Genome-Wide Identification of LBD Genes in Foxtail Millet (Setaria italica) and Functional Characterization of SiLBD21
Abstract
1. Introduction
2. Results
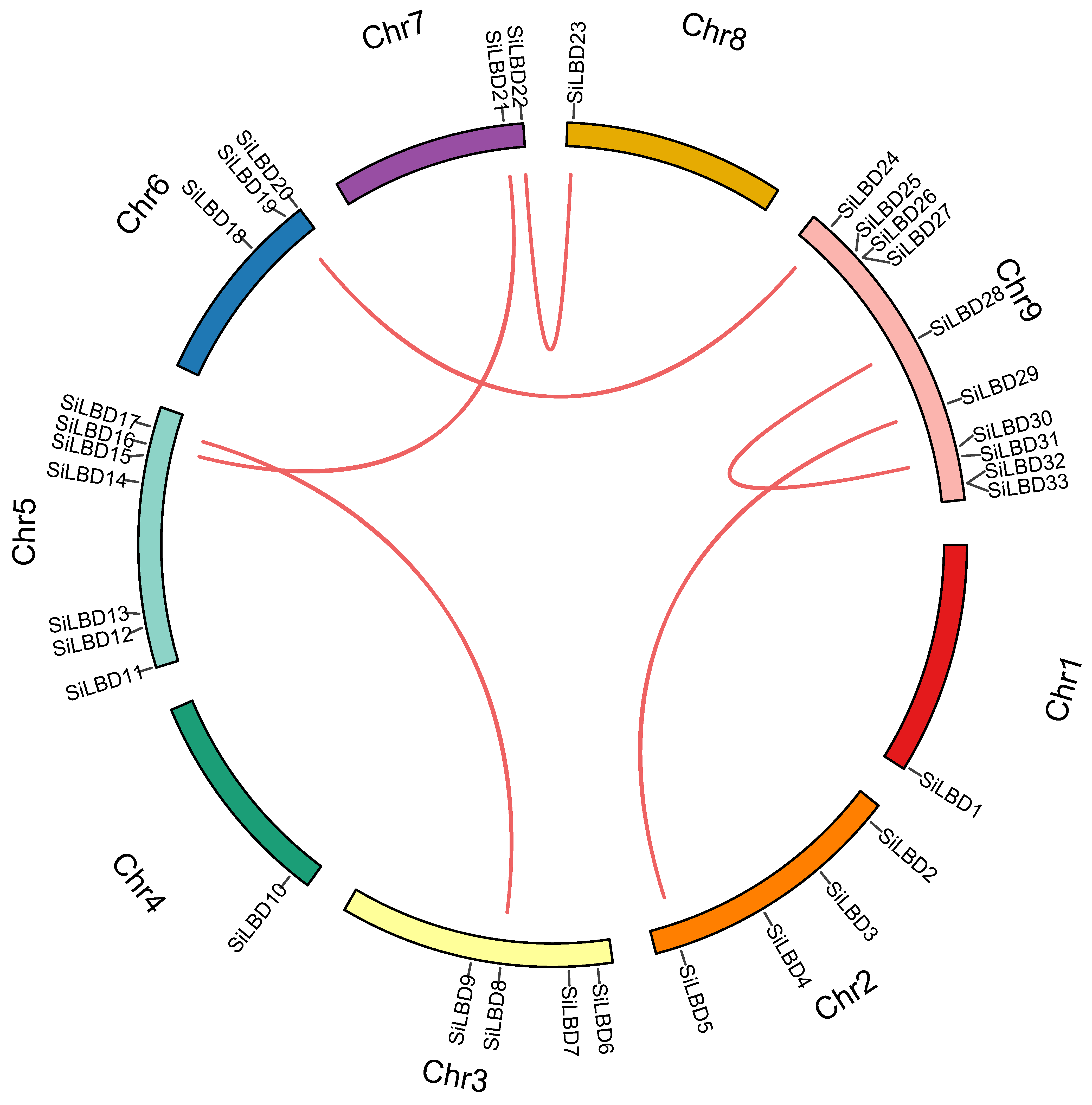
2.1. LBD TFs in Foxtail Millet
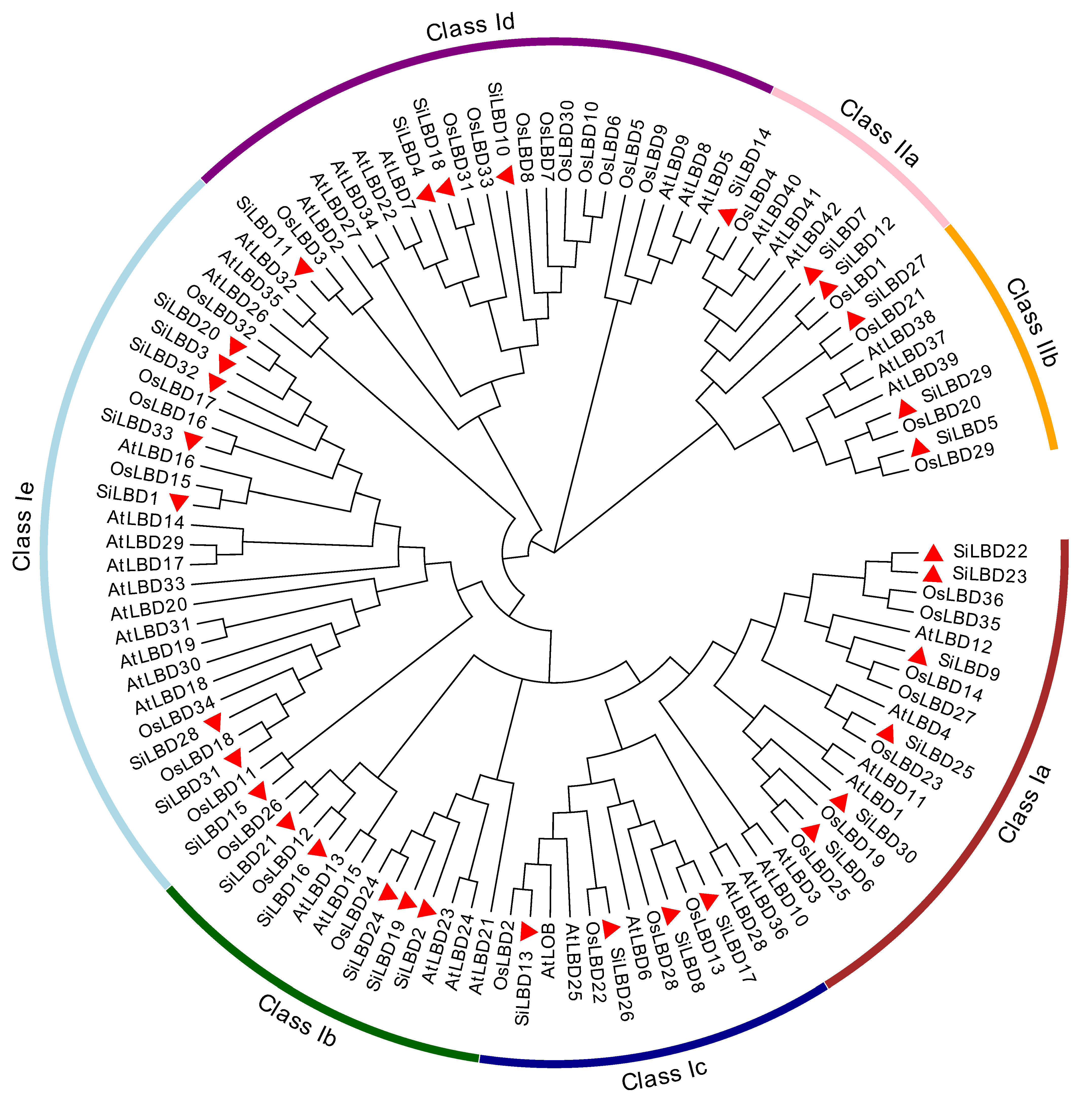
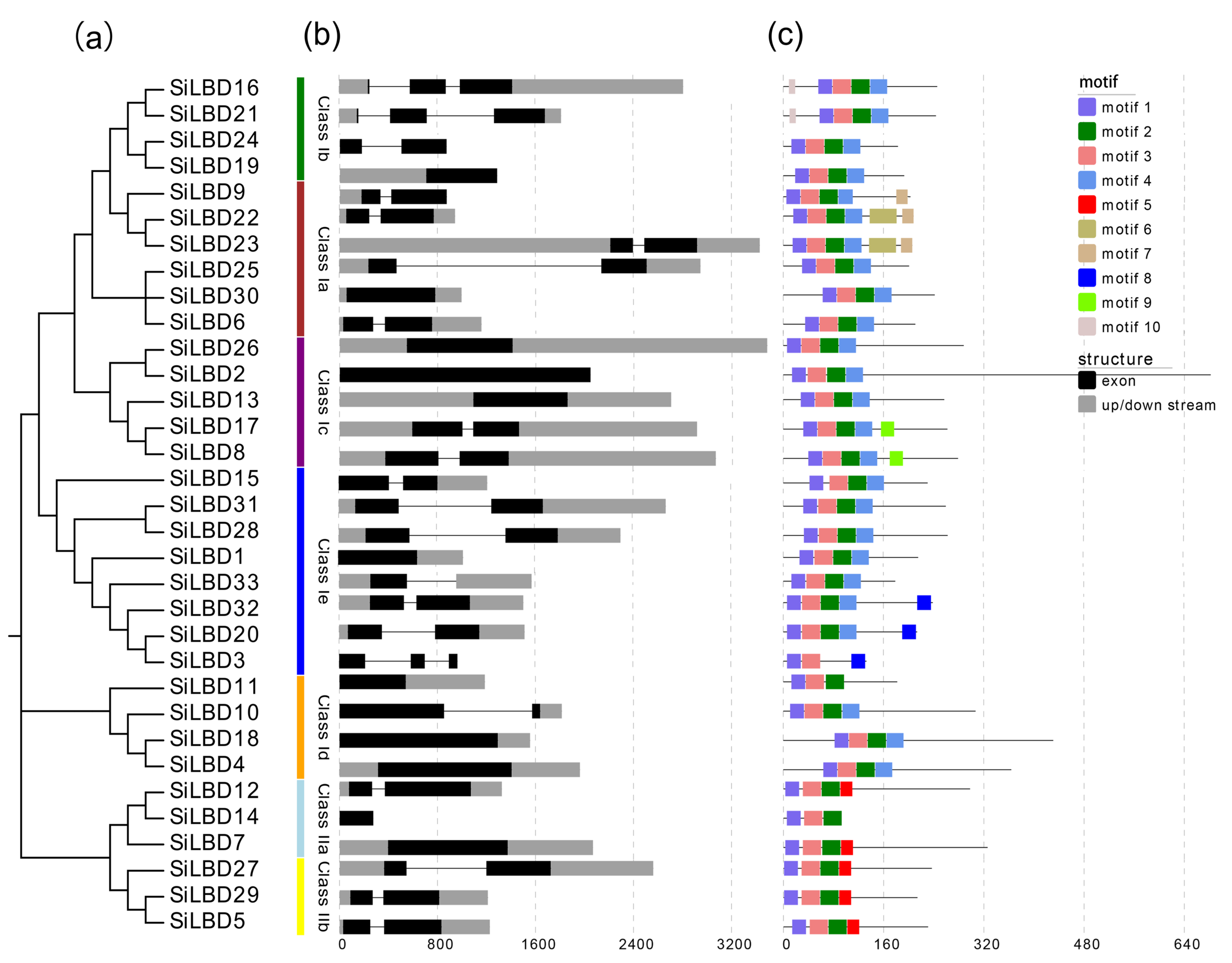
2.2. Phylogenetic Tree, Gene Structures, and Conserved Motifs
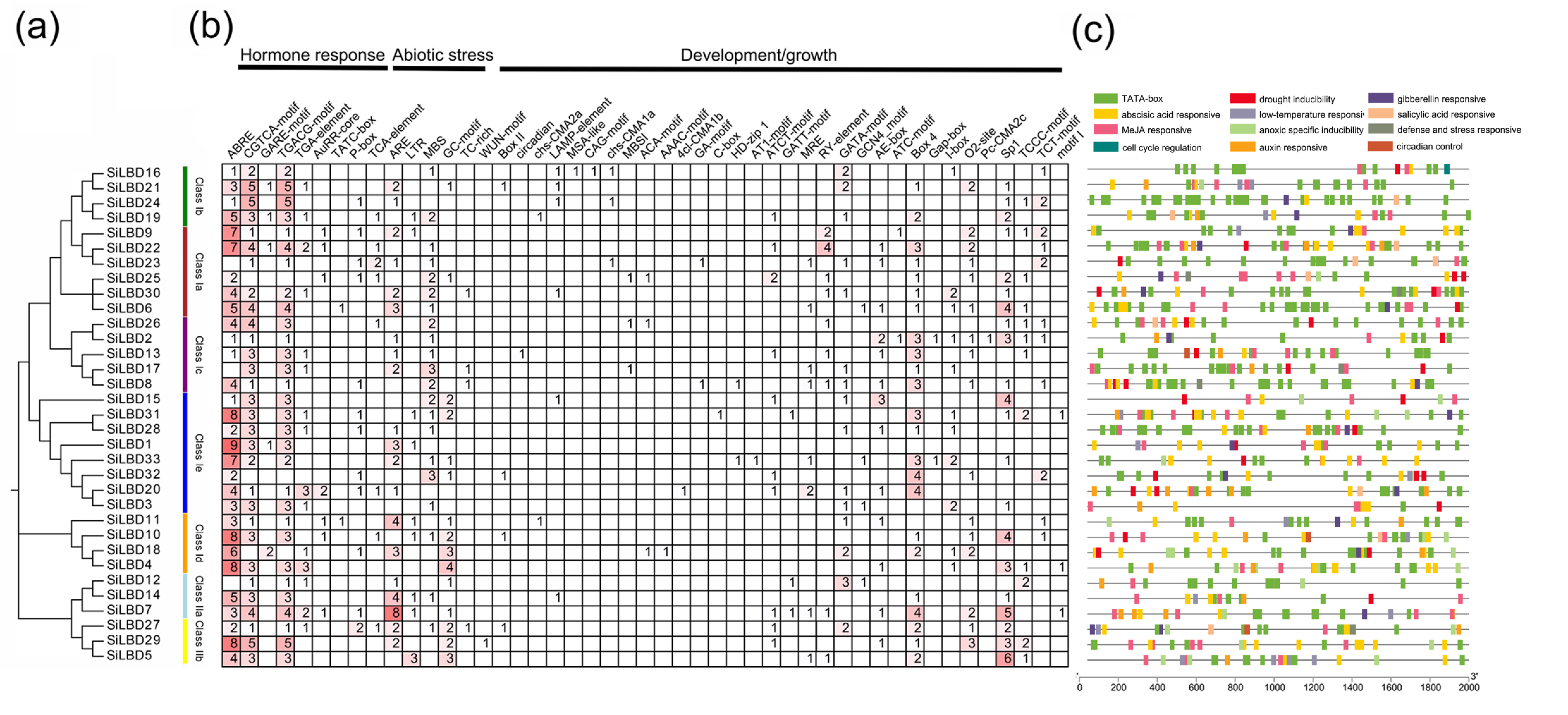
2.3. Cis-Elements
2.4. Expression Profiles
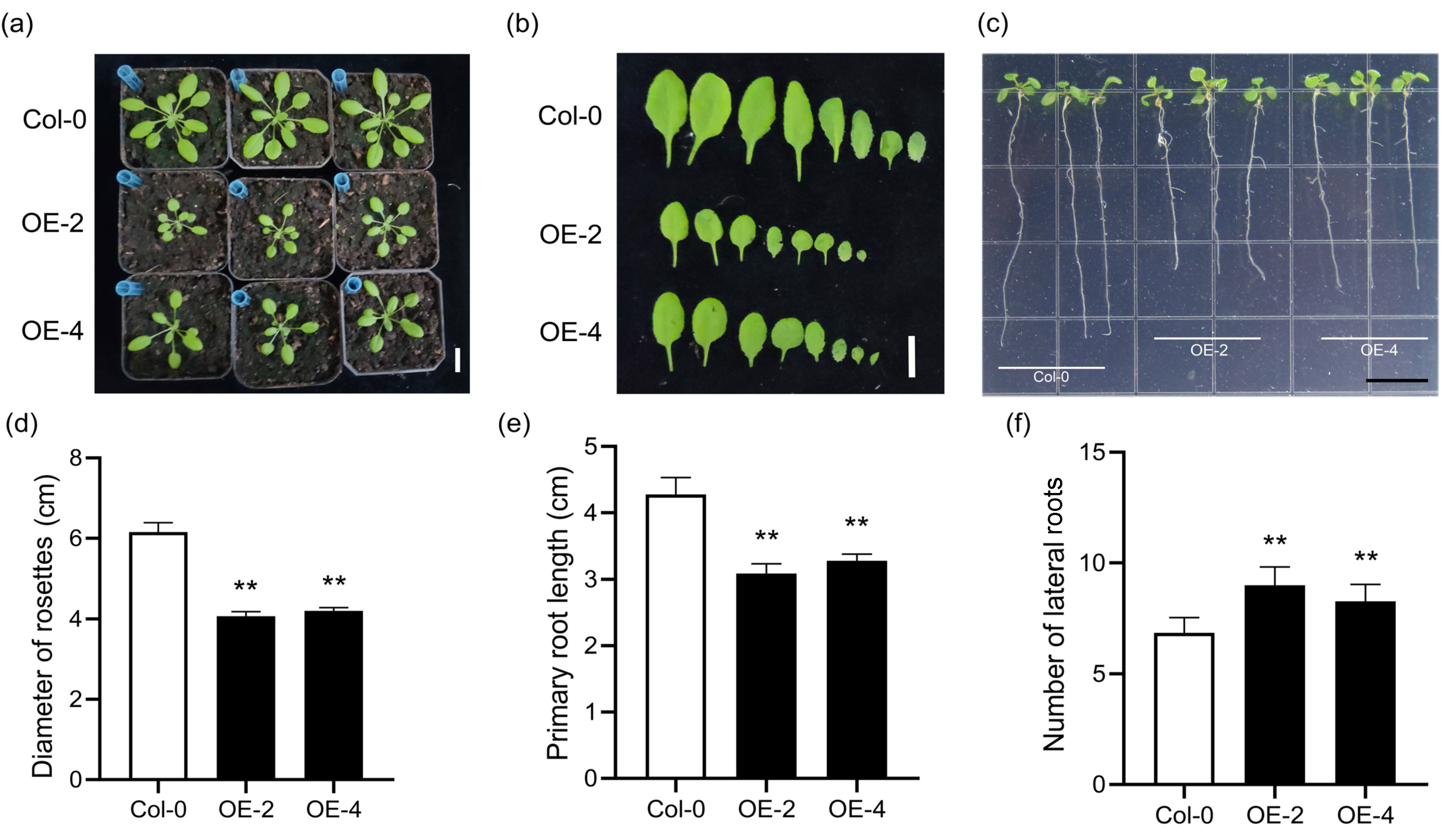
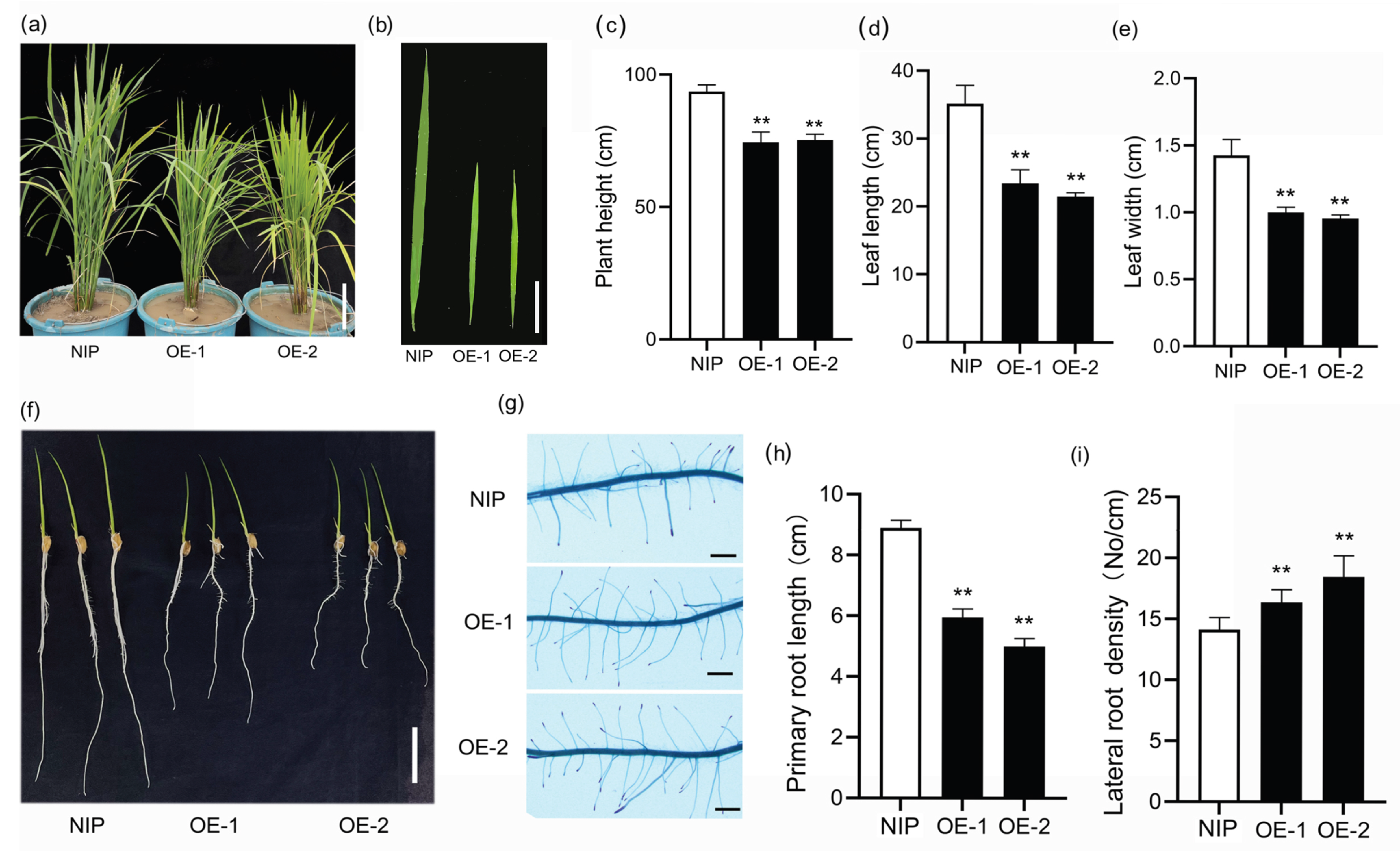
2.5. Phenotypes of Transgenic Arabidopsis and Rice Over-Expressing SiLBD21
3. Discussion
3.1. Monocot Plants Had Fewer LBD Genes
3.2. LBD TFs Display Extensive Functions
3.3. LBD TFs Play Important Roles in Root Development
4. Materials and Methods
4.1. Genome-Wide Identification of SiLBD Genes
4.2. Physicochemical Properties, Gene Duplication, Chromosome Distribution, and Phylogenetic Analyses
4.3. Gene Structure, Conserved Motif, and Cis-Element Analyses
4.4. Plant Growth Conditions and Treatments
4.5. RNA Extraction and qRT-PCR Analysis
4.6. Generation of Transgenic Plants
4.7. Phenotype Analysis of Transgenic Plants
4.8. Data Processing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shuai, B.; Reynaga-Peña, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Luo, F.; Hochholdinger, F. Lob domain proteins: Beyond lateral organ boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, X.; Wu, P. Comparison and evolution analysis of two rice subspecies lateral organ boundaries domain gene family and their evolutionary characterization from arabidopsis. Mol. Phylogenetics Evol. 2006, 39, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Chanderbali, A.S.; He, F.; Soltis, P.S.; Soltis, D.E. Out of the water: Origin and diversification of the lbd gene family. Mol. Biol. Evol. 2015, 32, 1996–2000. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Iwakawa, H.; Machida, Y.; Machida, C. Characterization of genes in the asymmetric leaves2/lateral organ boundaries (as2/lob) family in arabidopsis thaliana, and functional and molecular comparisons between as2 and other family members. Plant J. Cell Mol. Biol. 2009, 58, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C.; et al. The asymmetric leaves2 gene of arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef]
- Luo, L.; Ando, S.; Sakamoto, Y.; Suzuki, T.; Takahashi, H.; Ishibashi, N.; Kojima, S.; Kurihara, D.; Higashiyama, T.; Yamamoto, K.T.; et al. The formation of perinucleolar bodies is important for normal leaf development and requires the zinc-finger DNA-binding motif in arabidopsis asymmetric leaves2. Plant J. 2020, 101, 1118–1134. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, M.J.; Park, M.Y.; Han, K.H.; Kim, J. The conserved proline residue in the lob domain of lbd18 is critical for DNA-binding and biological function. Mol. Plant 2013, 6, 1722–1725. [Google Scholar] [CrossRef]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of lob domain proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef]
- Yang, H.; Shi, G.; Du, H.; Wang, H.; Zhang, Z.; Hu, D.; Wang, J.; Huang, F.; Yu, D. Genome-wide analysis of soybean lateral organ boundaries domain-containing genes: A functional investigation of gmlbd12. Plant Genome 2017, 10, plantgenome2016.07.0058. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Zhang, S.Z.; Zheng, C.C. Genomewide analysis of lateral organ boundaries domain gene family in zea mays. J. Genet. 2014, 93, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, R.; Cheng, Y.; Lei, P.; Song, W.; Zheng, W.; Nie, X. Genome-wide identification, evolution, and expression analysis of lbd transcription factor family in bread wheat (Triticum aestivum L.). Front. Plant Sci. 2021, 12, 721253. [Google Scholar] [CrossRef] [PubMed]
- Gombos, M.; Zombori, Z.; Szécsényi, M.; Sándor, G.; Kovács, H.; Györgyey, J. Characterization of the lbd gene family in brachypodium: A phylogenetic and transcriptional study. Plant Cell Rep. 2017, 36, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Cao, M.; Chen, X.; Ye, M.; Zhao, P.; Nan, Y.; Li, W.; Zhang, C.; Kong, L.; Kong, N.; et al. Genome-wide analysis of the lateral organ boundaries domain (lbd) gene family in solanum tuberosum. Int. J. Mol. Sci. 2019, 20, 5360. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Zeng, L.; Chen, X.; Rong, H.; Wu, J.; Batley, J. Genome-wide analysis of the lateral organ boundaries domain gene family in brassica napus. Genes 2020, 11, 280. [Google Scholar] [CrossRef]
- Hochholdinger, F.; Yu, P.; Marcon, C. Genetic control of root system development in maize. Trends Plant Sci. 2018, 23, 79–88. [Google Scholar] [CrossRef]
- Goh, T.; Toyokura, K.; Yamaguchi, N.; Okamoto, Y.; Uehara, T.; Kaneko, S.; Takebayashi, Y.; Kasahara, H.; Ikeyama, Y.; Okushima, Y.; et al. Lateral root initiation requires the sequential induction of transcription factors lbd16 and puchi in arabidopsis thaliana. New Phytol. 2019, 224, 749–760. [Google Scholar] [CrossRef]
- Berckmans, B.; Vassileva, V.; Schmid, S.P.; Maes, S.; Parizot, B.; Naramoto, S.; Magyar, Z.; Alvim Kamei, C.L.; Koncz, C.; Bögre, L.; et al. Auxin-dependent cell cycle reactivation through transcriptional regulation of arabidopsis e2fa by lateral organ boundary proteins. Plant Cell 2011, 23, 3671–3683. [Google Scholar] [CrossRef]
- Li, A.; Zhang, Y.; Wu, X.; Tang, W.; Wu, R.; Dai, Z.; Liu, G.; Zhang, H.; Wu, C.; Chen, G.; et al. Dh1, a lob domain-like protein required for glume formation in rice. Plant Mol. Biol. 2008, 66, 491–502. [Google Scholar] [CrossRef]
- Liu, H.; Wang, S.; Yu, X.; Yu, J.; He, X.; Zhang, S.; Shou, H.; Wu, P. Arl1, a lob-domain protein required for adventitious root formation in rice. Plant J. Cell Mol. Biol. 2005, 43, 47–56. [Google Scholar] [CrossRef]
- Li, C.; Wang, J.; Li, L.; Li, J.; Zhuang, M.; Li, B.; Li, Q.; Huang, J.; Du, Y.; Wang, J.; et al. Tamor is essential for root initiation and improvement of root system architecture in wheat. Plant Biotechnol. J. 2021, 20, 862–875. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Niu, C.; Li, K.; Fan, L.; Liu, Z.; Li, S.; Ma, D.; Tahir, M.M.; Xing, L.; Zhao, C.; et al. Cytokinin-responsive mdtcp17 interacts with mdwox11 to repress adventitious root primordium formation in apple rootstocks. Plant Cell 2022, 35, 1202–1221. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.; Wu, B.; Ma, S.; Zhang, J.; Liu, X.; Wang, H.; Zhang, J.; Gao, R.; Jiang, H.; Jia, C. Genome-wide characterization of lbd transcription factors in switchgrass (Panicum virgatum L.) and the involvement of pvlbd12 in salt tolerance. Plant Cell Rep. 2023. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; He, W.; Wang, L.; Zhang, X.; Wang, K.; Xiang, Y. Phelbd29, an lbd transcription factor from moso bamboo, causes leaf curvature and enhances tolerance to drought stress in transgenic arabidopsis. J. Plant Physiol. 2023, 280, 153865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Z.; Ma, B.; Hou, Q.; Wan, X. Phylogeny and functions of lob domain proteins in plants. Int. J. Mol. Sci. 2020, 21, 2278. [Google Scholar] [CrossRef]
- Albinsky, D.; Kusano, M.; Higuchi, M.; Hayashi, N.; Kobayashi, M.; Fukushima, A.; Mori, M.; Ichikawa, T.; Matsui, K.; Kuroda, H.; et al. Metabolomic screening applied to rice fox arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol. Plant 2010, 3, 125–142. [Google Scholar] [CrossRef]
- Li, C.; Zhu, S.; Zhang, H.; Chen, L.; Cai, M.; Wang, J.; Chai, J.; Wu, F.; Cheng, Z.; Guo, X.; et al. Oslbd37 and oslbd38, two class ii type lbd proteins, are involved in the regulation of heading date by controlling the expression of ehd1 in rice. Biochem. Biophys. Res. Commun. 2017, 486, 720–725. [Google Scholar] [CrossRef]
- Ariel, F.D.; Diet, A.; Crespi, M.; Chan, R.L. The lob-like transcription factor mt lbd1 controls medicago truncatula root architecture under salt stress. Plant Signal. Behav. 2010, 5, 1666–1668. [Google Scholar] [CrossRef]
- Teng, R.M.; Yang, N.; Li, J.W.; Liu, C.F.; Chen, Y.; Li, T.; Wang, Y.H.; Xiong, A.S.; Zhuang, J. Isolation and characterization of an lbd transcription factor cslbd39 from tea plant (camellia sinensis) and its roles in modulating nitrate content by regulating nitrate-metabolism-related genes. Int. J. Mol. Sci. 2022, 23, 9294. [Google Scholar] [CrossRef]
- Jiang, X.; Cui, H.; Wang, Z. Genome-wide analysis of the lateral organ boundaries domain (lbd) members in alfalfa and the involvement of mslbd48 in nitrogen assimilation. Int. J. Mol. Sci. 2023, 24, 4644. [Google Scholar] [CrossRef]
- Peng, R.; Zhang, B. Foxtail millet: A new model for c4 plants. Trends Plant Sci. 2021, 26, 199–201. [Google Scholar] [CrossRef] [PubMed]
- Muthamilarasan, M.; Prasad, M. Small millets for enduring food security amidst pandemics. Trends Plant Sci. 2021, 26, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, A.G.; Baldi, P.; Chauvin, Y.; Brunak, S. The biology of eukaryotic promoter prediction—a review. Comput. Chem. 1999, 23, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Greenblatt, J. Riding high on the TATA box. Nature 1992, 360, 16–17. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K. The tata box binding protein. Curr. Opin. Struct. Biol. 1996, 6, 69–75. [Google Scholar] [CrossRef]
- Mishal, R.; Luna-Arias, J.P. Role of the TATA-box binding protein (TBP) and associated family members in transcription regulation. Gene 2022, 833, 146581. [Google Scholar] [CrossRef]
- Sutoh, K.; Yamauchi, D. Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J. Cell Mol. Biol. 2003, 34, 635–645. [Google Scholar] [CrossRef]
- Nelson, C.C.; Hendy, S.C.; Romaniuk, P.J. Relationship between p-box amino acid sequence and DNA binding specificity of the thyroid hormone receptor. The effects of half-site sequence in everted repeats. J. Biol. Chem. 1995, 270, 16981–16987. [Google Scholar] [CrossRef]
- Liu, Z.B.; Ulmasov, T.; Shi, X.; Hagen, G.; Guilfoyle, T.J. Soybean gh3 promoter contains multiple auxin-inducible elements. Plant Cell 1994, 6, 645–657. [Google Scholar]
- Ballas, N.; Wong, L.M.; Theologis, A. Identification of the auxin-responsive element, auxre, in the primary indoleacetic acid-inducible gene, ps-iaa4/5, of pea (Pisum sativum). J. Mol. Biol. 1993, 233, 580–596. [Google Scholar] [CrossRef]
- Zheng, Z.; Yang, X.; Fu, Y.; Zhu, L.; Wei, H.; Lin, X. Overexpression of pvpin1, a bamboo homolog of pin1-type parvulin 1, delays flowering time in transgenic arabidopsis and rice. Front. Plant Sci. 2017, 8, 1526. [Google Scholar] [CrossRef] [PubMed]
- Goldsbrough, A.P.; Albrecht, H.; Stratford, R. Salicylic acid-inducible binding of a tobacco nuclear protein to a 10 bp sequence which is highly conserved amongst stress-inducible genes. Plant J. Cell Mol. Biol. 1993, 3, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Obayashi, T.; Masuda, T. Role of the g-box element in regulation of chlorophyll biosynthesis in arabidopsis roots. Plant Signal. Behav. 2012, 7, 922–926. [Google Scholar] [CrossRef] [PubMed]
- Carlini, L.E.; Ketudat, M.; Parsons, R.L.; Prabhakar, S.; Schmidt, R.J.; Guiltinan, M.J. The maize embp-1 orthologue differentially regulates opaque2-dependent gene expression in yeast and cultured maize endosperm cells. Plant Mol. Biol. 1999, 41, 339–349. [Google Scholar] [CrossRef]
- Dunn, M.A.; White, A.J.; Vural, S.; Hughes, M.A. Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.). Plant Mol. Biol. 1998, 38, 551–564. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Kazan, K.; Manners, J.M. Lateral organ boundaries domain transcription factors: New roles in plant defense. Plant Signal. Behav. 2012, 7, 1702–1704. [Google Scholar] [CrossRef]
- Zhang, C.; Zhu, P.; Zhang, M.; Huang, Z.; Hippolyte, A.R.; Hou, Y.; Lou, X.; Ji, K. Identification, classification and characterization of lbd transcription factor family genes in pinus massoniana. Int. J. Mol. Sci. 2022, 23, 13215. [Google Scholar] [CrossRef]
- Tian, Y.; Han, X.; Qu, Y.; Zhang, Y.; Rong, H.; Wu, K.; Xu, L.A. Genome-wide identification of the ginkgo (ginkgo biloba l.) lbd transcription factor gene and characterization of its expression. Int. J. Mol. Sci. 2022, 23, 5474. [Google Scholar] [CrossRef]
- Liang, J.; Hou, Z.; Liao, J.; Qin, Y.; Wang, L.; Wang, X.; Su, W.; Cai, Z.; Fang, Y.; Aslam, M.; et al. Genome-wide identification and expression analysis of lbd transcription factor genes in passion fruit (passiflora edulis). Int. J. Mol. Sci. 2022, 23, 4700. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the lbd family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef]
- Goff, S.A.; Ricke, D.; Lan, T.H.; Presting, G.; Wang, R.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. Japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The arabidopsis information resource (tair): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Chung, C.Y.-L.; Li, M.-W.; Wong, F.-L.; Wang, X.; Liu, A.; Wang, Z.; Leung, A.K.-Y.; Wong, T.-H.; Tong, S.-W.; et al. A reference-grade wild soybean genome. Nat. Commun. 2019, 10, 1216. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Plant genetics. Early allopolyploid evolution in the post-neolithic brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, gossypium hirsutum and gossypium barbadense. Nat. Genet. 2019, 51, 224–229. [Google Scholar] [CrossRef]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome sequence of cultivated upland cotton (gossypium hirsutum tm-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The b73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar]
- Vogel, J.P.; Garvin, D.F.; Mockler, T.C.; Schmutz, J.; Rokhsar, D.; Bevan, M.W.; Barry, K.; Lucas, S.; Harmon-Smith, M.; Lail, K.; et al. Genome sequencing and analysis of the model grass brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar]
- Taylor, J.S.; Raes, J. Duplication and divergence: The evolution of new genes and old ideas. Annu. Rev. Genet. 2004, 38, 615–643. [Google Scholar] [CrossRef]
- Bell, E.M.; Lin, W.C.; Husbands, A.Y.; Yu, L.; Jaganatha, V.; Jablonska, B.; Mangeon, A.; Neff, M.M.; Girke, T.; Springer, P.S. Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA 2012, 109, 21146–21151. [Google Scholar] [CrossRef] [PubMed]
- Naito, T.; Yamashino, T.; Kiba, T.; Koizumi, N.; Kojima, M.; Sakakibara, H.; Mizuno, T. A link between cytokinin and asl9 (asymmetric leaves 2 like 9) that belongs to the as2/lob (lateral organ boundaries) family genes in arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2007, 71, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Scheible, W.R.; Morcuende, R.; Czechowski, T.; Fritz, C.; Osuna, D.; Palacios-Rojas, N.; Schindelasch, D.; Thimm, O.; Udvardi, M.K.; Stitt, M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of arabidopsis in response to nitrogen. Plant Physiol. 2004, 136, 2483–2499. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.J.; Tao, Y.; Zhao, X.Y.; Zhang, X.S. Wheat taas2, a member of lob family, affects the adaxial–abaxial polarity of leaves in transgenic arabidopsis. Plant Sci. 2007, 172, 181–188. [Google Scholar] [CrossRef]
- Lin, W.C.; Shuai, B.; Springer, P.S. The arabidopsis lateral organ boundaries-domain gene asymmetric leaves2 functions in the repression of knox gene expression and in adaxial-abaxial patterning. Plant Cell 2003, 15, 2241–2252. [Google Scholar] [CrossRef]
- Guo, M.; Thomas, J.; Collins, G.; Timmermans, M.C. Direct repression of knox loci by the asymmetric leaves1 complex of arabidopsis. Plant Cell 2008, 20, 48–58. [Google Scholar] [CrossRef]
- Feng, X.; Xiong, J.; Zhang, W.; Guan, H.; Zheng, D.; Xiong, H.; Jia, L.; Hu, Y.; Zhou, H.; Wen, Y.; et al. Zmlbd5, a class-ii lbd gene, negatively regulates drought tolerance by impairing abscisic acid synthesis. Plant J. 2022, 112, 6. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, J.; Jia, H.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B.; White, F.F.; Wang, N.; Jones, J.B. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, E521–E529. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, N.Y.; Lee, D.J.; Kim, J. Lbd18/asl20 regulates lateral root formation in combination with lbd16/asl18 downstream of arf7 and arf19 in arabidopsis. Plant Physiol. 2009, 151, 1377–1389. [Google Scholar] [CrossRef]
- Jeon, B.W.; Kim, J. Role of lbd14 during aba-mediated control of root system architecture in arabidopsis. Plant Signal. Behav. 2018, 13, e1507405. [Google Scholar]
- Inukai, Y.; Sakamoto, T.; Ueguchi-Tanaka, M.; Shibata, Y.; Gomi, K.; Umemura, I.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M. Crown rootless1, which is essential for crown root formation in rice, is a target of an auxin response factor in auxin signaling. Plant Cell 2005, 17, 1387–1396. [Google Scholar] [CrossRef] [PubMed]
- Taramino, G.; Sauer, M.; Stauffer, J.L., Jr.; Multani, D.; Niu, X.; Sakai, H.; Hochholdinger, F. The maize (Zea mays L.) rtcs gene encodes a lob domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. Cell Mol. Biol. 2007, 50, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, D.; Li, Q.; Mao, X.; Li, A.; Wang, J.; Chang, X.; Jing, R. Overexpression of wheat gene tamor improves root system architecture and grain yield in oryza sativa. J. Exp. Bot. 2016, 67, 4155–4167. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zheng, C.; Liu, R.; Song, A.; Zhang, Z.; Xin, J.; Jiang, J.; Chen, S.; Zhang, F.; Fang, W.; et al. Chrysanthemum transcription factor cmlbd1 direct lateral root formation in arabidopsis thaliana. Sci. Rep. 2016, 6, 20009. [Google Scholar] [CrossRef]
- Cho, C.; Jeon, E.; Pandey, S.K.; Ha, S.H.; Kim, J. Lbd13 positively regulates lateral root formation in arabidopsis. Planta 2019, 249, 1251–1258. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C. Expasy, the swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mploc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Gu, Z.; Cavalcanti, A.; Chen, F.C.; Bouman, P.; Li, W.H. Extent of gene duplication in the genomes of drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. [gsds: A gene structure display server]. Yi Chuan = Hereditas 2007, 29, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012, 40, W569–W572. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Sallaud, C.; Meynard, D.; van Boxtel, J.; Gay, C.; Bès, M.; Brizard, J.P.; Larmande, P.; Ortega, D.; Raynal, M.; Portefaix, M.; et al. Highly efficient production and characterization of t-DNA plants for rice (Oryza sativa L.) functional genomics. TAG. Theor. Appl. Genet. Theor. Und Angew. Genet. 2003, 106, 1396–1408. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Wei, Y.; Wang, Y.; Tan, B.; Chen, S.; Li, H. Genome-Wide Identification of LBD Genes in Foxtail Millet (Setaria italica) and Functional Characterization of SiLBD21. Int. J. Mol. Sci. 2023, 24, 7110. https://doi.org/10.3390/ijms24087110
Li K, Wei Y, Wang Y, Tan B, Chen S, Li H. Genome-Wide Identification of LBD Genes in Foxtail Millet (Setaria italica) and Functional Characterization of SiLBD21. International Journal of Molecular Sciences. 2023; 24(8):7110. https://doi.org/10.3390/ijms24087110
Chicago/Turabian StyleLi, Kunjie, Yaning Wei, Yimin Wang, Bin Tan, Shoukun Chen, and Haifeng Li. 2023. "Genome-Wide Identification of LBD Genes in Foxtail Millet (Setaria italica) and Functional Characterization of SiLBD21" International Journal of Molecular Sciences 24, no. 8: 7110. https://doi.org/10.3390/ijms24087110
APA StyleLi, K., Wei, Y., Wang, Y., Tan, B., Chen, S., & Li, H. (2023). Genome-Wide Identification of LBD Genes in Foxtail Millet (Setaria italica) and Functional Characterization of SiLBD21. International Journal of Molecular Sciences, 24(8), 7110. https://doi.org/10.3390/ijms24087110




