Gamma-Hemolysin Components: Computational Strategies for LukF-Hlg2 Dimer Reconstruction on a Model Membrane
Abstract
1. Introduction
2. Results
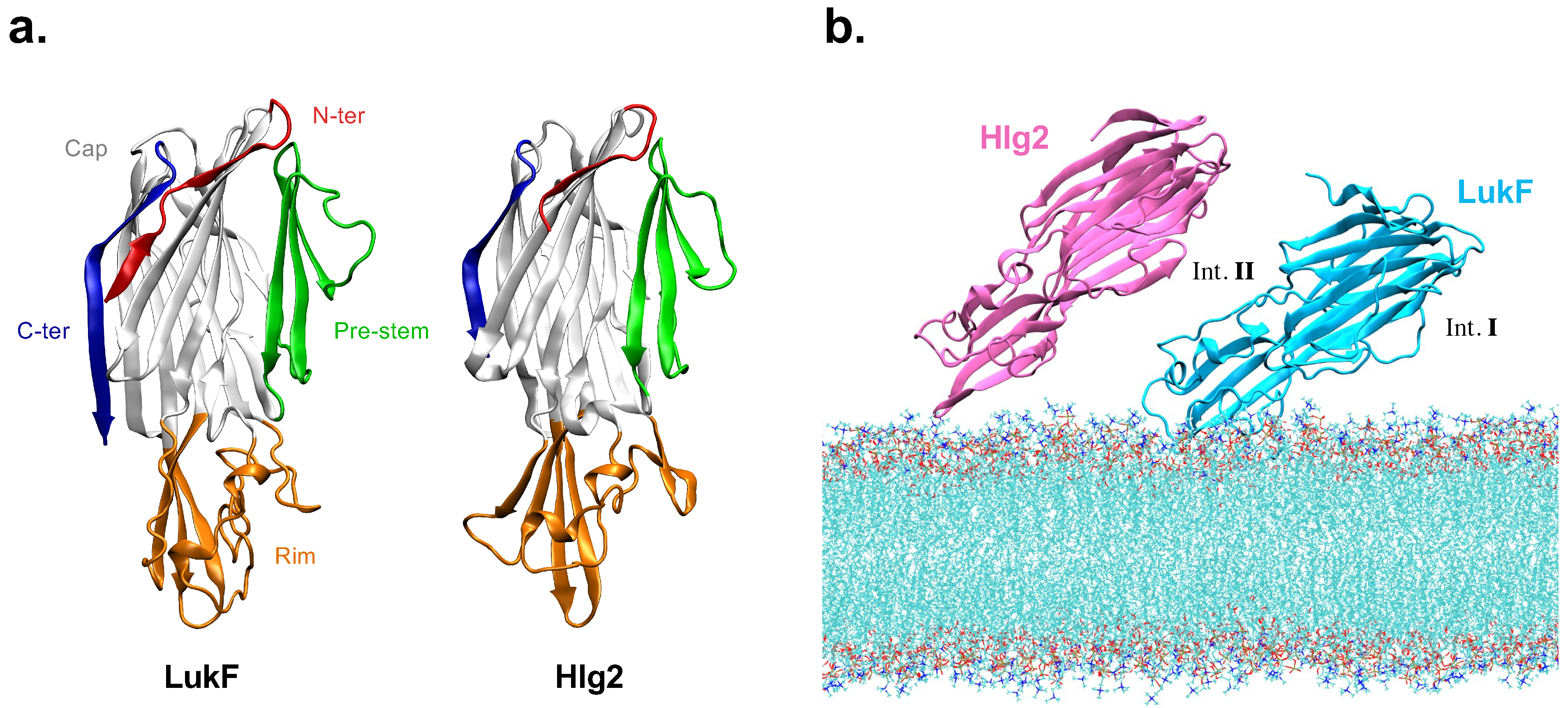
2.1. Spontaneous LukF-Hlg2 Dimerization on the Membrane Shows a Large Contact Interface through the Rim Domains
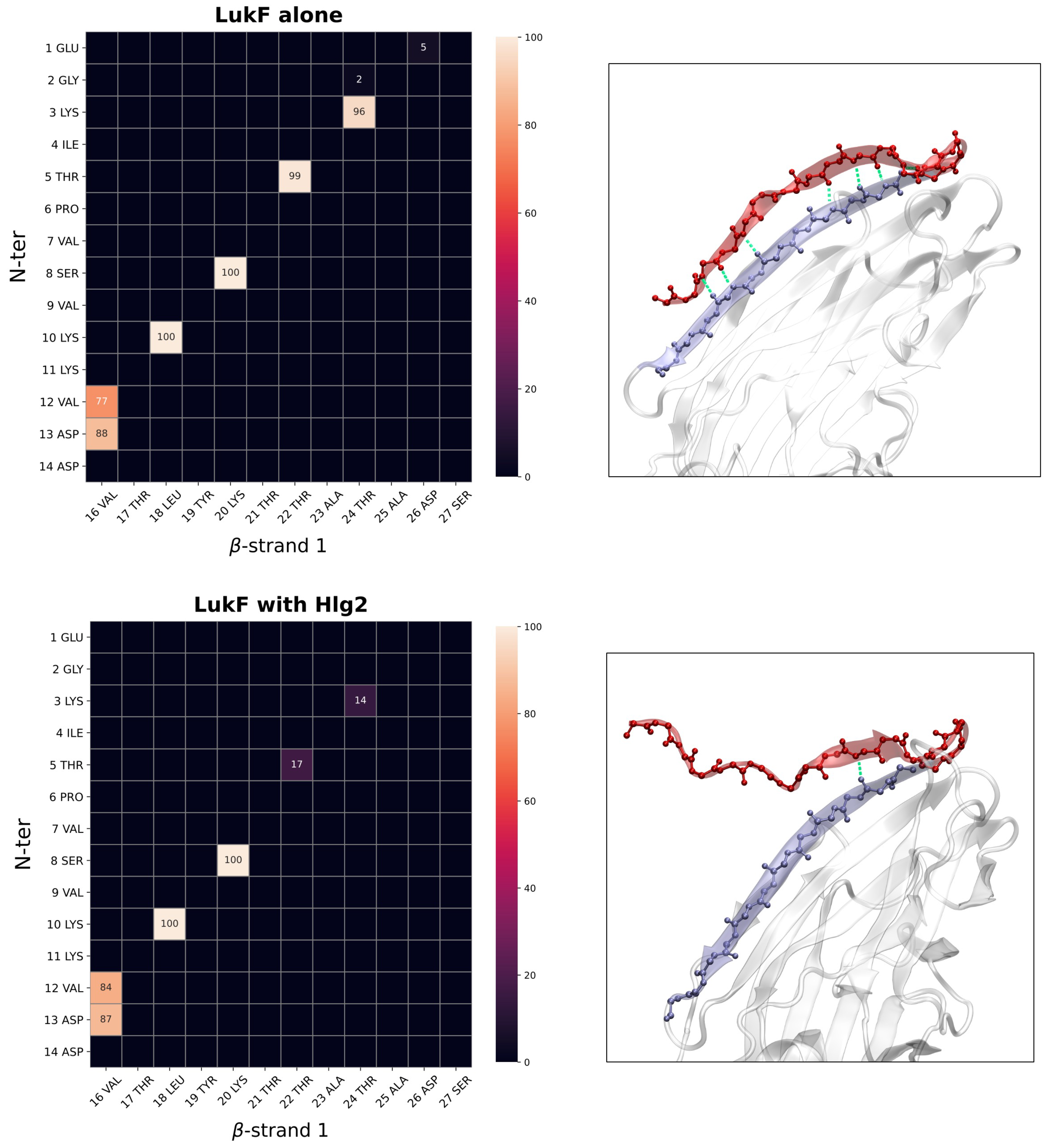
2.2. Contacts Formed by LukF N-Terminus upon Dimerization
2.3. Protein–Lipid Interactions and Reorientation of the Dimer with Respect to the Membrane Plane
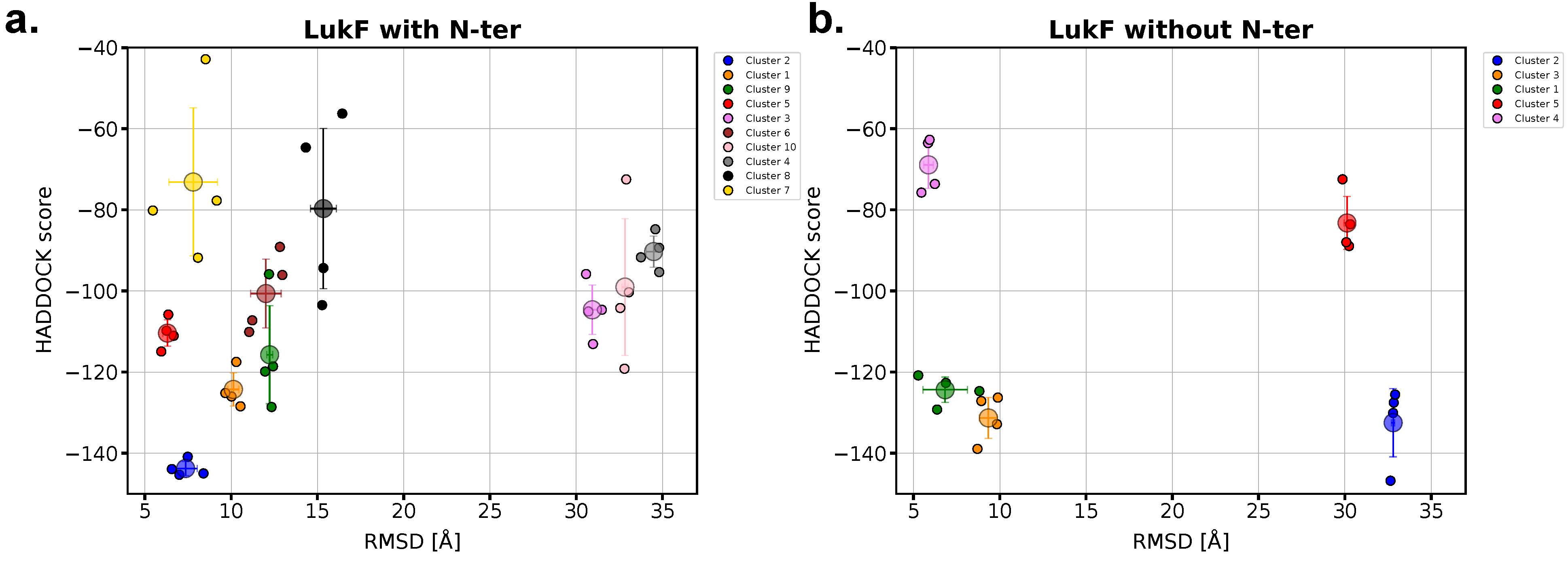
2.4. The Presence of LukF N-Terminus Impacts the Modeling of the Dimer with HADDOCK
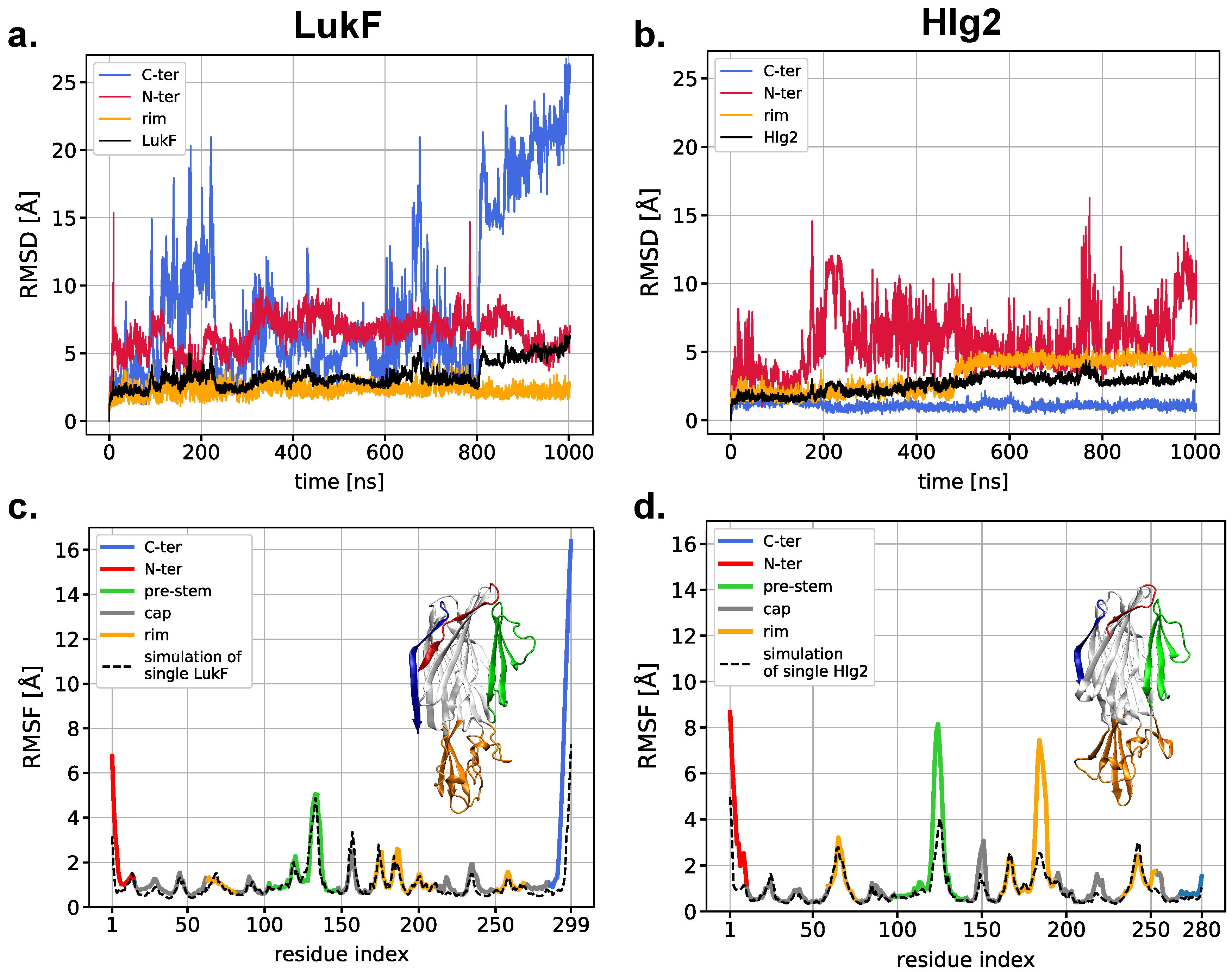
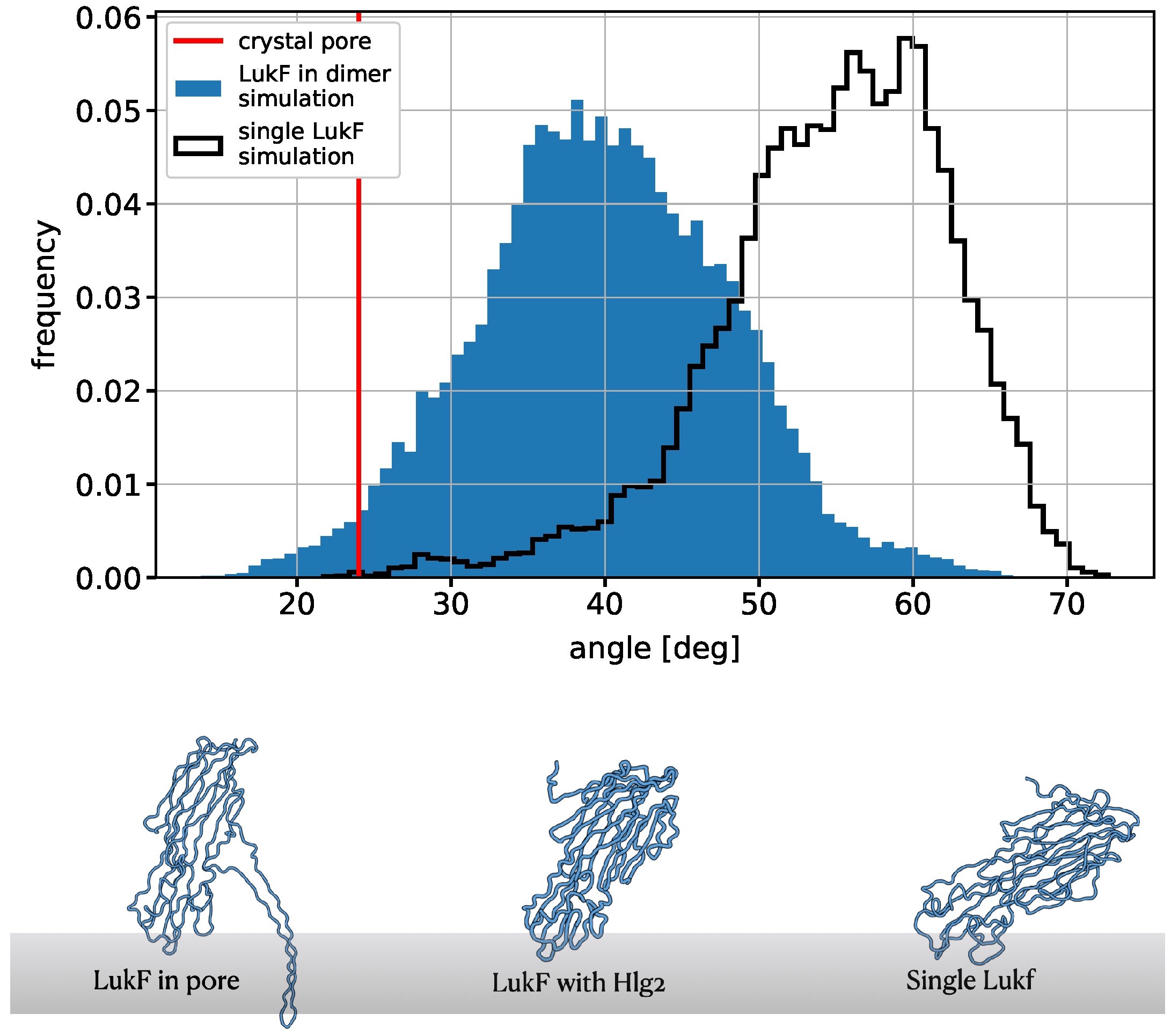
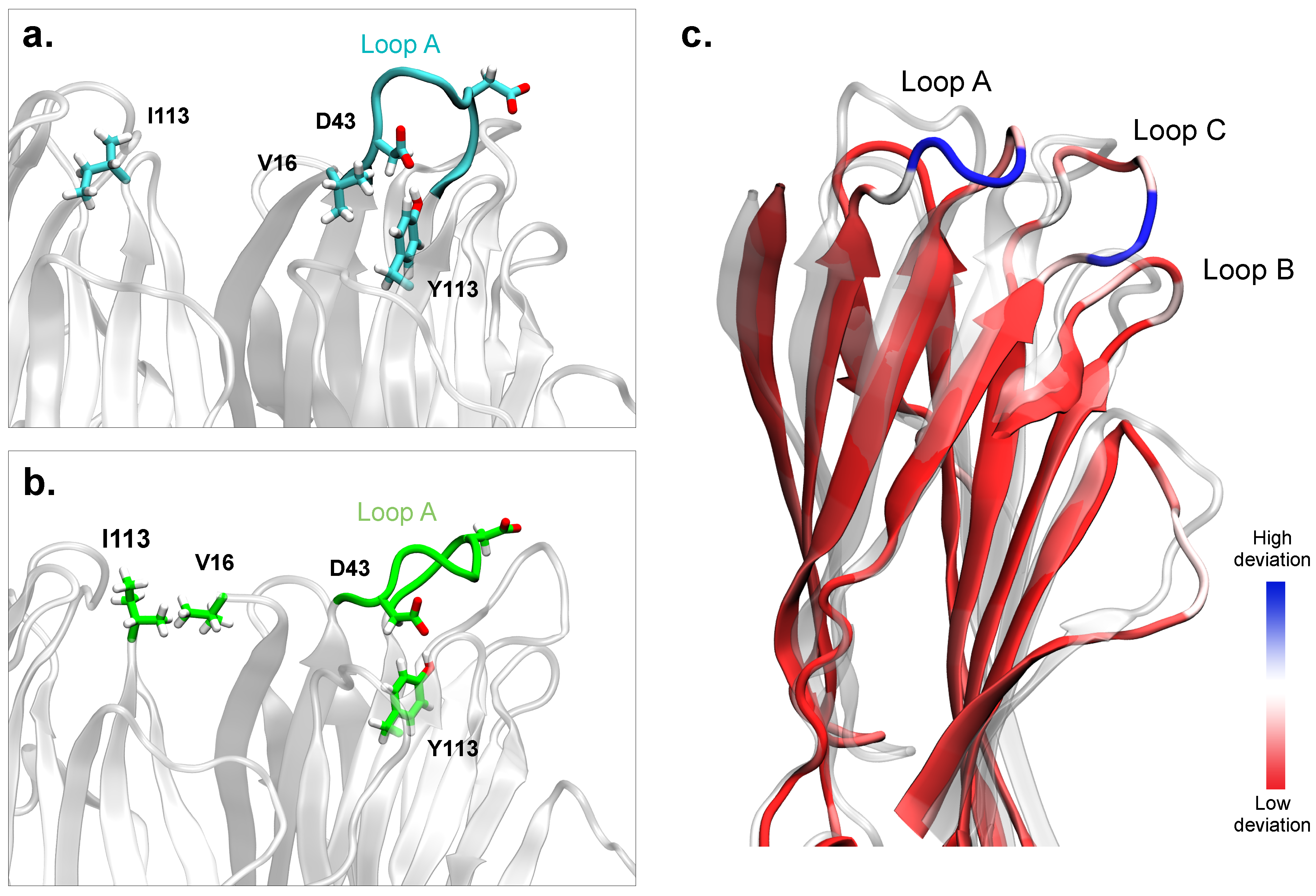
2.5. Simulations of the Modeled Dimers Reveal Conformational Changes of LukF in Absence of the N-Terminal
3. Discussion and Conclusions
4. Materials and Methods
4.1. System Setup
4.2. Protein–Protein Docking
4.3. Simulation Details
4.4. Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dal Peraro, M.; Van Der Goot, F.G. Pore-forming toxins: Ancient, but never really out of fashion. Nat. Rev. Microbiol. 2016, 14, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Tromp, A.T.; van Strijp, J.A. Studying staphylococcal leukocidins: A challenging endeavor. Front. Microb. 2020, 11, 611. [Google Scholar] [CrossRef] [PubMed]
- Ulhuq, F.R.; Mariano, G. Bacterial pore-forming toxins. Microbiology 2022, 168, 001154. [Google Scholar] [CrossRef] [PubMed]
- Willyard, C. The drug-resistant bacteria that pose the greatest health threats. Nat. News 2017, 543, 15. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Qiu, J.; Zhang, Y.; Lu, C.; Dai, X.; Wang, J.; Li, H.; Wang, X.; Tan, W.; Luo, M.; et al. Oroxylin A inhibits hemolysis via hindering the self-assembly of α-hemolysin heptameric transmembrane pore. PLoS Comput. Biol. 2013, 9, e1002869. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Wang, D.; Zhang, Y.; Dong, J.; Wang, J.; Niu, X. Molecular modeling reveals the novel inhibition mechanism and binding mode of three natural compounds to staphylococcal α-hemolysin. PLoS ONE 2013, 8, e80197. [Google Scholar] [CrossRef]
- Prevost, G.; Couppie, P.; Prevost, P.; Gayet, S.; Petiau, P.; Cribier, B.; Monteil, H.; Piemont, Y. Epidemiological data on Staphylococcus aureus strains producing synergohymenotropic toxins. J. Med. Microb. 1995, 42, 237–245. [Google Scholar] [CrossRef]
- Yamashita, D.; Sugawara, T.; Takeshita, M.; Kaneko, J.; Kamio, Y.; Tanaka, I.; Tanaka, Y.; Yao, M. Molecular basis of transmembrane beta-barrel formation of staphylococcal pore-forming toxins. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Olson, R.; Nariya, H.; Yokota, K.; Kamio, Y.; Gouaux, E. Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel. Nat. Struct. Biol. 1999, 6, 134–140. [Google Scholar]
- Yamashita, K.; Kawai, Y.; Tanaka, Y.; Hirano, N.; Kaneko, J.; Tomita, N.; Ohta, M.; Kamio, Y.; Yao, M.; Tanaka, I. Crystal structure of the octameric pore of staphylococcal γ-hemolysin reveals the β-barrel pore formation mechanism by two components. Proc. Natl. Acad. Sci. USA 2011, 108, 17314–17319. [Google Scholar] [CrossRef]
- Mishra, S.; Roy, A.K.; Dutta, S. Cryo-EM-based structural insights into supramolecular assemblies of γ-Hemolysin from Staphylococcus aureus reveal the pore formation mechanism. bioRxiv 2022. [Google Scholar] [CrossRef]
- Roblin, P.; Guillet, V.; Joubert, O.; Keller, D.; Erard, M.; Maveyraud, L.; Prévost, G.; Mourey, L. A covalent S-F heterodimer of leucotoxin reveals molecular plasticity of β-barrel pore-forming toxins. Proteins Struct. Funct. Bioinf. 2008, 71, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, N.; Kanagami, N.; Matsui, T.; Takeda, K.; Kaneko, J.; Shiraishi, Y.; Choe, C.A.; Uchikubo-Kamo, T.; Shirouzu, M.; Hashimoto, T.; et al. Chimeric mutants of staphylococcal hemolysin, which act as both one-component and two-component hemolysin, created by grafting the stem domain. FEBS J. 2022, 289, 3505–3520. [Google Scholar] [CrossRef] [PubMed]
- Vögele, M.; Bhaskara, R.M.; Mulvihill, E.; van Pee, K.; Yildiz, Ö.; Kühlbrandt, W.; Müller, D.J.; Hummer, G. Membrane perforation by the pore-forming toxin pneumolysin. Proc. Natl. Acad. Sci. USA 2019, 116, 13352–13357. [Google Scholar] [CrossRef]
- Ponmalar, I.I.; Cheerla, R.; Ayappa, K.G.; Basu, J.K. Correlated protein conformational states and membrane dynamics during attack by pore-forming toxins. Proc. Natl. Acad. Sci. USA 2019, 116, 12839–12844. [Google Scholar] [CrossRef]
- Sathyanarayana, P.; Visweswariah, S.S.; Ayappa, K.G. Mechanistic insights into pore Formation by an α-Pore forming toxin: Protein and lipid bilayer interactions of cytolysin A. Acc. Chem. Res. 2020, 54, 120–131. [Google Scholar] [CrossRef]
- Desikan, R.; Behera, A.; Maiti, P.K.; Ayappa, K.G. Using multiscale molecular dynamics simulations to obtain insights into pore forming toxin mechanisms. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 649, pp. 461–502. [Google Scholar]
- Herrera, A.; Kim, Y.; Chen, J.; Jedrzejczak, R.; Shukla, S.; Maltseva, N.; Joachimiak, G.; Welk, L.; Wiersum, G.; Jaroszewski, L.; et al. A genomic island of Vibrio cholerae encodes a three-component cytotoxin with monomer and protomer forms structurally similar to alpha-pore-forming toxins. J. Bacteriol. 2022, 204, e00555-21. [Google Scholar] [CrossRef]
- Jiao, F.; Dehez, F.; Ni, T.; Yu, X.; Dittman, J.S.; Gilbert, R.; Chipot, C.; Scheuring, S. Perforin-2 clockwise hand-over-hand pre-pore to pore transition mechanism. Nat. Commun. 2022, 13, 5039. [Google Scholar] [CrossRef]
- Russell, C.M.; Schaefer, K.G.; Dixson, A.; Gray, A.L.; Pyron, R.J.; Alves, D.S.; Moore, N.; Conley, E.A.; Schuck, R.J.; White, T.A.; et al. The Candida albicans virulence factor candidalysin polymerizes in solution to form membrane pores and damage epithelial cells. eLife 2022, 11, e75490. [Google Scholar] [CrossRef]
- Tarenzi, T.; Lattanzi, G.; Potestio, R. Membrane binding of pore-forming γ-hemolysin components studied at different lipid compositions. Biochim. Biophys. Acta Biomembr. 2022, 1864, 183970. [Google Scholar] [CrossRef]
- Potrich, C.; Bastiani, H.; Colin, D.; Huck, S.; Prevost, G.; Dalla Serra, M. The influence of membrane lipids in Staphylococcus aureus gamma-hemolysins pore formation. J. Membr. Biol. 2009, 227, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Laventie, B.J.; Potrich, C.; Atmanene, C.; Saleh, M.; Joubert, O.; Viero, G.; Bachmeyer, C.; Antonini, V.; Mancini, I.; Cianferani-Sanglier, S.; et al. p-Sulfonato-calix [n] arenes inhibit staphylococcal bicomponent leukotoxins by supramolecular interactions. Biochem. J. 2013, 450, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.T.; Higuchi, H.; Kamio, Y. Controlling pore assembly of staphylococcal γ-haemolysin by low temperature and by disulphide bond formation in double-cysteine LukF mutants. Mol. Microb. 2002, 45, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Monma, N.; Nguyen, V.T.; Kaneko, J.; Higuchi, H.; Kamio, Y. Essential residues, W177 and R198, of LukF for phosphatidylcholine-binding and pore-formation by staphylococcal γ-hemolysin on human erythrocyte membranes. J. Biochem. 2004, 136, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Yokota, K.; Kamio, Y. Tyrosine72 residue at the bottom of rim domain in LukF crucial for the sequential binding of the staphylococcal γ-hemolysin to human erythrocytes. Biosci. Biotechnol. Biochem. 2000, 64, 2744–2747. [Google Scholar] [CrossRef] [PubMed]
- Van Zundert, G.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.; Karaca, E.; Melquiond, A.; van Dijk, M.; De Vries, S.; Bonvin, A. The HADDOCK2. 2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Honorato, R.; Koukos, P.; Jiménez-García, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A. Structural biology in the clouds: The WeNMR-EOSC ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef]
- Grison, C.M.; Lambey, P.; Jeannot, S.; Del Nero, E.; Fontanel, S.; Peysson, F.; Heuninck, J.; Sounier, R.; Durroux, T.; Leyrat, C.; et al. Molecular insights into mechanisms of GPCR hijacking by Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2021, 118, e2108856118. [Google Scholar] [CrossRef]
- Takeda, K.; Tanaka, Y.; Kaneko, J. The N-terminal amino-latch region of Hlg2 component of staphylococcal bi-component γ-haemolysin is dispensable for prestem release to form β-barrel pores. J. Biochem. 2020, 168, 349–354. [Google Scholar] [CrossRef]
- Kaneko, J.; Mascarenas, M.A.L.; Huda, M.N.; Tomita, T.; Kamio, Y. An N-terminal region of LukF of staphylococcal leukocidin/γ-hemolysin crucial for the biological activity of the toxin. Biosci. Biotechnol. Biochem. 1998, 62, 1465–1467. [Google Scholar] [CrossRef]
- Lambey, P.; Otun, O.; Cong, X.; Hoh, F.; Brunel, L.; Verdié, P.; Grison, C.M.; Peysson, F.; Jeannot, S.; Durroux, T.; et al. Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins. eLife 2022, 11, e72555. [Google Scholar] [CrossRef] [PubMed]
- Choorit, W.; Kaneko, J.; Muramoto, K.; Kamio, Y. Existence of a new protein component with the same function as the LukF component of leukocidin or γ-hemolysin and its gene in Staphylococcus aureus P83. FEBS Lett. 1995, 357, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Hirano, N.; Kaneko, J.; Kamio, Y.; Yao, M.; Tanaka, I. 2-Methyl-2, 4-pentanediol induces spontaneous assembly of staphylococcal α-hemolysin into heptameric pore structure. Protein Sci. 2011, 20, 448–456. [Google Scholar] [CrossRef]
- Sugawara-Tomita, N.; Tomita, T.; Kamio, Y. Stochastic assembly of two-component staphylococcal γ-hemolysin into heteroheptameric transmembrane pores with alternate subunit arrangements in ratios of 3:4 and 4:3. J. Bacteriol. 2002, 184, 4747–4756. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Tanaka, Y.; Abe, N.; Kaneko, J. Intermolecular ionic interactions serve as a possible switch for stem release in the staphylococcal bi-component toxin for β-barrel pore assembly. Toxicon 2018, 155, 43–48. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protocol. Bioinf. 2016, 54, 5–6. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; De Groot, B.L.; Grubmüller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Meth. 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D., Jr.; Pastor, R.W. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [PubMed]
- Nosé, S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984, 52, 255–268. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Tiberti, M.; Invernizzi, G.; Lambrughi, M.; Inbar, Y.; Schreiber, G.; Papaleo, E. PyInteraph: A framework for the analysis of interaction networks in structural ensembles of proteins. J. Chem. Inf. Model. 2014, 54, 1537–1551. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Corey, R.A.; Ansell, T.B.; Cassidy, C.K.; Horrell, M.R.; Duncan, A.L.; Stansfeld, P.J.; Sansom, M.S. PyLipID: A Python package for analysis of protein–lipid interactions from molecular dynamics simulations. J. Chem. Theory Comput. 2022, 18, 1188–1201. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. Vmd: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 27–38. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paternoster, C.; Tarenzi, T.; Potestio, R.; Lattanzi, G. Gamma-Hemolysin Components: Computational Strategies for LukF-Hlg2 Dimer Reconstruction on a Model Membrane. Int. J. Mol. Sci. 2023, 24, 7113. https://doi.org/10.3390/ijms24087113
Paternoster C, Tarenzi T, Potestio R, Lattanzi G. Gamma-Hemolysin Components: Computational Strategies for LukF-Hlg2 Dimer Reconstruction on a Model Membrane. International Journal of Molecular Sciences. 2023; 24(8):7113. https://doi.org/10.3390/ijms24087113
Chicago/Turabian StylePaternoster, Costanza, Thomas Tarenzi, Raffaello Potestio, and Gianluca Lattanzi. 2023. "Gamma-Hemolysin Components: Computational Strategies for LukF-Hlg2 Dimer Reconstruction on a Model Membrane" International Journal of Molecular Sciences 24, no. 8: 7113. https://doi.org/10.3390/ijms24087113
APA StylePaternoster, C., Tarenzi, T., Potestio, R., & Lattanzi, G. (2023). Gamma-Hemolysin Components: Computational Strategies for LukF-Hlg2 Dimer Reconstruction on a Model Membrane. International Journal of Molecular Sciences, 24(8), 7113. https://doi.org/10.3390/ijms24087113





