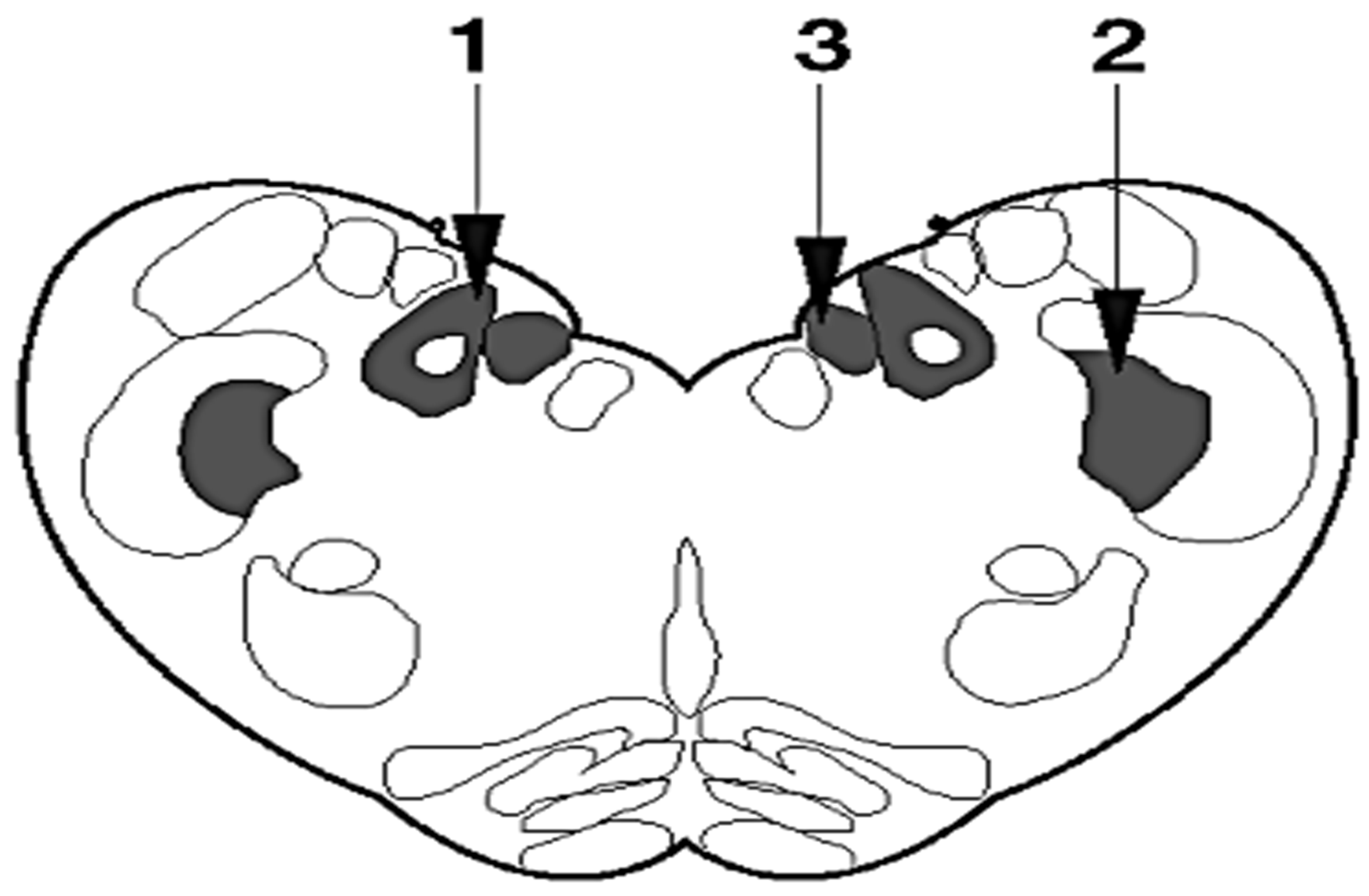



| Classical BSE (C-Type) | Atypical BSE (H-Type and L-Type/BASE) | |
|---|---|---|
| Source of infection | through an animal’s consumption of prion-contaminated feed | occur spontaneously at a very low rate in all cattle populations, does not appear to be associated with contaminated feed |
| Clinical signs | changes in behavior and temperament, hyperreactivity, hind-limb incoordination, weakness, and loss of body condition | some of the clinical signs of the disease include dullness, low head carriage, inactivity, or lack of nervousness |
| Transmission | efficiently transmitted orally to other species (e.g., sheep, goats, macaques) | low risk for oral transmission |
| Zoonotic potential | can be transmitted to humans through the consumption of contaminated meat causing variant Creutzfeldt–Jakob disease | transmission to humans has never been reported; the full risks presented to humans remain unknown, but some data suggesting that L-type BSE may be zoonotic |
| First diagnosis | 1986, Great Britain | 2004, Italy and France |
| Occurrence | worldwide distribution, cases have been reported in more than 20 countries, the implementation of appropriate control measures resulted in its decline, can affect cattle of any age | very rare disease, worldwide distribution, even in countries where no classical BSE has been reported, is expected to occur in all cattle populations, regardless of the control measures in place, occurs particularly in cattle older than 8 years |
| World Organisation for Animal Health (WOAH) | its occurrence may be considered for the purpose of the official BSE risk status recognition by the WOAH | its occurrence is not considered for the purpose of the official BSE risk status recognition by the WOAH |
| Biochemical characteristics of PrPSc | extremely resistant to degradation by proteinase digestion | less resistant to degradation by proteinase digestion, can be degraded by stringent proteinase K digestion |
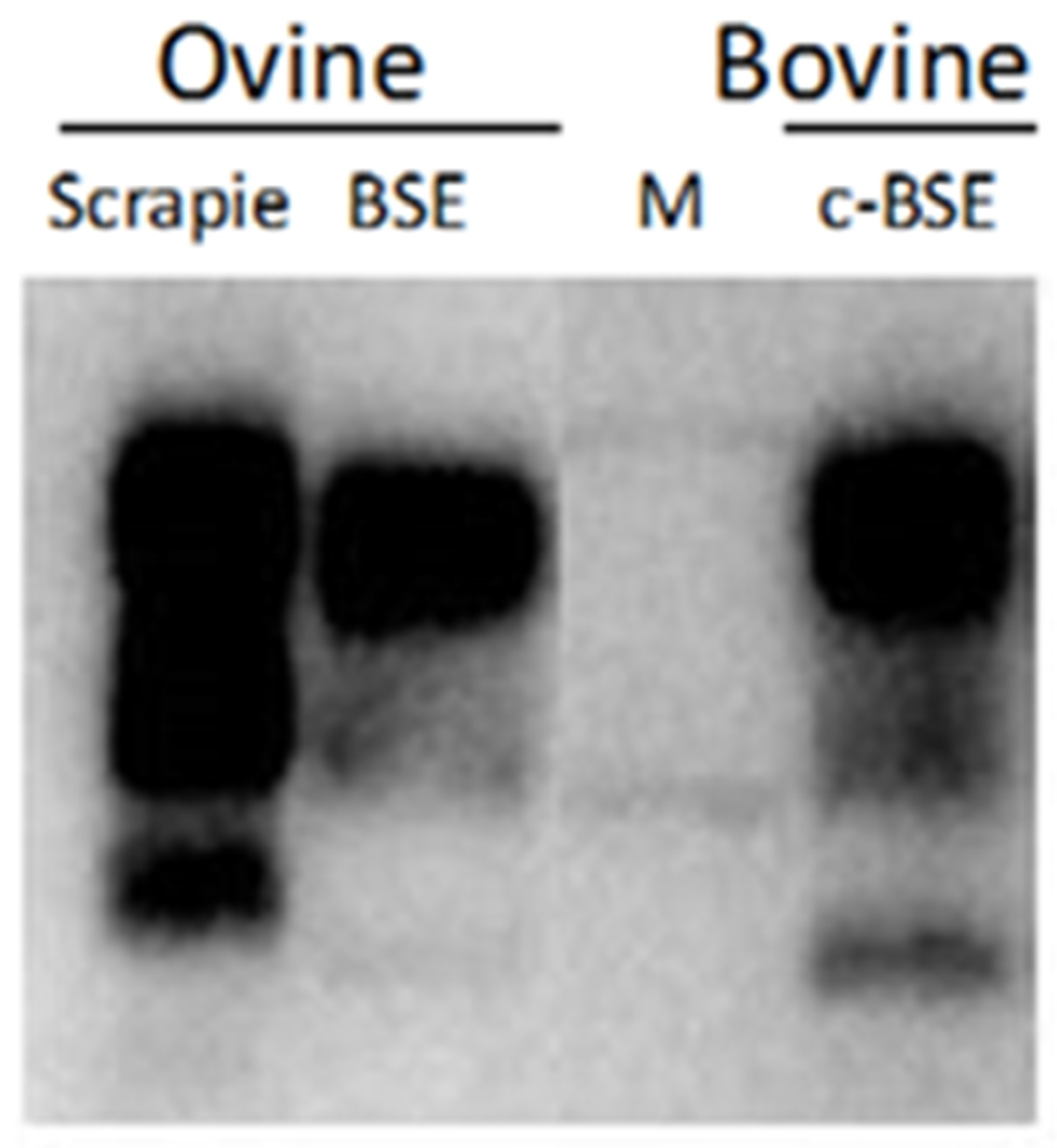
| WB pattern | Three bands correspond to the diglycosylated (~28 kDa), monoglycosylated (~22 kDa), and unglycosylated (~18 kDa) forms | L-type form has a lower while H-type higher molecular mass of the unglycosylated PrPSc when compared with classical BSE |
| Antibody | Epitope | Type | Application | Reference | ||
|---|---|---|---|---|---|---|
| Region | Name | Amino-Acid Sequence | Position | |||
| N-terminal region | SAF32 | QPHGGGW a | 62–92 b | Monoclonal | EIA, FC, IHC, WB | [43] |
| B103 | QGGTHGQWNKPSKPKTNMK | 103–121 b | Polyclonal | IHC | [44] | |
| P4 | WGQGGSH | 101–107 b | Monoclonal | WB, ELISA, IHC, PET-blot | [45] | |
| BG4 | SPGGNRYPP | 46–54 c | Monoclonal | IHC | [16] | |
| 8G8 | SQWNKPSK | 100–107 c | Monoclonal | WB, IHC | [46] | |
| R521 | GQGGSHSQWNKPGGC | 94–105 c | Polyclonal | WB | [47] | |
| R505 | CSQWNKPSKPKTN | 100–111 c | Polyclonal | WB | [47] | |
| 12B2 | WGQGG | 101–105 b | Monoclonal | IHC, WB, IFMA | [48] | |
| Core/globularregion | SHa31 | YEDRYYRE | 156–163 b | Monoclonal | IFMA, WB, IHC | [43] |
| F89/160.1.5 | SRPLIHFGSDYEDR | 146–159 b | Monoclonal | IHC, WB, ICC/IF, ELISA | [49] | |
| 12F10 | SDYEDRYYRE | 154–163 b | Monoclonal | IHC | [43] | |
| 6H4 | YEDRYYREN | 156–164 b | Monoclonal | WB, IHC, | [50,51] | |
| T1 | LIHFGND | 141–147 c | Monoclonal | IHC | [52] | |
| F89 | IHFG | 142–145 | Monoclonal | IHC, | [49] | |
| L42 | YEDRYY | 156–161 b | Monoclonal | WB, IHC, ELISA | [48] | |
| 2G11 | YRENMY | 153–158 c | Monoclonal | IHC | [53] | |
| 132 | AVVGGLGGY | 119–127 d | Monoclonal | ELISA, IFA | [54] | |
| 9A2 | WNK | 110–112 b | Monoclonal | WB, IFMA | [48] | |
| C-terminal region | F99/97.6.1 | QYQRES | 228–233 b | Monoclonal | IHC | [55] |
| 31C6 | DWEDRYY | 143–149 d | Monoclonal | ELISA, IHC | [54] | |
| R145 | YQRESQAYYQRGA | 221–233 b | Monoclonal | IHC, PET-blot | [48,56] | |
| 44B1 | 155–231 | Monoclonal | IHC, ELISA | [54] | ||
| SAF84 | RPVDQY | 175–180 b | Monoclonal | IHC, WB | [43,56] | |
| 94B4 | HTVTTTTK | 198–205 b | Monoclonal | WB, IFMA | [57] | |
| 6H10 | NA | NA | Monoclonal | IHC | [58] | |
| Method | Advantages | Disadvantages |
|---|---|---|
| HA * | it is one of the least expensive morphological methods; preserves tissue morphology; paraffin-embedded and frozen tissue samples can be stored and accessed when required; it does not require any special equipment, results can be viewed using a conventional bright-field microscope | particular areas of the brain are needed; inaccurate hemisectioning could result in the complete loss of a target area for testing; multi-step procedure; tissue is highly processed and may lead to loss of information; subjective interpretation of results; requires good sample preservation; has lower sensitivity than other methods; time-consuming and laborious; requires specialized reagents and qualified personnel; less specific than IHC; low throughput; qualitative method |
| IHC * | high specific and sensitive; fresh, frozen, and autolyzed samples can be used; paraffin-embedded and frozen tissue samples can be stored and accessed when required; allows cellular localization of protein; relatively inexpensive; widely used; it does not require any special equipment, results can be viewed using a conventional bright-field microscope; verify the results obtained by HA | semi-quantitative; multi-step procedure; variability depends on the fixation procedure, staining protocol, and antibody selection; particular areas of the brain are needed; requires specialized reagents and qualified personnel; subjective interpretation of results; low throughput; non-specific reactions can occur, time-consuming and laborious |
| PET-blot | highly sensitive and specific; can be used to discriminate TSE strains, macroscopic observation possible | not a rapid tool; not suitable for large-scale screening; time-consuming and laborious; requires paraffin-embedded tissues, specialized equipment, reagents, and qualified personnel; has lower microscopic resolution than IHC; multi-step procedure; it is not commonly used; lack of standardization |
| WB * | very sensitive and specific; fresh, frozen, and autolyzed samples can be used; widely used; provide valuable information about the biochemical properties of PrPSc; can be used to discriminate TSE strains; can be quantitative or qualitative; similar sensitivity to IHC | give no information on the neuroanatomical location of PrPSc; requires specialized equipment, reagents, and qualified personnel; multi-step procedure; non-specific reactions can occur; subjective interpretation of results; paraffin-embedded tissues cannot be used; sensitivity varies between methods and laboratories |
| Rapid tests * | highly sensitive and specific; rapid; suitable for large-scale screening; full automation is possible; based on WB, immunochromatography, and ELISA; does not need ultracentrifugation steeps needed to concentrate the PrPSc; can be quantitative or qualitative; fresh, frozen, and autolyzed samples can be used; can be used to discriminate TSE strains; most of them are commercially available | particular areas of the brain are needed; occasionally give false positive results; require specialized reagents and qualified personnel; some tests require specialized equipment; paraffin-embedded tissues cannot be used |
| IFA | cells or frozen tissue can be used; highly sensitive and specific; rapid, | expensive; requires specialized equipment (fluorescence or laser microscope), reagents, and qualified personnel; lack of standardization; frozen sections have poor morphology; non-specific results can occur; low throughput, |
| IFMA | extremely sensitive; allow simultaneous detection of multiple analytes; amenable to high-volume testing; can be used to discriminate TSE strains; automated | expensive; requires specialized equipment (cytometer), reagents, and qualified personnel; non-specific results can occur; rarely used; lack of standardization |
| SAF | highly specific; autolyzed sample can be used | results depend on the region used and sample purification scheme; labor-intensive; requires specialized equipment (electron microscope), reagents, and qualified personnel; less sensitive than WB; rarely used; only qualitative |
| Bioassay | the most sensitive and specific; provide information about PrPSc infectivity; strain-typing method | labor-intensive and time-consuming; not suitable for large-scale screening; have the highest sensitivity only when performed in homologous species |
| PMCA | extremely sensitive, can be used to understand the biology of prion proteins, can be quantitative or qualitative; can be automated; can be used for antemortem TSE diagnosis | requires specialized equipment (sonicator), reagents, and qualified personnel; no real-time detection; time-consuming; false-positive results may occur; lack of standardization; requires additional substrate purification step |
| RT-QuIC | extremely sensitivity, fast; most straightforward; practicable; relatively inexpensive; less labor-intensive and time-consuming than PMCA; automated; spontaneous fibrilization is minimized, real-time detection; can be quantitative or qualitative; can be used to discriminate TSE strains; suitable for large-scale screening; can be used for antemortem TSE diagnosis | no infectivity propagation; lack of standardization |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olech, M. Conventional and State-of-the-Art Detection Methods of Bovine Spongiform Encephalopathy (BSE). Int. J. Mol. Sci. 2023, 24, 7135. https://doi.org/10.3390/ijms24087135
Olech M. Conventional and State-of-the-Art Detection Methods of Bovine Spongiform Encephalopathy (BSE). International Journal of Molecular Sciences. 2023; 24(8):7135. https://doi.org/10.3390/ijms24087135
Chicago/Turabian StyleOlech, Monika. 2023. "Conventional and State-of-the-Art Detection Methods of Bovine Spongiform Encephalopathy (BSE)" International Journal of Molecular Sciences 24, no. 8: 7135. https://doi.org/10.3390/ijms24087135
APA StyleOlech, M. (2023). Conventional and State-of-the-Art Detection Methods of Bovine Spongiform Encephalopathy (BSE). International Journal of Molecular Sciences, 24(8), 7135. https://doi.org/10.3390/ijms24087135





