A Biochemical Deconstruction-Based Strategy to Assist the Characterization of Bacterial Electric Conductive Filaments
Abstract
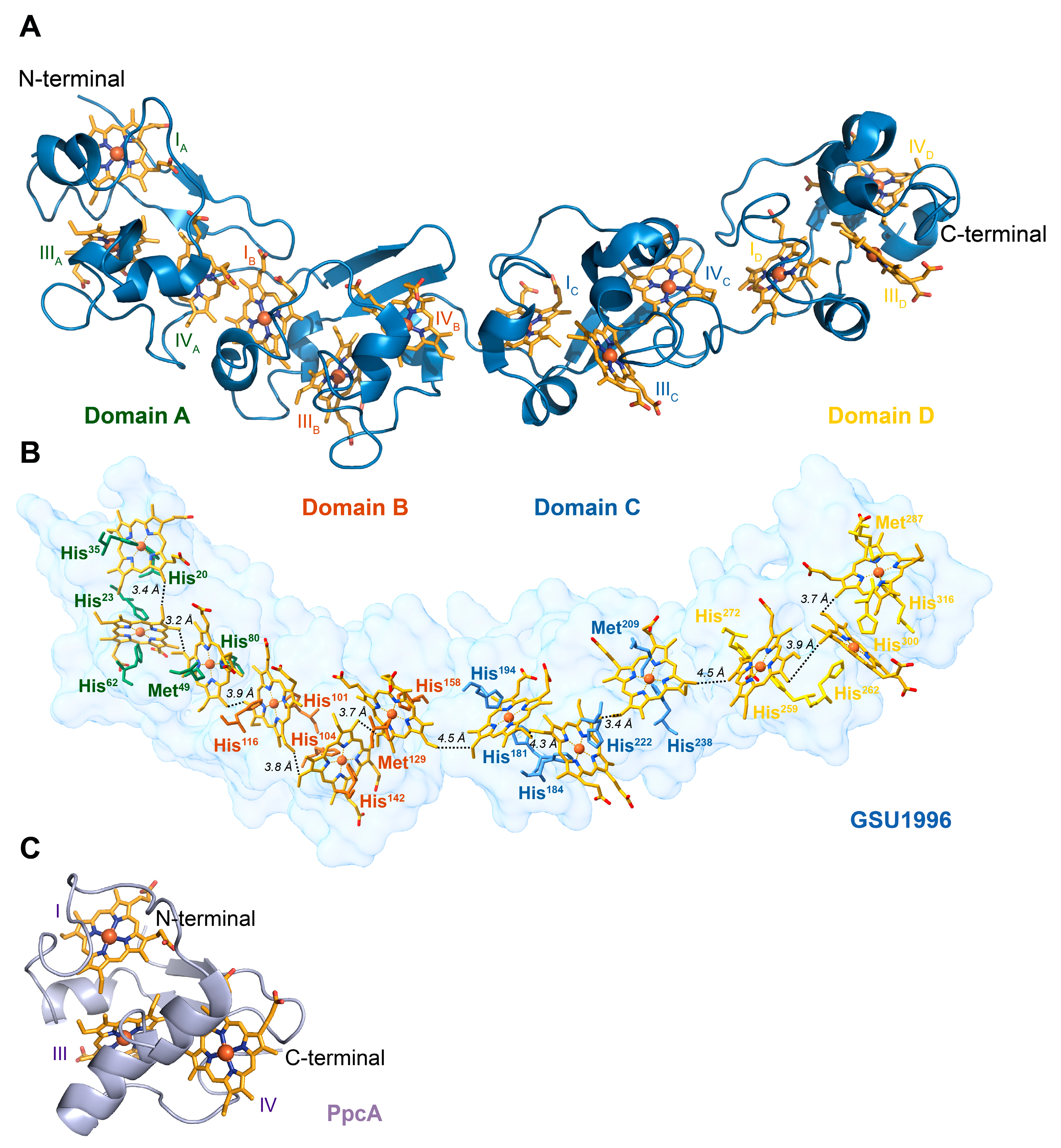
1. Introduction
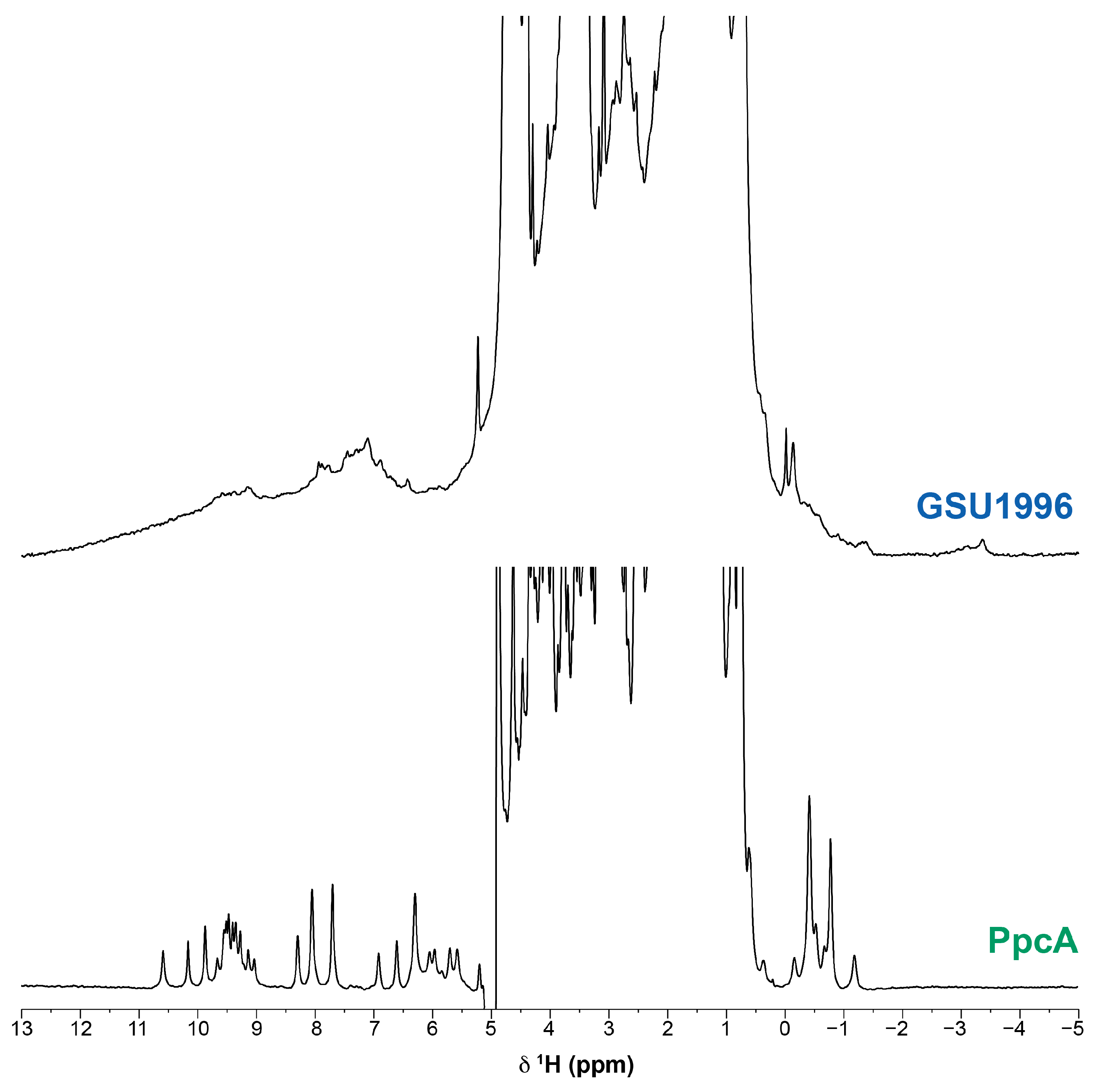
2. Results and Discussion
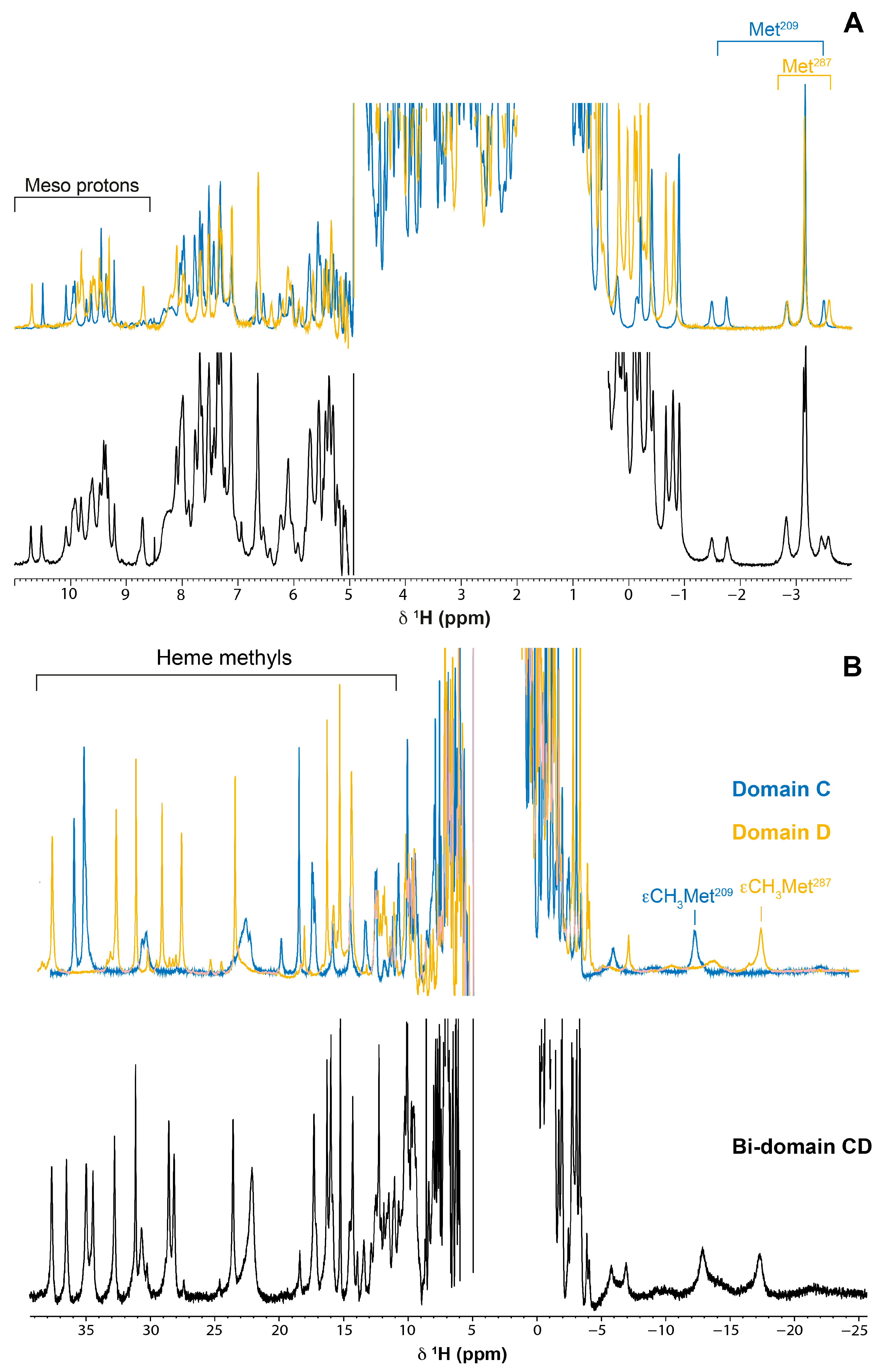
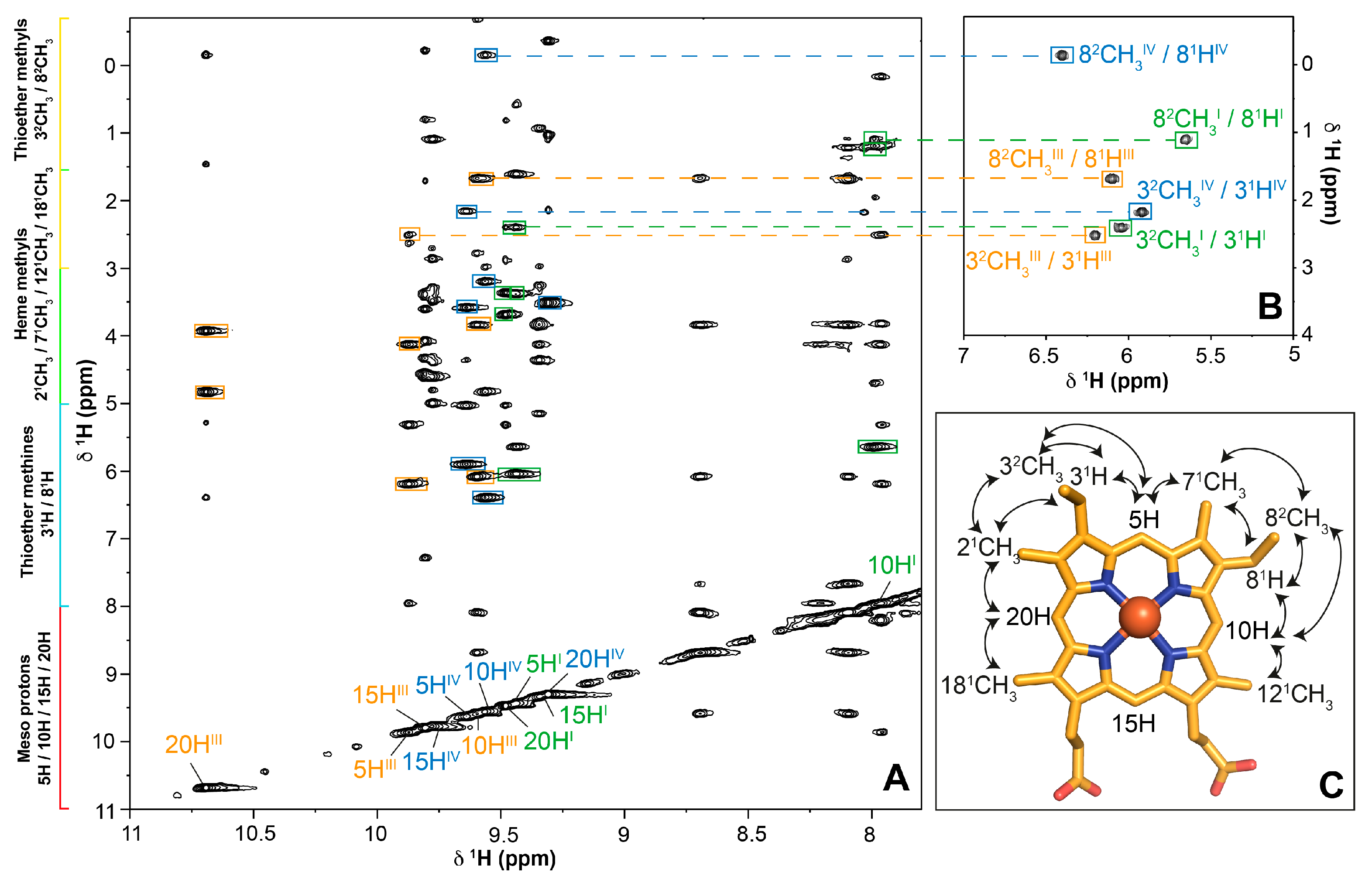
2.1. Assignment of the Heme Proton’s NMR Signals of Domain D in the Reduced State
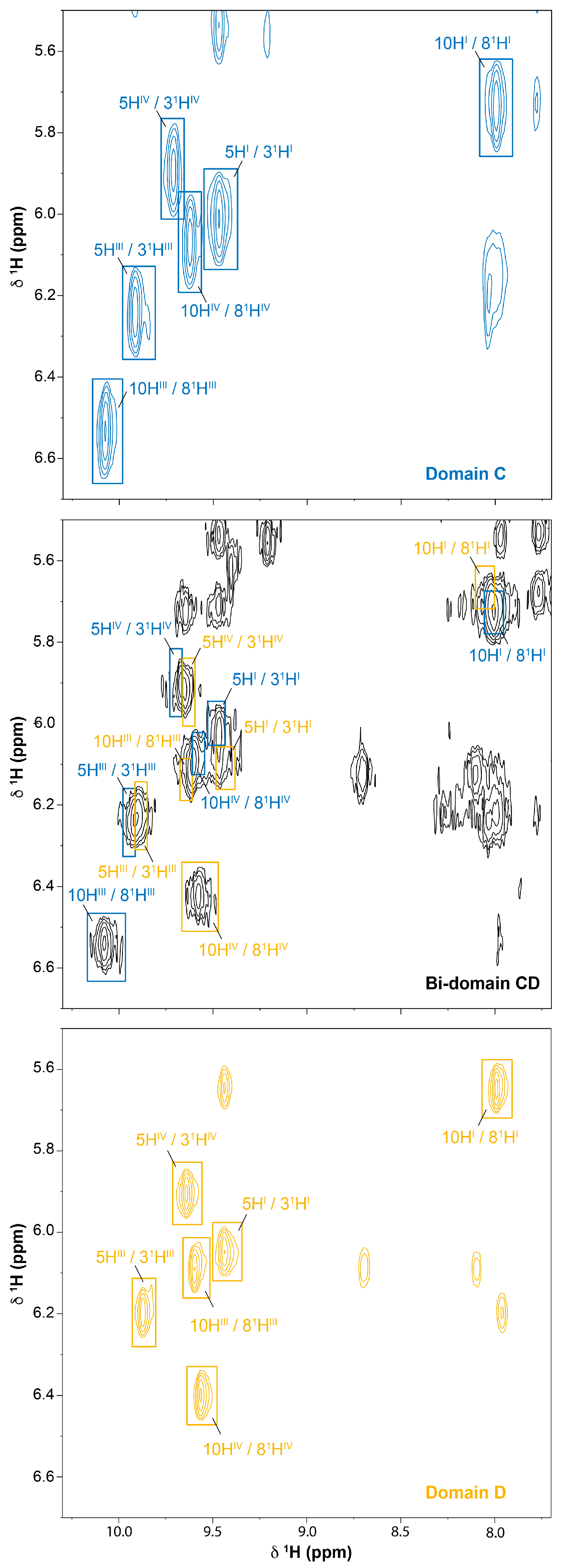
2.2. Cross-Assignment of the Hemes to the Structure of Domain D
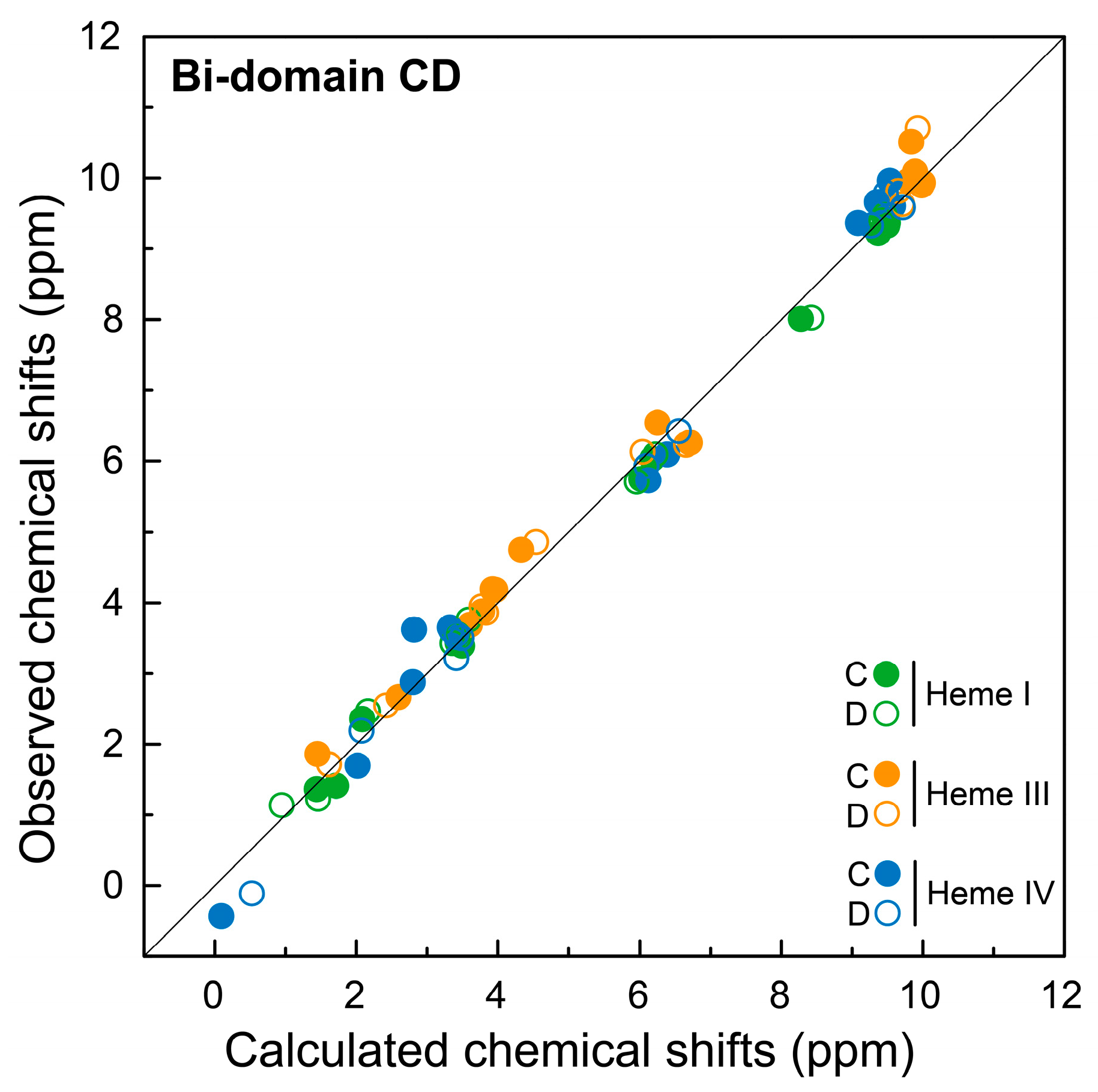
2.3. Assignment of the Heme Signals of Bi-Domain CD in the Reduced State
3. Materials and Methods
3.1. Expression and Purification of Proteins
3.2. Sample Preparation for NMR Studies
3.3. NMR Experiments
3.4. Calculation of Ring-Current Shifts
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lloyd, J.R.; Lovley, D.R. Microbial detoxification of metals and radionuclides. Curr. Opin. Biotechnol 2001, 12, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Holmes, D.E.; Finneran, K.T.; O’Neil, R.A.; Lovley, D.R. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 2002, 68, 2300–2306. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R.; Ueki, T.; Zhang, T.; Malvankar, N.S.; Shrestha, P.M.; Flanagan, K.A.; Aklujkar, M.; Butler, J.E.; Giloteaux, L.; Rotaru, A.E.; et al. Geobacter: The microbe electric’s physiology, ecology, and practical applications. Adv. Microb. Physiol. 2011, 59, 1–100. [Google Scholar] [PubMed]
- Lovley, D.R. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol. Rev. 1991, 55, 259–287. [Google Scholar] [CrossRef]
- Lovley, D.R. Bioremediation of organic and metal contaminants with dissimilatory metal reduction. J. Ind. Microbiol. 1995, 14, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Bond, D.R.; Lovley, D.R. Electricity production by Geobacter sulfurreducens attached to electrodes. Appl. Environ. Microbiol. 2003, 69, 1548–1555. [Google Scholar] [CrossRef]
- Malvankar, N.S.; Lovley, D.R. Microbial nanowires for bioenergy applications. Curr. Opin. Biotechnol. 2014, 27, 88–95. [Google Scholar] [CrossRef]
- Lovley, D.R. Powering microbes with electricity: Direct electron transfer from electrodes to microbes. Environ. Microbiol. Rep. 2011, 3, 27–35. [Google Scholar] [CrossRef]
- Wang, L.Y.; Nevin, K.P.; Woodard, T.L.; Mu, B.Z.; Lovley, D.R. Expanding the Diet for DIET: Electron Donors Supporting Direct Interspecies Electron Transfer (DIET) in Defined Co-Cultures. Front. Microbiol. 2016, 7, 236. [Google Scholar] [CrossRef]
- Lee, K.Y.; Bosch, J.; Meckenstock, R.U. Use of metal-reducing bacteria for bioremediation of soil contaminated with mixed organic and inorganic pollutants. Environ. Geochem. Health 2012, 34 (Suppl. S1), 135–142. [Google Scholar] [CrossRef]
- Salgueiro, C.A.; Morgado, L.; Silva, M.A.; Ferreira, M.R.; Fernandes, T.M.; Portela, P.C. From iron to bacterial electroconductive filaments: Exploring cytochrome diversity using Geobacter bacteria. Coord. Chem. Rev. 2022, 452, 214284. [Google Scholar] [CrossRef]
- Wang, F.; Mustafa, K.; Suciu, V.; Joshi, K.; Chan, C.H.; Choi, S.; Su, Z.; Si, D.; Hochbaum, A.I.; Egelman, E.H.; et al. Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing. Nat. Microbiol. 2022, 7, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Gu, Y.; O’Brien, J.P.; Yi, S.M.; Yalcin, S.E.; Srikanth, V.; Shen, C.; Vu, D.; Ing, N.L.; Hochbaum, A.I.; et al. Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers. Cell 2019, 177, 361–369.e310. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chan, C.H.; Suciu, V.; Mustafa, K.; Ammend, M.; Si, D.; Hochbaum, A.I.; Egelman, E.H.; Bond, D.R. Structure of Geobacter OmcZ filaments suggests extracellular cytochrome polymers evolved independently multiple times. Elife 2022, 11, e81551. [Google Scholar] [CrossRef] [PubMed]
- Pokkuluri, P.R.; Londer, Y.Y.; Duke, N.E.; Pessanha, M.; Yang, X.; Orshonsky, V.; Orshonsky, L.; Erickson, J.; Zagyanskiy, Y.; Salgueiro, C.A.; et al. Structure of a novel dodecaheme cytochrome c from Geobacter sulfurreducens reveals an extended 12 nm protein with interacting hemes. J. Struct Biol. 2011, 174, 223–233. [Google Scholar] [CrossRef]
- Pokkuluri, P.R.; Londer, Y.Y.; Duke, N.E.; Erickson, J.; Pessanha, M.; Salgueiro, C.A.; Schiffer, M. Structure of a novel c7-type three-heme cytochrome domain from a multidomain cytochrome c polymer. Protein. Sci. 2004, 13, 1684–1692. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.E.; Young, N.D.; Lovley, D.R. Evolution of electron transfer out of the cell: Comparative genomics of six Geobacter genomes. BMC Genom. 2010, 11, 40. [Google Scholar] [CrossRef]
- Esteve-Nunez, A.; Sosnik, J.; Visconti, P.; Lovley, D.R. Fluorescent properties of c-type cytochromes reveal their potential role as an extracytoplasmic electron sink in Geobacter sulfurreducens. Environ. Microbiol. 2008, 10, 497–505. [Google Scholar] [CrossRef]
- Alves, M.N.; Fernandes, A.P.; Salgueiro, C.A.; Paquete, C.M. Unraveling the electron transfer processes of a nanowire protein from Geobacter sulfurreducens. Biochim. Biophys. Acta 2016, 1857, 7–13. [Google Scholar] [CrossRef]
- Page, C.C.; Moser, C.C.; Chen, X.; Dutton, P.L. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 1999, 402, 47–52. [Google Scholar] [CrossRef]
- Morgado, L.; Paixao, V.B.; Schiffer, M.; Pokkuluri, P.R.; Bruix, M.; Salgueiro, C.A. Revealing the structural origin of the redox-Bohr effect: The first solution structure of a cytochrome from Geobacter sulfurreducens. Biochem. J. 2012, 441, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Morgado, L.; Fernandes, A.P.; Londer, Y.Y.; Pokkuluri, P.R.; Schiffer, M.; Salgueiro, C.A. Thermodynamic characterization of the redox centres in a representative domain of a novel c-type multihaem cytochrome. Biochem. J. 2009, 420, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.M.; Morgado, L.; Turner, D.L.; Salgueiro, C.A. Protein Engineering of Electron Transfer Components from Electroactive Geobacter Bacteria. Antioxidants 2021, 10, 844. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, B.M.; Paquete, C.M.; Salgueiro, C.A.; Louro, R.O. The role of intramolecular interactions in the functional control of multiheme cytochromes c. FEBS Lett. 2012, 586, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Santos, H.; Moura, J.J.; Moura, I.; LeGall, J.; Xavier, A.V. NMR studies of electron transfer mechanisms in a protein with interacting redox centres: Desulfovibrio gigas cytochrome c3. Eur. J. Biochem. 1984, 141, 283–296. [Google Scholar] [CrossRef]
- Salgueiro, C.A.; Turner, D.L.; Santos, H.; LeGall, J.; Xavier, A.V. Assignment of the redox potentials to the four haems in Desulfovibrio vulgaris cytochrome c3 by 2D-NMR. FEBS Lett. 1992, 314, 155–158. [Google Scholar] [CrossRef]
- Turner, D.L.; Salgueiro, C.A.; LeGall, J.; Xavier, A.V. Structural studies of Desulfovibrio vulgaris ferrocytochrome c3 by two-dimensional NMR. Eur. J. Biochem. 1992, 210, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.L.; Salgueiro, C.A.; Schenkels, P.; LeGall, J.; Xavier, A.V. Carbon-13NMR studies of the influence of axial ligand orientation on haem electronic structure. Biochim. Biophys. Acta 1995, 1246, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Dantas, J.M.; Saraiva, I.H.; Morgado, L.; Silva, M.A.; Schiffer, M.; Salgueiro, C.A.; Louro, R.O. Orientation of the axial ligands and magnetic properties of the hemes in the cytochrome c7 family from Geobacter sulfurreducens determined by paramagnetic NMR. Dalton. Trans. 2011, 40, 12713–12718. [Google Scholar] [CrossRef]
- Morgado, L.; Fernandes, A.P.; Londer, Y.Y.; Bruix, M.; Salgueiro, C.A. One simple step in the identification of the cofactors signals, one giant leap for the solution structure determination of multiheme proteins. Biochem. Biophys. Res. Commun. 2010, 393, 466–470. [Google Scholar] [CrossRef]
- Keller, R.M.; Wuthrich, K. Assignment of the heme c resonances in the 360 MHz H NMR spectra of cytochrome c. Biochim. Biophys. Acta 1978, 533, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Rivera, M.; Stark, R.E. An 1H-13C-13C-edited 1H NMR experiment for making resonance assignments in the active site of heme proteins. J. Magn. Reson. 1998, 130, 76–81. [Google Scholar] [CrossRef]
- Ghini, V.; Chevance, S.; Turano, P. About the use of (13)C-(13)C NOESY in bioinorganic chemistry. J. Inorg. Biochem. 2019, 192, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Dantas, J.M.; Morgado, L.; Aklujkar, M.; Bruix, M.; Londer, Y.Y.; Schiffer, M.; Pokkuluri, P.R.; Salgueiro, C.A. Rational engineering of Geobacter sulfurreducens electron transfer components: A foundation for building improved Geobacter-based bioelectrochemical technologies. Front. Microbiol. 2015, 6, 752. [Google Scholar] [CrossRef]
- McDonald, C.C.; Phillips, W.D.; Vinogradov, S.N. Proton magnetic resonance evidence for methionine-iron coordination in mammalian-type ferrocytochrome c. Biochem. Biophys. Res. Commun. 1969, 36, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.R.; Pettigrew, G.W. Cytochromes C: Evolutionary, Structural and Physicochemical Aspects; Springer Science & Business Media: Berlin, Germany, 1990. [Google Scholar]
- Pessanha, M.; Brennan, L.; Xavier, A.V.; Cuthbertson, P.M.; Reid, G.A.; Chapman, S.K.; Turner, D.L.; Salgueiro, C.A. NMR structure of the haem core of a novel tetrahaem cytochrome isolated from Shewanella frigidimarina: Identification of the haem-specific axial ligands and order of oxidation. FEBS Lett. 2001, 489, 8–13. [Google Scholar] [CrossRef]
- Pessanha, M.; Londer, Y.Y.; Long, W.C.; Erickson, J.; Pokkuluri, P.R.; Schiffer, M.; Salgueiro, C.A. Redox characterization of Geobacter sulfurreducens cytochrome c7: Physiological relevance of the conserved residue F15 probed by site-specific mutagenesis. Biochemistry 2004, 43, 9909–9917. [Google Scholar] [CrossRef]
- Harada, E.; Fukuoka, Y.; Ohmura, T.; Fukunishi, A.; Kawai, G.; Fujiwara, T.; Akutsu, H. Redox-coupled conformational alternations in cytochrome c3 from D. vulgaris Miyazaki F on the basis of its reduced solution structure. J. Mol. Biol. 2002, 319, 767–778. [Google Scholar] [CrossRef]
- Harada, E.; Kumagai, J.; Ozawa, K.; Imabayashi, S.; Tsapin, A.S.; Nealson, K.H.; Meyer, T.E.; Cusanovich, M.A.; Akutsu, H. A directional electron transfer regulator based on heme-chain architecture in the small tetraheme cytochrome c from Shewanella oneidensis. FEBS Lett. 2002, 532, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Louro, R.O.; Pacheco, I.; Turner, D.L.; LeGall, J.; Xavier, A.V. Structural and functional characterization of cytochrome c3 from D. desulfuricans ATCC 27774 by 1H-NMR. FEBS Lett. 1996, 390, 59–62. [Google Scholar] [CrossRef]
- Picarra-Pereira, M.A.; Turner, D.L.; LeGall, J.; Xavier, A.V. Structural studies on Desulfovibrio gigas cytochrome c3 by two-dimensional 1H-nuclear-magnetic-resonance spectroscopy. Biochem. J. 1993, 294 Pt 3, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, I.B.; Turner, D.L.; LeGall, J.; Xavier, A.V. Characterization of the structure and redox behaviour of cytochrome c3 from Desulfovibrio baculatus by 1H-nuclear-magnetic-resonance spectroscopy. Biochem. J. 1993, 294 Pt 3, 899–908. [Google Scholar] [CrossRef]
- Pereira, P.M.; Pacheco, I.; Turner, D.L.; Louro, R.O. Structure-function relationship in type II cytochrome c3 from Desulfovibrio africanus: A novel function in a familiar heme core. J. Biol. Inorg. Chem. 2002, 7, 815–822. [Google Scholar] [CrossRef]
- Morgado, L.; Bruix, M.; Orshonsky, V.; Londer, Y.Y.; Duke, N.E.; Yang, X.; Pokkuluri, P.R.; Schiffer, M.; Salgueiro, C.A. Structural insights into the modulation of the redox properties of two Geobacter sulfurreducens homologous triheme cytochromes. Biochim. Biophys. Acta 2008, 1777, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Moss, G.P. Nomenclature of tetrapyrroles. Recommendations 1986 IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Eur. J. Biochem. 1988, 178, 277–328. [Google Scholar] [CrossRef]
- Londer, Y.Y.; Pokkuluri, P.R.; Orshonsky, V.; Orshonsky, L.; Schiffer, M. Heterologous expression of dodecaheme “nanowire” cytochromes c from Geobacter sulfurreducens. Protein. Expr. Purif. 2006, 47, 241–248. [Google Scholar] [CrossRef]
- Londer, Y.Y.; Pokkuluri, P.R.; Erickson, J.; Orshonsky, V.; Schiffer, M. Heterologous expression of hexaheme fragments of a multidomain cytochrome from Geobacter sulfurreducens representing a novel class of cytochromes c. Protein. Expr. Purif. 2005, 39, 254–260. [Google Scholar] [CrossRef]
- Fernandes, A.P.; Nunes, T.C.; Paquete, C.M.; Salgueiro, C.A. Interaction studies between periplasmic cytochromes provide insights into extracellular electron transfer pathways of Geobacter sulfurreducens. Biochem. J. 2017, 474, 797–808. [Google Scholar] [CrossRef]
- Gordon, E.H.; Steensma, E.; Ferguson, S.J. The cytochrome c domain of dimeric cytochrome cd1 of Paracoccus pantotrophus can be produced at high levels as a monomeric holoprotein using an improved c-type cytochrome expression system in Escherichia coli. Biochem. Biophys. Res. Commun. 2001, 281, 788–794. [Google Scholar] [CrossRef]
- Arslan, E.; Schulz, H.; Zufferey, R.; Kunzler, P.; Thony-Meyer, L. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 1998, 251, 744–747. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, S.; Cord-Ruwisch, R.; Schink, B. A periplasmic and extracellular c-type cytochrome of Geobacter sulfurreducens acts as a ferric iron reductase and as an electron carrier to other acceptors or to partner bacteria. J. Bacteriol. 1998, 180, 3686–3691. [Google Scholar] [CrossRef] [PubMed]
- Pierattelli, R.; Banci, L.; Turner, D.L. Indirect determination of magnetic susceptibility tensors in peroxidases: A novel approach to structure elucidation by NMR. J. Biol. Inorg. Chem. 1996, 1, 320–329. [Google Scholar] [CrossRef]
- Pessanha, M.; Rothery, E.L.; Miles, C.S.; Reid, G.A.; Chapman, S.K.; Louro, R.O.; Turner, D.L.; Salgueiro, C.A.; Xavier, A.V. Tuning of functional heme reduction potentials in Shewanella fumarate reductases. Biochim. Biophys. Acta 2009, 1787, 113–120. [Google Scholar] [CrossRef] [PubMed]

| Chemical Shifts (ppm) | |||||
|---|---|---|---|---|---|
| Heme Substituent | Domain | Heme I | Heme III | Heme IV | |
| 5H | C | 9.49 (−0.13) | 9.94 (−0.58) | 9.73 (−0.37) | |
| D | 9.44 (−0.08) | 9.87 (−0.51) | 9.64 (−0.28) | ||
| CD | C | 9.47 (−0.11) | 9.93 (−0.57) | 9.65 (−0.29) | |
| D | 9.47 (−0.11) | 9.89 (−0.53) | 9.65 (−0.29) | ||
| 10H | C | 8.01 (1.35) | 10.09 (−0.73) | 9.64 (−0.28) | |
| D | 7.99 (1.37) | 9.59 (−0.23) | 9.56 (−0.20) | ||
| CD | C | 8.00 (1.36) | 10.08 (−0.72) | 9.60 (−0.24) | |
| D | 8.02 (1.34) | 9.63 (−0.27) | 9.58 (−0.22) | ||
| 15H | C | 9.37 (−0.01) | 9.96 (−0.60) | 9.99 (−0.63) | |
| D | 9.34 (0.02) | 9.80 (−0.44) | 9.77 (−0.41) | ||
| CD | C | 9.36 (0.00) | 9.96 (−0.60) | 9.96 (−0.60) | |
| D | 9.32 (0.04) | 9.81 (−0.45) | 9.78 (−0.42) | ||
| 20H | C | 9.23 (0.13) | 10.51 (−1.15) | 9.46 (−0.10) | |
| D | 9.48 (−0.12) | 10.69 (−1.33) | 9.31 (0.05) | ||
| CD | C | 9.21 (0.15) | 10.51 (−1.15) | 9.36 (0.00) | |
| D | 9.40 (−0.04) | 10.70 (−1.34) | 9.32 (0.04) | ||
| 21CH3 | C | 3.59 (−0.11) | 4.74 (−1.26) | 3.65 (−0.17) | |
| D | 3.69 (−0.21) | 4.83 (−1.35) | 3.55 (−0.07) | ||
| CD | C | 3.56 (−0.08) | 4.74 (−1.26) | 3.61 (−0.13) | |
| D | 3.75 (−0.27) | 4.85 (−1.37) | 3.55 (−0.07) | ||
| 71CH3 | C | 3.45 (0.03) | 4.18 (−0.70) | 3.64 (−0.16) | |
| D | 3.38 (0.10) | 4.13 (−0.65) | 3.59 (−0.11) | ||
| CD | C | 3.43 (0.05) | 4.17 (−0.69) | 3.64 (−0.16) | |
| D | 3.42 (0.06) | 4.18 (−0.70) | 3.60 (−0.12) | ||
| 121CH3 | C | 1.42 (2.06) | 3.88 (−0.40) | 2.91 (0.57) | |
| D | 1.20 (2.28) | 3.84 (−0.36) | 3.20 (0.28) | ||
| CD | C | 1.40 (2.08) | 3.87 (−0.39) | 2.87 (0.61) | |
| D | 1.22 (2.26) | 3.85 (−0.37) | 3.21 (0.27) | ||
| 181CH3 | C | 3.39 (0.09) | 3.68 (−0.20) | 3.51 (−0.03) | |
| D | 3.37 (0.11) | 3.93 (−0.45) | 3.49 (−0.01) | ||
| CD | C | 3.38 (0.10) | 3.68 (−0.20) | 3.51 (−0.03) | |
| D | 3.55 (−0.07) | 3.94 (−0.46) | 3.49 (−0.01) | ||
| 31H | C | 6.03 (0.10) | 6.26 (−0.13) | 5.91 (0.22) | |
| D | 6.04 (0.09) | 6.19 (−0.06) | 5.91 (0.22) | ||
| CD | C | 6.02 (0.11) | 6.25 (−0.12) | 5.72 (0.41) | |
| D | 6.09 (0.04) | 6.22 (−0.09) | 5.92 (0.21) | ||
| 81H | C | 5.75 (0.38) | 6.55 (−0.42) | 6.09 (0.04) | |
| D | 5.65 (0.48) | 6.09 (0.04) | 6.40 (−0.27) | ||
| CD | C | 5.74 (0.39) | 6.54 (−0.41) | 6.09 (0.04) | |
| D | 5.70 (0.43) | 6.12 (0.01) | 6.42 (−0.29) | ||
| 32CH3 | C | 2.37 (−0.25) | 2.69 (−0.57) | 2.16 (−0.04) | |
| D | 2.40 (−0.28) | 2.51 (−0.39) | 2.17 (−0.05) | ||
| CD | C | 2.35 (−0.23) | 2.66 (−0.54) | 1.69 (0.43) | |
| D | 2.46 (−0.34) | 2.54 (−0.42) | 2.18 (−0.06) | ||
| 82CH3 | C | 1.36 (0.76) | 1.86 (0.26) | −0.41 (2.53) | |
| D | 1.10 (1.02) | 1.67 (0.45) | −0.15 (2.27) | ||
| CD | C | 1.36 (0.76) | 1.85 (0.27) | −0.44 (2.56) | |
| D | 1.13 (0.99) | 1.70 (0.42) | −0.12 (2.24) | ||
| Chemical Shifts (ppm) | ||||
|---|---|---|---|---|
| Heme Substituent | Domain | Heme I | Heme III | Heme IV |
| 5H | C | 9.45 (−0.09) | 10.01 (−0.65) | 9.39 (−0.03) |
| D | 9.47 (−0.11) | 9.99 (−0.63) | 9.36 (0.00) | |
| 10H | C | 8.29 (1.07) | 9.90 (−0.54) | 9.59 (−0.23) |
| D | 8.44 (0.93) | 9.71 (−0.35) | 9.72 (−0.36) | |
| 15H | C | 9.52 (−0.16) | 9.81 (−0.45) | 9.54 (−0.18) |
| D | 9.50 (−0.14) | 9.67 (−0.31) | 9.49 (−0.13) | |
| 20H | C | 9.38 (−0.02) | 9.85 (−0.49) | 9.10 (0.26) |
| D | 9.42 (−0.06) | 9.94 (−0.58) | 9.28 (0.08) | |
| 21CH3 | C | 3.44 (0.04) | 4.34 (−0.86) | 2.83 (0.66) |
| D | 3.60 (−0.12) | 4.55 (−1.07) | 3.42 (0.06) | |
| 71CH3 | C | 3.44 (0.04) | 3.97 (−0.49) | 3.32 (0.16) |
| D | 3.37 (0.11) | 3.93 (−0.45) | 3.34 (0.14) | |
| 121CH3 | C | 1.73 (1.75) | 3.79 (−0.31) | 2.81 (0.67) |
| D | 1.47 (2.01) | 3.84 (−0.36) | 3.42 (0.06) | |
| 181CH3 | C | 3.51 (−0.03) | 3.61 (−0.13) | 3.42 (0.06) |
| D | 3.44 (0.04) | 3.78 (−0.30) | 3.49 (−0.01) | |
| 31H | C | 6.19 (−0.06) | 6.72 (−0.59) | 6.14 (−0.01) |
| D | 6.23 (−0.10) | 6.67 (−0.54) | 6.10 (0.03) | |
| 81H | C | 6.03 (0.10) | 6.26 (−0.13) | 6.40 (−0.27) |
| D | 5.97 (0.16) | 6.05 (0.08) | 6.56 (−0.43) | |
| 32CH3 | C | 2.09 (0.03) | 2.61 (−0.49) | 2.03 (0.10) |
| D | 2.17 (−0.05) | 2.43 (−0.31) | 2.08 (0.04) | |
| 82CH3 | C | 1.45 (0.67) | 1.45 (0.67) | 0.10 (2.02) |
| D | 0.95 (1.17) | 1.62 (0.50) | 0.53 (1.59) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, M.A.; Fernandes, A.P.; Turner, D.L.; Salgueiro, C.A. A Biochemical Deconstruction-Based Strategy to Assist the Characterization of Bacterial Electric Conductive Filaments. Int. J. Mol. Sci. 2023, 24, 7032. https://doi.org/10.3390/ijms24087032
Silva MA, Fernandes AP, Turner DL, Salgueiro CA. A Biochemical Deconstruction-Based Strategy to Assist the Characterization of Bacterial Electric Conductive Filaments. International Journal of Molecular Sciences. 2023; 24(8):7032. https://doi.org/10.3390/ijms24087032
Chicago/Turabian StyleSilva, Marta A., Ana P. Fernandes, David L. Turner, and Carlos A. Salgueiro. 2023. "A Biochemical Deconstruction-Based Strategy to Assist the Characterization of Bacterial Electric Conductive Filaments" International Journal of Molecular Sciences 24, no. 8: 7032. https://doi.org/10.3390/ijms24087032
APA StyleSilva, M. A., Fernandes, A. P., Turner, D. L., & Salgueiro, C. A. (2023). A Biochemical Deconstruction-Based Strategy to Assist the Characterization of Bacterial Electric Conductive Filaments. International Journal of Molecular Sciences, 24(8), 7032. https://doi.org/10.3390/ijms24087032







