Surveillance of cfDNA Hot Spot Mutations in NSCLC Patients during Disease Progression
Abstract
1. Introduction
2. Results
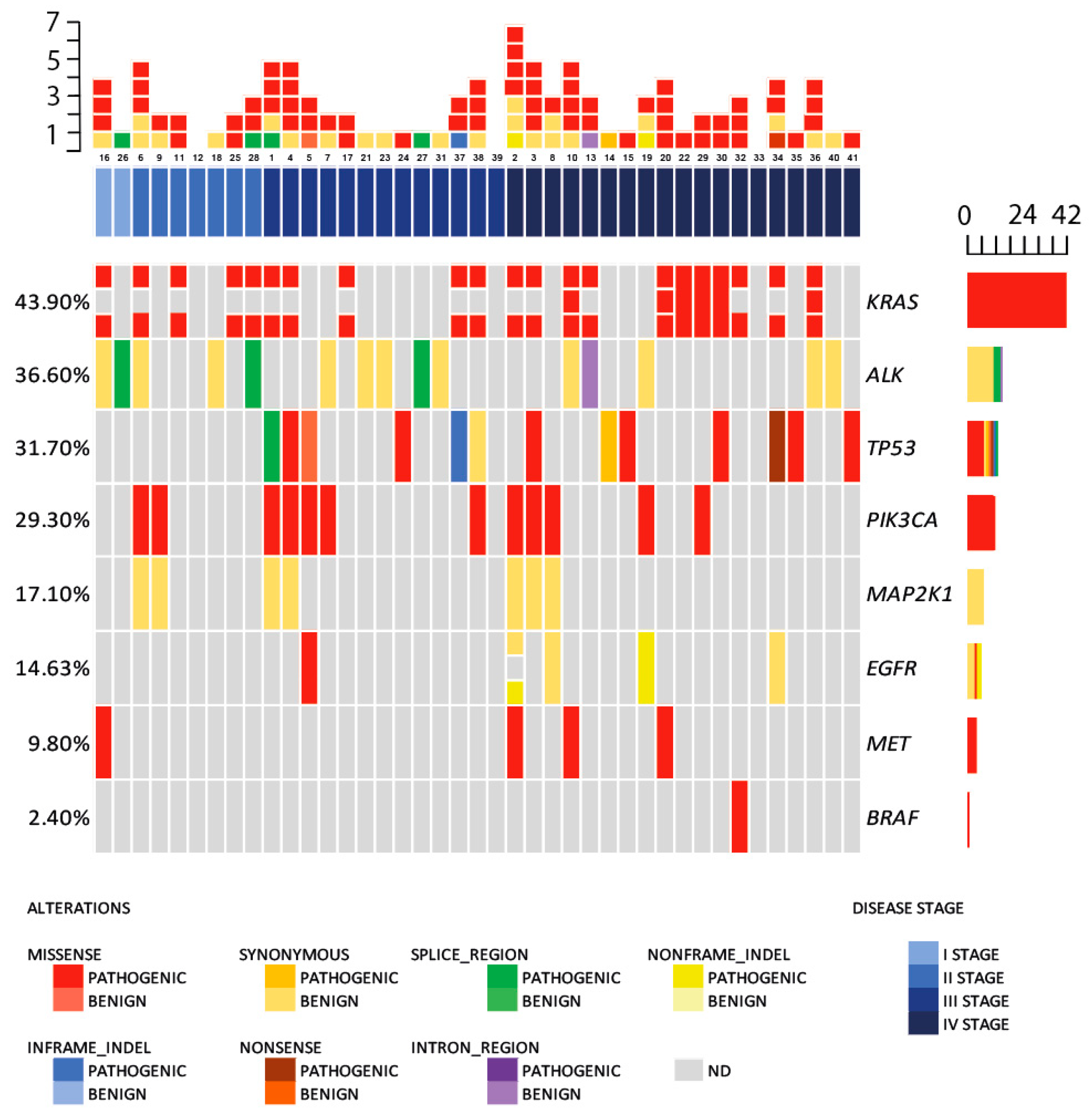
2.1. Circulating DNA Somatic Mutations
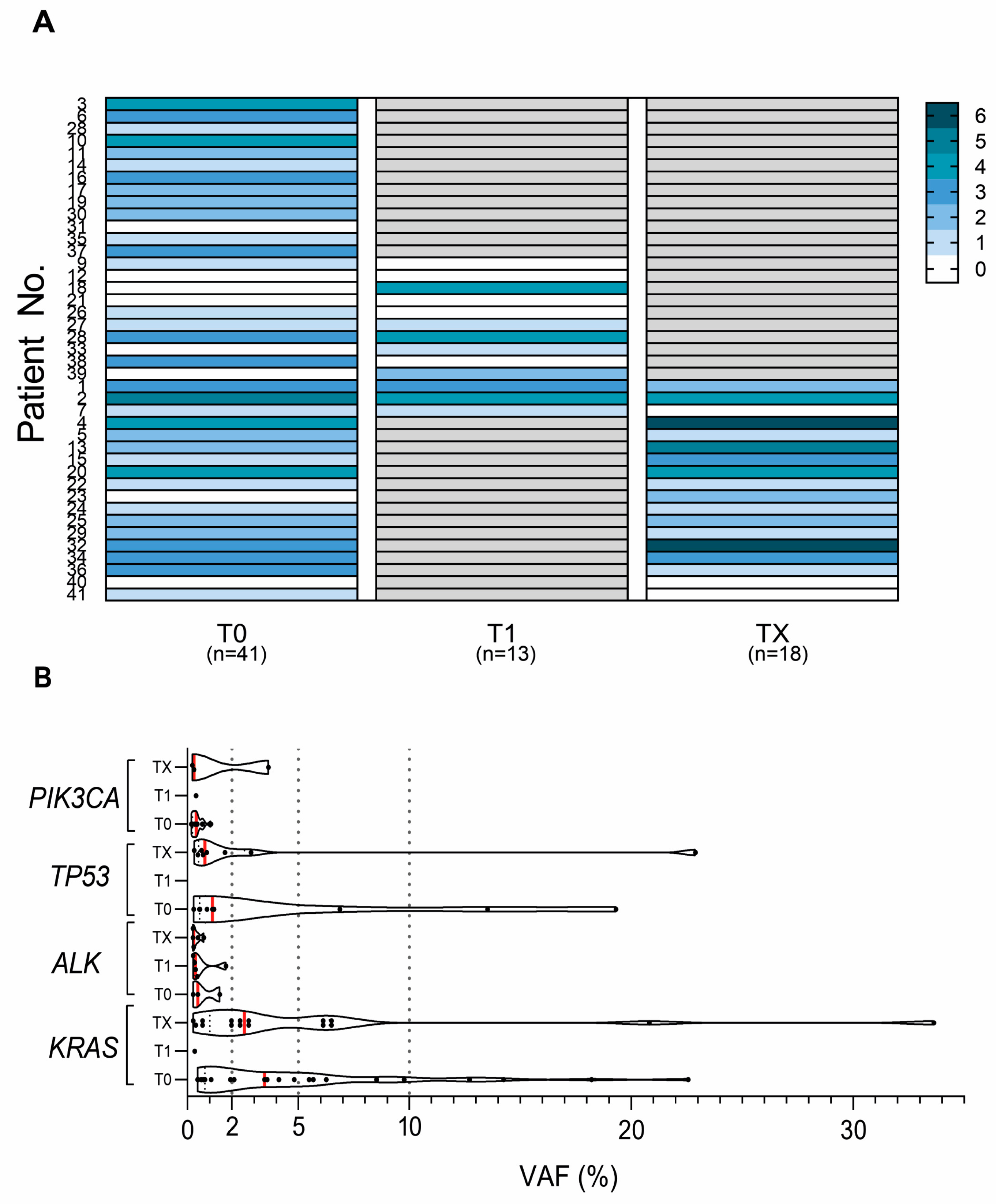
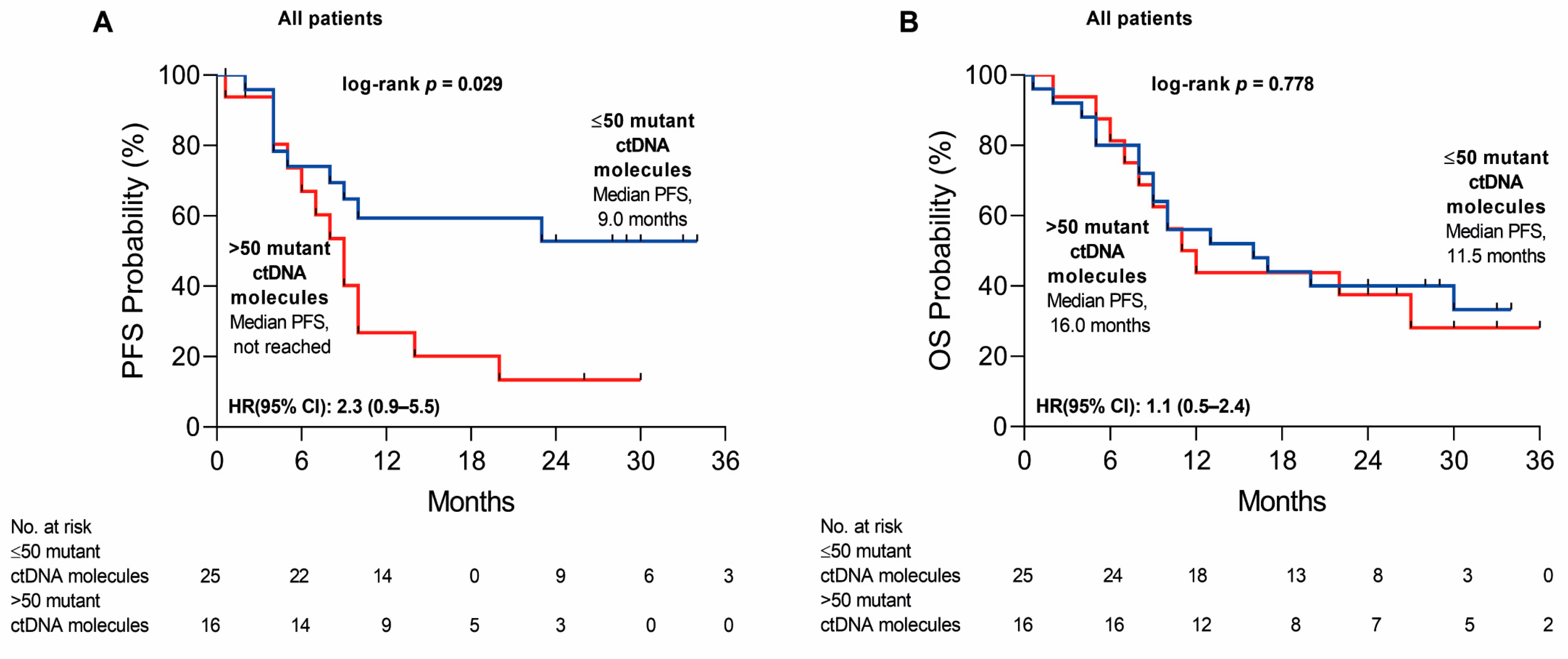
2.2. Variation in cfDNA Mutation Count and VAF during the Disease Progression
2.3. Variation in cfDNA Mutation Count and VAF during the Disease Progression
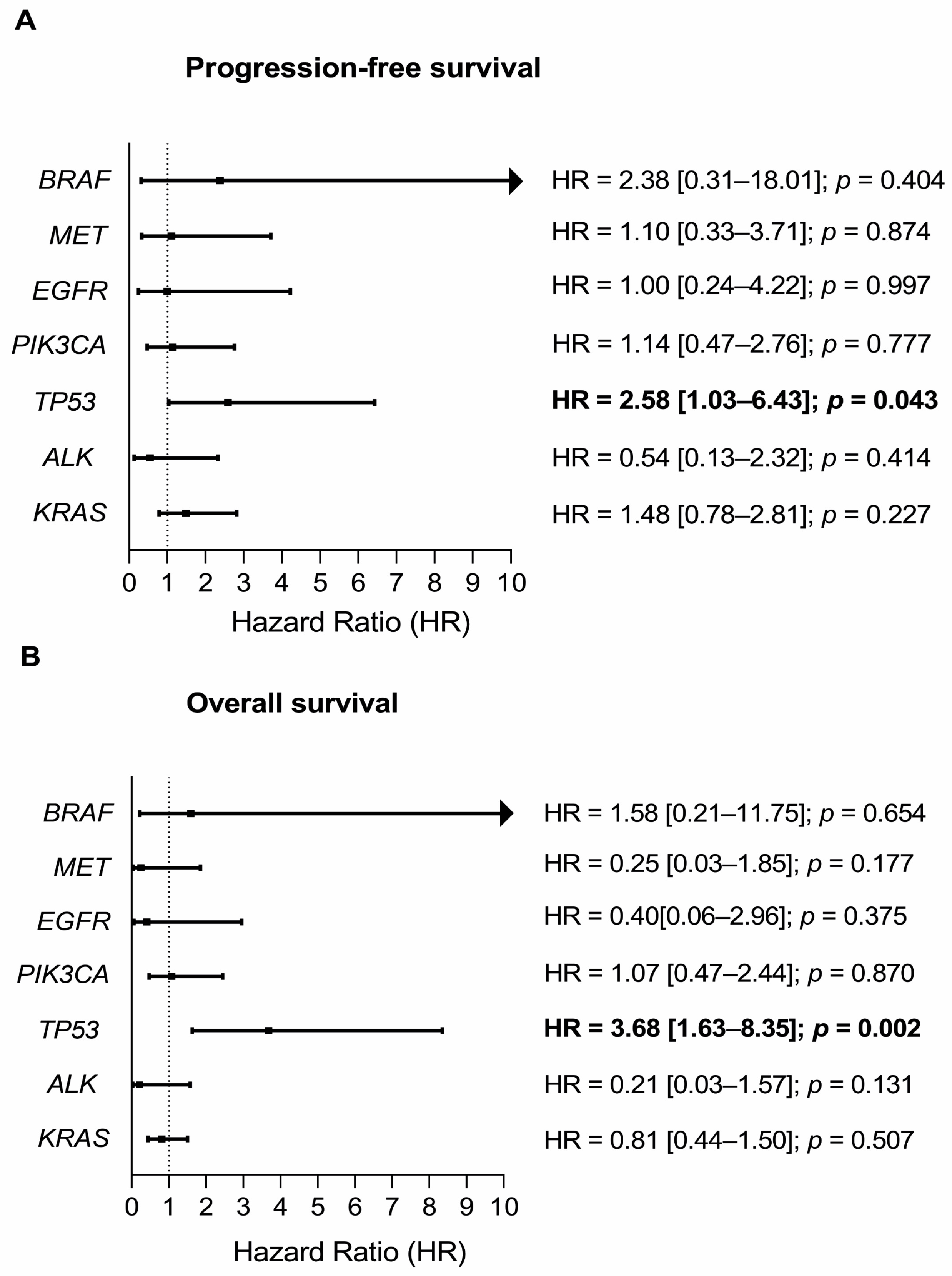
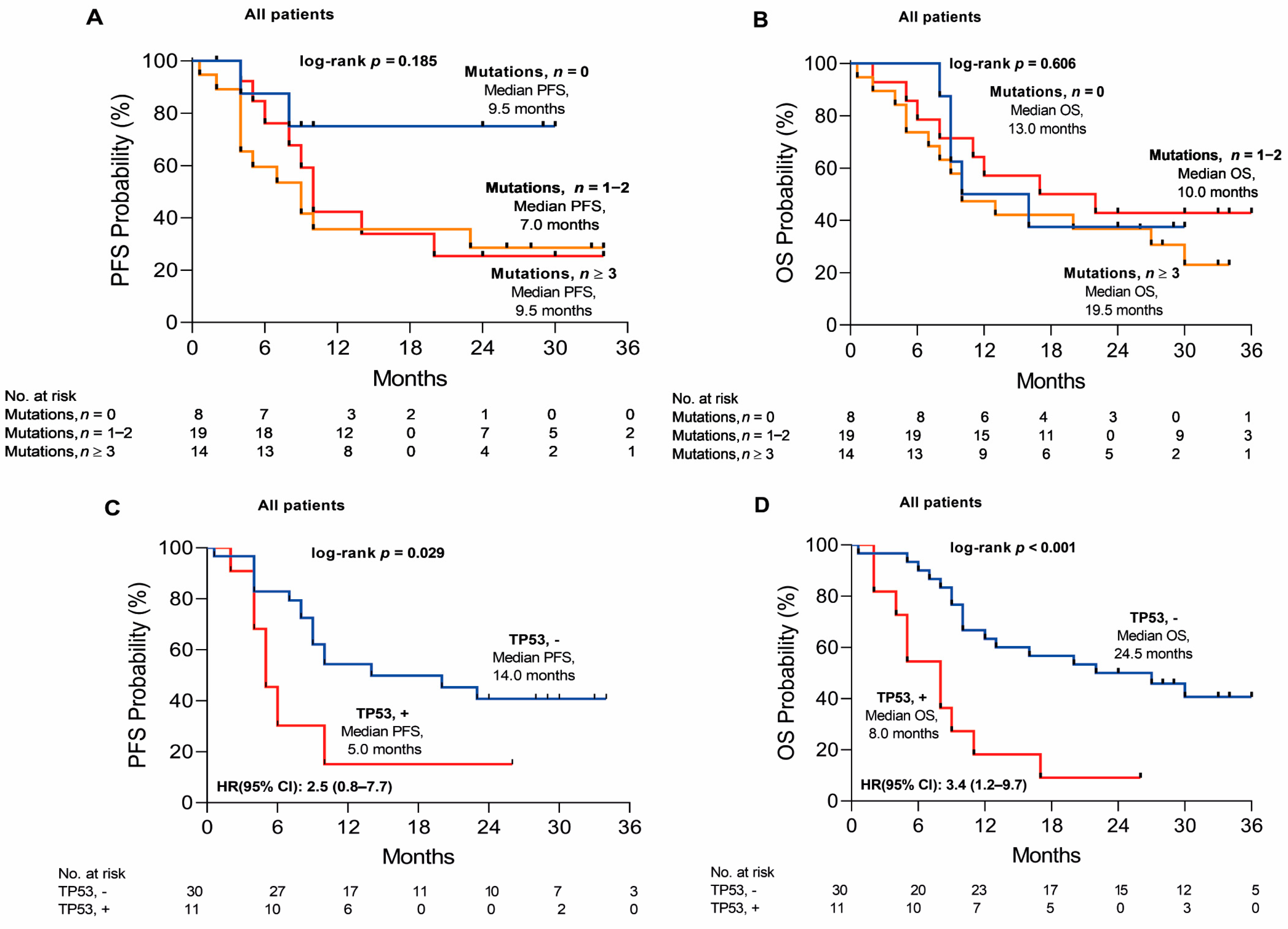
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Specimen Collection and Study Design
4.3. Mutational Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Cancer Today. Available online: http://gco.iarc.fr/today/home (accessed on 7 November 2022).
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-Small Cell Lung Cancer: Epidemiology, Risk Factors, Treatment, and Survivorship. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2008; Volume 83, pp. 584–594. [Google Scholar]
- Gridelli, C.; Rossi, A.; Carbone, D.P.; Guarize, J.; Karachaliou, N.; Mok, T.; Petrella, F.; Spaggiari, L.; Rosell, R. Non-small-cell lung cancer. Nat. Rev. Dis. Primer. 2015, 1, 1727–1740. [Google Scholar] [CrossRef]
- Bollig-Fischer, A.; Chen, W.; Gadgeel, S.M.; Wenzlaff, A.S.; Cote, M.L.; Schwartz, A.G.; Bepler, G. Racial Diversity of Actionable Mutations in Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2015, 10, 250–255. [Google Scholar] [CrossRef]
- Hill, A.; Gupta, R.; Zhao, D.; Vankina, R.; Amanam, I.; Salgia, R. Targeted Therapies in Non-small-Cell Lung Cancer. In Precision Medicine in Cancer Therapy; Cancer Treatment and Research; von Hoff, D.D., Han, H., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 3–43. Available online: https://doi.org/10.1007/978-3-030-16391-4_1 (accessed on 7 November 2022).
- Garrido-Navas, M.C.; García-Díaz, A.; Molina-Vallejo, M.P.; González-Martínez, C.; Alcaide Lucena, M.; Cañas-García, I.; Bayarri, C.; Delgado, J.R.; González, E.; Lorente, J.A.; et al. The Polemic Diagnostic Role of TP53 Mutations in Liquid Biopsies from Breast, Colon and Lung Cancers. Cancers 2020, 12, 3343. [Google Scholar] [CrossRef]
- Wadowska, K.; Bil-Lula, I.; Trembecki, Ł.; Śliwińska-Mossoń, M. Genetic Markers in Lung Cancer Diagnosis: A Review. Int. J. Mol. Sci. 2020, 21, 4569. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour, DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef]
- Johann, D.J.; Steliga, M.; Shin, I.J.; Yoon, D.; Arnaoutakis, K.; Hutchins, L.; Meeiyueh, L.; Liem, J.; Walker, K.; Pereira, A.; et al. Liquid biopsy and its role in an advanced clinical trial for lung cancer. Exp. Biol. Med. 2018, 243, 262–271. [Google Scholar] [CrossRef]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef]
- Papadopoulou, E.; Tsoulos, N.; Tsantikidi, K.; Metaxa-Mariatou, V.; Stamou, P.E.; Kladi-Skandali, A.; Kapeni, E.; Tsaousis, G.; Pentheroudakis, G.; Petrakis, D.; et al. Clinical feasibility of NGS liquid biopsy analysis in NSCLC patients. PLoS ONE 2019, 14, e0226853. [Google Scholar] [CrossRef]
- Guo, D.; Yang, L.; Yang, J.; Shi, K. Plasma cell-free DNA Methylation combined with tumor mutation detection in prognostic prediction of patients with non-small cell lung cancer (NSCLC). Medicine 2020, 99, e20431. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Adams, H.P.; Lange, M.; Siemann, S.; Feldkamp, M.; McNamara, S.; Froehler, S.; Yaung, S.J.; Yao, L.; Balasubramanyam, A.; et al. Plasma-based longitudinal mutation monitoring as a potential predictor of disease progression in subjects with adenocarcinoma in advanced non-small cell lung cancer. BMC Cancer 2020, 20, 885. [Google Scholar] [CrossRef] [PubMed]
- Mack, P.C.; Banks, K.C.; Espenschied, C.R.; Burich, R.A.; Zill, O.A.; Lee, C.E.; Riess, J.W.; Mortimer, S.A.; Talasaz, A.; Lanman, R.B.; et al. Spectrum of driver mutations and clinical impact of circulating tumor DNA analysis in non–small cell lung cancer: Analysis of over 8000 cases. Cancer 2020, 126, 3219–3228. [Google Scholar] [CrossRef] [PubMed]
- Jee, J.; Lebow, E.S.; Yeh, R.; Das, J.P.; Namakydoust, A.; Paik, P.K.; Chaft, J.E.; Jayakumaran, G.; Brannon, A.R.; Benayed, R.; et al. Overall survival with circulating tumor DNA-guided therapy in advanced non-small-cell lung cancer. Nat. Med. 2022, 28, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Waldeck, S.; Mitschke, J.; Wiesemann, S.; Rassner, M.; Andrieux, G.; Deuter, M.; Mutter, J.; Lüchtenborg, A.M.; Kottmann, D.; Titze, L.; et al. Early assessment of circulating tumor DNA after curative-intent resection predicts tumor recurrence in early-stage and locally advanced non-small-cell lung cancer. Mol. Oncol. 2022, 16, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, B.X.; Li, J.; Shao, Y.; Li, M.T.; Li, J.J.; Kuang, P.P.; Liu, Z.; Sun, T.Y.; Wu, H.Q.; et al. Perioperative circulating tumor DNA as a potential prognostic marker for operable stage I to IIIA non-small cell lung cancer. Cancer 2022, 128, 708–718. [Google Scholar] [CrossRef]
- Xia, L.; Mei, J.; Kang, R.; Deng, S.; Chen, Y.; Yang, Y.; Feng, G.; Deng, Y.; Gan, F.; Lin, Y.; et al. Perioperative ctDNA-Based Molecular Residual Disease Detection for Non-Small Cell Lung Cancer: A Prospective Multicenter Cohort Study (LUNGCA-1). Clin. Cancer Res. 2022, 28, 3308–3317. [Google Scholar] [CrossRef]
- Yang, N.; Li, Y.; Liu, Z.; Qin, H.; Du, D.; Cao, X.; Cao, X.; Li, J.; Li, D.; Jiang, B.; et al. The characteristics of ctDNA reveal the high complexity in matching the corresponding tumor tissues. BMC Cancer 2018, 18, 319. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, Z.; Li, C.S.; Zhao, W.; Liang, Z.X.; Dai, Y.; Zhu, Q.; Miao, K.L.; Cui, D.H.; Chen, L.A. Differences in the genomic profiles of cell-free DNA between plasma, sputum, urine, tumor tissue in advanced NSCLC. Cancer Med. 2019, 8, 910–919. [Google Scholar] [CrossRef]
- Majeed, U.; Manochakian, R.; Zhao, Y.; Lou, Y. Targeted therapy in advanced non-small cell lung cancer: Current advances and future trends. J. Hematol. Oncol. 2021, 14, 108. [Google Scholar] [CrossRef]
- Forthun, R.B.; Hovland, R.; Schuster, C.; Puntervoll, H.; Brodal, H.P.; Namløs, H.M.; Aasheim, L.B.; Meza-Zepeda, L.A.; Gjertsen, B.T.; Knappskog, S.; et al. ctDNA detected by ddPCR reveals changes in tumour load in Metastatic malignant melanoma treated with bevacizumab. Sci. Rep. 2019, 9, 17471. [Google Scholar] [CrossRef]
- Kang, J.; Luo, Y.; Wang, D.; Men, Y.; Wang, J.; Che, Y.Q.; Hui, Z. Tumor Mutation Load: A Novel Independent Prognostic Factor in Stage IIIA-N2 Non-Small-Cell Lung Cancer. Dis. Markers 2019, 2019, e3837687. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lv, J.; Wan, S.; Xin, J.; Xie, T.; Li, T.; Zhu, W.; Zhang, G.; Wang, Y.; Tang, Y.; et al. High Sensitive and Non-invasive ctDNAs Sequencing Facilitate Clinical Diagnosis And Clinical Guidance of Non-small Cell Lung Cancer Patient: A Time Course Study. Front. Oncol. 2018, 8, 491. Available online: https://www.frontiersin.org/articles/10.3389/fonc.2018.00491 (accessed on 7 November 2022). [CrossRef] [PubMed]
- Cai, X.; Sheng, J.; Tang, C.; Nandakumar, V.; Ye, H.; Ji, H.; Tang, H.; Qin, Y.; Guan, H.; Lou, F.; et al. Frequent Mutations in EGFR, KRAS and TP53 Genes in Human Lung Cancer Tumors Detected by Ion Torrent DNA Sequencing. PLoS ONE 2014, 9, e95228. [Google Scholar] [CrossRef]
- Negrao, M.V.; Raymond, V.M.; Lanman, R.B.; Robichaux, J.P.; He, J.; Nilsson, M.B.; Ng, P.K.S.; Amador, B.E.; Roarty, E.B.; Nagy, R.J.; et al. Molecular Landscape of BRAF-Mutant NSCLC Reveals an Association Between Clonality and Driver Mutations and Identifies Targetable Non-V600 Driver Mutations. J. Thorac. Oncol. 2020, 15, 1611–1623. [Google Scholar] [CrossRef]
- Mäki-Nevala, S.; Sarhadi, V.K.; Rönty, M.; Kettunen, E.; Husgafvel-Pursiainen, K.; Wolff, H.; Knuuttila, H.; Knuutila, A. Hot spot mutations in Finnish non-small cell lung cancers. Lung Cancer 2016, 99, 102–110. [Google Scholar] [CrossRef]
- Aggarwal, C.; Davis, C.W.; Mick, R.; Thompson, J.C.; Ahmed, S.; Jeffries, S.; Bagley, S.; Gabriel, P.; Evans, T.L.; Bauml, J.M.; et al. Influence of TP53 Mutation on Survival in Patients with Advanced EGFR-Mutant Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2018, 2, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Mayo-de-las-Casas, C.; Garzón Ibáñez, M.; Jordana-Ariza, N.; García-Peláez, B.; Balada-Bel, A.; Villatoro, S.; Malapelle, U.; Karachaliou, N.; Troncone, G.; Rosell, R.; et al. An update on liquid biopsy analysis for diagnostic and monitoring applications in non-small cell lung cancer. Expert Rev. Mol. Diagn. 2018, 18, 35–45. [Google Scholar] [CrossRef]
- Qin, K.; Hou, H.; Liang, Y.; Zhang, X. Prognostic value of TP53 concurrent mutations for EGFR-TKIs and ALK-TKIs based targeted therapy in advanced non-small cell lung cancer: A meta-analysis. BMC Cancer 2020, 20, 328. [Google Scholar] [CrossRef]
- Schwaederlé, M.C.; Patel, S.P.; Husain, H.; Ikeda, M.; Lanman, R.B.; Banks, K.C.; Talasaz, A.; Bazhenova, L.; Kurzrock, R. Utility of Genomic Assessment of Blood-Derived Circulating Tumor DNA (ctDNA) in Patients with Advanced Lung Adenocarcinoma. Clin. Cancer Res. 2017, 23, 5101–5111. [Google Scholar] [CrossRef]
- Yang, S.; Yu, X.; Fan, Y.; Shi, X.; Jin, Y. Clinicopathologic characteristics and survival outcome in patients with advanced lung adenocarcinoma and KRAS mutation. J. Cancer 2018, 9, 2930–2937. [Google Scholar] [CrossRef]
- Skoulidis, F.; Heymach, J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 2019, 19, 495–509. [Google Scholar] [CrossRef]
- Tomasini, P.; Mascaux, C.; Jao, K.; Labbe, C.; Kamel-Reid, S.; Stockley, T.; Hwang, D.M.; Leighl, N.B.; Liu, G.; Bradbury, P.A.; et al. Effect of Coexisting KRAS and TP53 Mutations in Patients Treated with Chemotherapy for Non–small-cell Lung Cancer. Clin. Lung Cancer 2019, 20, e338–e345. [Google Scholar] [CrossRef] [PubMed]
- Bange, E.; Marmarelis, M.E.; Hwang, W.T.; Yang, Y.X.; Thompson, J.C.; Rosenbaum, J.; Bauml, J.; Ciunci, C.; Alley, E.W.; Cohen, R.B.; et al. Impact of KRAS and TP53 Co-Mutations on Outcomes After First-Line Systemic Therapy Among Patients with STK11-Mutated Advanced Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2019, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Uras, I.Z.; Moll, H.P.; Casanova, E. Targeting KRAS Mutant Non-Small-Cell Lung Cancer: Past, Present and Future. Int. J. Mol. Sci. 2020, 21, 4325. [Google Scholar] [CrossRef]
- Gamerith, G.; Kocher, F.; Rudzki, J.; Pircher, A. ASCO 2018 NSCLC highlights—Combination therapy is key. Memo-Mag. Eur. Med. Oncol. 2018, 11, 266–271. [Google Scholar] [CrossRef]
- Cai, D.; Hu, C.; Li, L.; Deng, S.; Yang, J.; Han-Zhang, H.; Li, M. The prevalence and prognostic value of KRAS co-mutation subtypes in Chinese advanced non-small cell lung cancer patients. Cancer Med. 2020, 9, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Wang, M.; Zhu, X.; Sun, Z.; Jiang, A.; Yao, H. Therapeutic effects of lenvatinib in combination with rAd-p53 for the treatment of non-small cell lung cancer. Oncol. Lett. 2018, 16, 6573–6581. [Google Scholar] [CrossRef]
- Nenkov, M.; Ma, Y.; Haase, D.; Zhou, Z.; Li, Y.; Petersen, I.; Lu, G.; Chen, Y. Growth inhibitory role of the p53 activator SCH 529074 in non-small cell lung cancer cells expressing mutant p53. Oncol. Rep. 2020, 43, 2073–2082. [Google Scholar] [CrossRef]
- Green, S.; Trejo, C.L.; McMahon, M. PIK3CAH1047R Accelerates and Enhances KRASG12D-Driven Lung Tumorigenesis. Cancer Res. 2015, 75, 5378–5391. [Google Scholar] [CrossRef] [PubMed]
- Scheffler, M.; Ihle, M.A.; Hein, R.; Merkelbach-Bruse, S.; Scheel, A.H.; Siemanowski, J.; Brägelmann, J.; Kron, A.; Adebpour, N.; Ueckeroth, F.; et al. K-ras Mutation Subtypes in NSCLC and Associated Co-occuring Mutations in Other Oncogenic Pathways. J. Thorac. Oncol. 2019, 14, 606–616. [Google Scholar] [CrossRef]
- Zhao, J.; Han, Y.; Li, J.; Chai, R.; Bai, C. Prognostic value of KRAS/TP53/PIK3CA in non-small cell lung cancer. Oncol. Lett. 2019, 17, 3233–3240. [Google Scholar] [CrossRef]
- Arcila, M.E.; Nafa, K.; Chaft, J.E.; Rekhtman, N.; Lau, C.; Reva, B.A.; Zakowski, M.F.; Kris, M.G.; Ladanyi, M. EGFR Exon 20 Insertion Mutations in Lung Adenocarcinomas: Prevalence, Molecular Heterogeneity, and Clinicopathologic Characteristics. Mol. Cancer Ther. 2013, 12, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Feldman, R.; Wistuba, I.I. Update on EGFR Mutational Testing and the Potential of Noninvasive Liquid Biopsy in Non–Small-cell Lung Cancer. Clin. Lung Cancer 2018, 19, 105–114. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Y.; Qi, C.; Zhang, C.; Liu, D.; Deng, Y.; Fu, Y.; Khadka, V.S.; Wang, D.D.; Tan, S.; et al. Dysregulated KRAS gene-signaling axis and abnormal chromatin remodeling drive therapeutic resistance in heterogeneous-sized circulating tumor cells in gastric cancer patients. Cancer Lett. 2021, 517, 78–87. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): AStatement Paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Kunimasa, K.; Hirotsu, Y.; Nakagomi, T.; Yokoyama, Y.; Higuchi, R.; Otake, S.; Oyama, T.; Amemiya, K.; Mochizuki, H.; et al. Association of Mutation Profiles with Postoperative Survival in Patients with Non–Small Cell Lung Cancer. Cancers 2020, 12, 3472. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Lou, F.; Ma, Y.; Li, J.; Yang, B.; Chen, W.; Ye, H.; Zhang, J.B.; Zhao, M.Y.; Wu, W.J.; et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci. Rep. 2016, 6, 33519. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Liu, J.; Li, L.; Yuan, Y.; Nan, K.; Wu, X.; Zhang, Z.; Wu, Y.; Li, X.; Zhu, J.; et al. Detection of circulating tumor DNA in patients with advanced non-small cell lung cancer. Oncotarget 2017, 8, 2130–2140. [Google Scholar] [CrossRef] [PubMed]
- Oellerich, M.; Schütz, E.; Beck, J.; Kanzow, P.; Plowman, P.N.; Weiss, G.J.; Walson, P.D. Using circulating cell-free DNA to monitor personalized cancer therapy. Crit. Rev. Clin. Lab. Sci. 2017, 54, 205–218. [Google Scholar] [CrossRef]
- Wang, S.; Li, M.; Zhang, J.; Xiang, P.; Wu, M.; Meng, F.; Jiang, F.; Wang, J.; Bao, H.; Huang, J.; et al. Circulating tumor DNA integrating tissue clonality detects minimal residual disease in resectable non-small-cell lung cancer. J. Hematol. Oncol. 2022, 15, 137. [Google Scholar] [CrossRef] [PubMed]
- Moding, E.J.; Liu, Y.; Nabet, B.Y.; Chabon, J.J.; Chaudhuri, A.A.; Hui, A.B.; Bonilla, R.F.; Ko, R.B.; Yoo, C.H.; Gojenola, L.; et al. Circulating Tumor DNA Dynamics Predict Benefit from Consolidation Immunotherapy in Locally Advanced Non-Small Cell Lung Cancer. Nat. Cancer 2020, 1, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Gale, D.; Heider, K.; Ruiz-Valdepenas, A.; Hackinger, S.; Perry, M.; Marsico, G.; Rundell, V.; Wulff, J.; Sharma, G.; Knock, H.; et al. Residual ctDNA after treatment predicts early relapse in patients with early-stage non-small cell lung cancer. Ann. Oncol. 2022, 33, 500–510. [Google Scholar] [CrossRef]
- Hsu, Y.C.; Chang, Y.H.; Chang, G.C.; Ho, B.C.; Yuan, S.S.; Li, Y.C.; Zeng, J.W.; Yu, S.L.; Li, K.C.; Yang, P.C.; et al. Tumor mutation burden and recurrent tumors in hereditary lung cancer. Cancer Med. 2019, 8, 2179–2187. [Google Scholar] [CrossRef]
- Lyu, G.Y.; Yeh, Y.H.; Yeh, Y.C.; Wang, Y.C. Mutation load estimation model as a predictor of the response to cancer immunotherapy. NPJ Genom. Med. 2018, 3, 12. [Google Scholar] [CrossRef]
- Gao, G.; Liao, W.; Ma, Q.; Zhang, B.; Chen, Y.; Wang, Y. KRAS G12D mutation predicts lower TMB and drives immune suppression in lung adenocarcinoma. Lung Cancer 2020, 149, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.T.; Choi, Y.L.; Yun, J.W.; Kim, N.K.D.; Kim, S.Y.; Jeon, H.J.; Nam, J.J.; Lee, C.; Ryu, D.; Kim, S.C.; et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat. Commun. 2017, 8, 1377. [Google Scholar] [CrossRef] [PubMed]
- Kruger, D.T.; Jansen, M.P.H.M.; Konings, I.R.H.M.; Dercksen, W.M.; Jager, A.; Oulad Hadj, J.; Sleijfer, S.; Martens, J.W.M.; Boven, E. High ctDNA molecule numbers relate with poor outcome in advanced ER+, HER2− postmenopausal breast cancer patients treated with everolimus and exemestane. Mol. Oncol. 2020, 14, 490–503. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef]
| NSCLC Patients (n = 41) | |
|---|---|
| Age, median (range) | 63 (44–78) |
| Sex, n (%) | |
| Male | 31 (76) |
| Female | 10 (24) |
| Histology, n (%) | |
| Adenocarcinoma | 17 (41) |
| Squamous | 17 (41) |
| Other | 7 (17) |
| Disease stage, n (%) | |
| Ib | 2 (5) |
| II | 7 (17) |
| III | 13 (32) |
| IV | 19 (46) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sestokaite, A.; Gedvilaite, V.; Cicenas, S.; Sabaliauskaite, R.; Jarmalaite, S. Surveillance of cfDNA Hot Spot Mutations in NSCLC Patients during Disease Progression. Int. J. Mol. Sci. 2023, 24, 6958. https://doi.org/10.3390/ijms24086958
Sestokaite A, Gedvilaite V, Cicenas S, Sabaliauskaite R, Jarmalaite S. Surveillance of cfDNA Hot Spot Mutations in NSCLC Patients during Disease Progression. International Journal of Molecular Sciences. 2023; 24(8):6958. https://doi.org/10.3390/ijms24086958
Chicago/Turabian StyleSestokaite, Agne, Vaida Gedvilaite, Saulius Cicenas, Rasa Sabaliauskaite, and Sonata Jarmalaite. 2023. "Surveillance of cfDNA Hot Spot Mutations in NSCLC Patients during Disease Progression" International Journal of Molecular Sciences 24, no. 8: 6958. https://doi.org/10.3390/ijms24086958
APA StyleSestokaite, A., Gedvilaite, V., Cicenas, S., Sabaliauskaite, R., & Jarmalaite, S. (2023). Surveillance of cfDNA Hot Spot Mutations in NSCLC Patients during Disease Progression. International Journal of Molecular Sciences, 24(8), 6958. https://doi.org/10.3390/ijms24086958





