A Computational Biology Study on the Structure and Dynamics Determinants of Thermal Stability of the Chitosanase from Aspergillus fumigatus
Abstract
1. Introduction
2. Results and Discussion
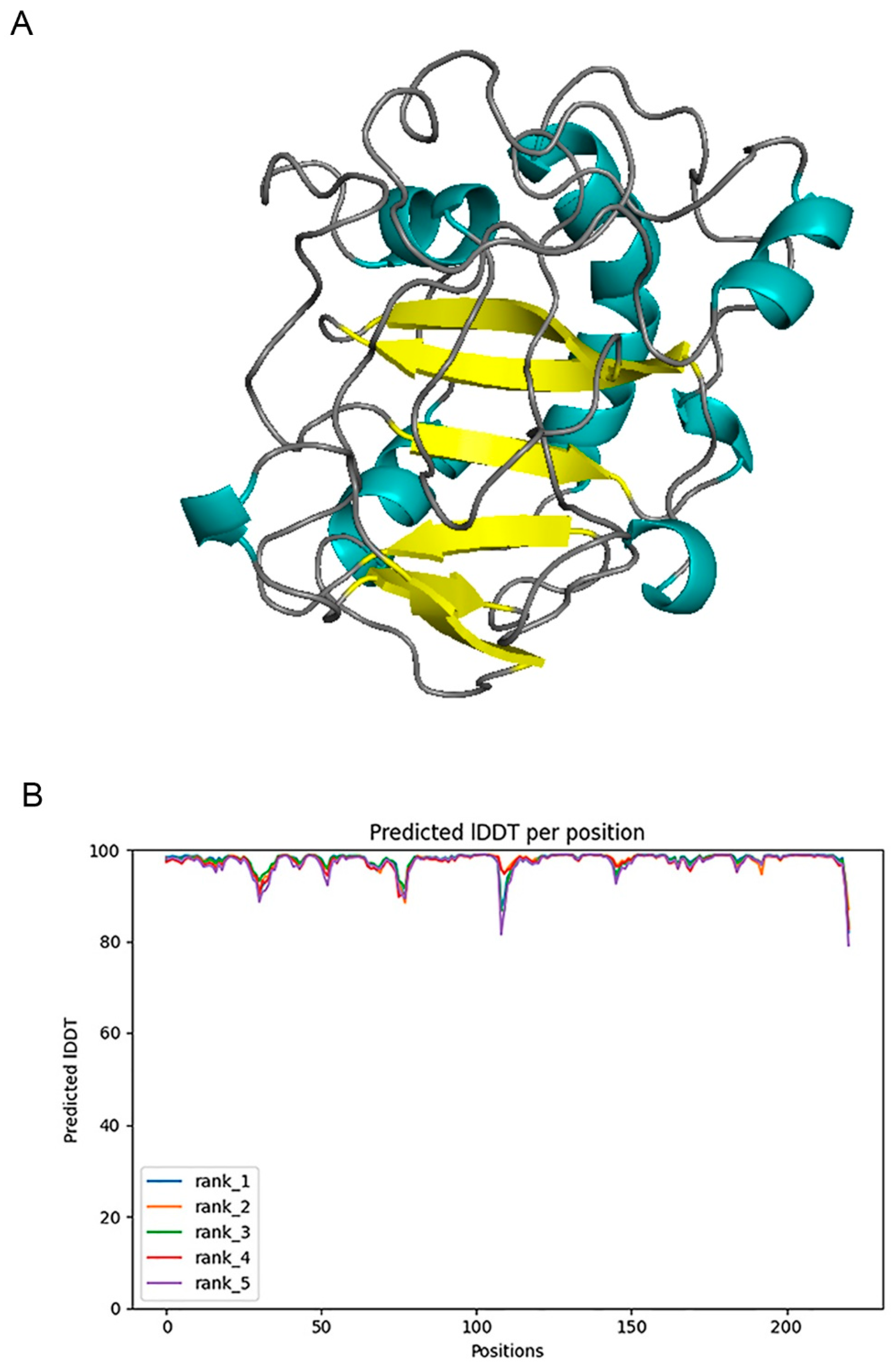
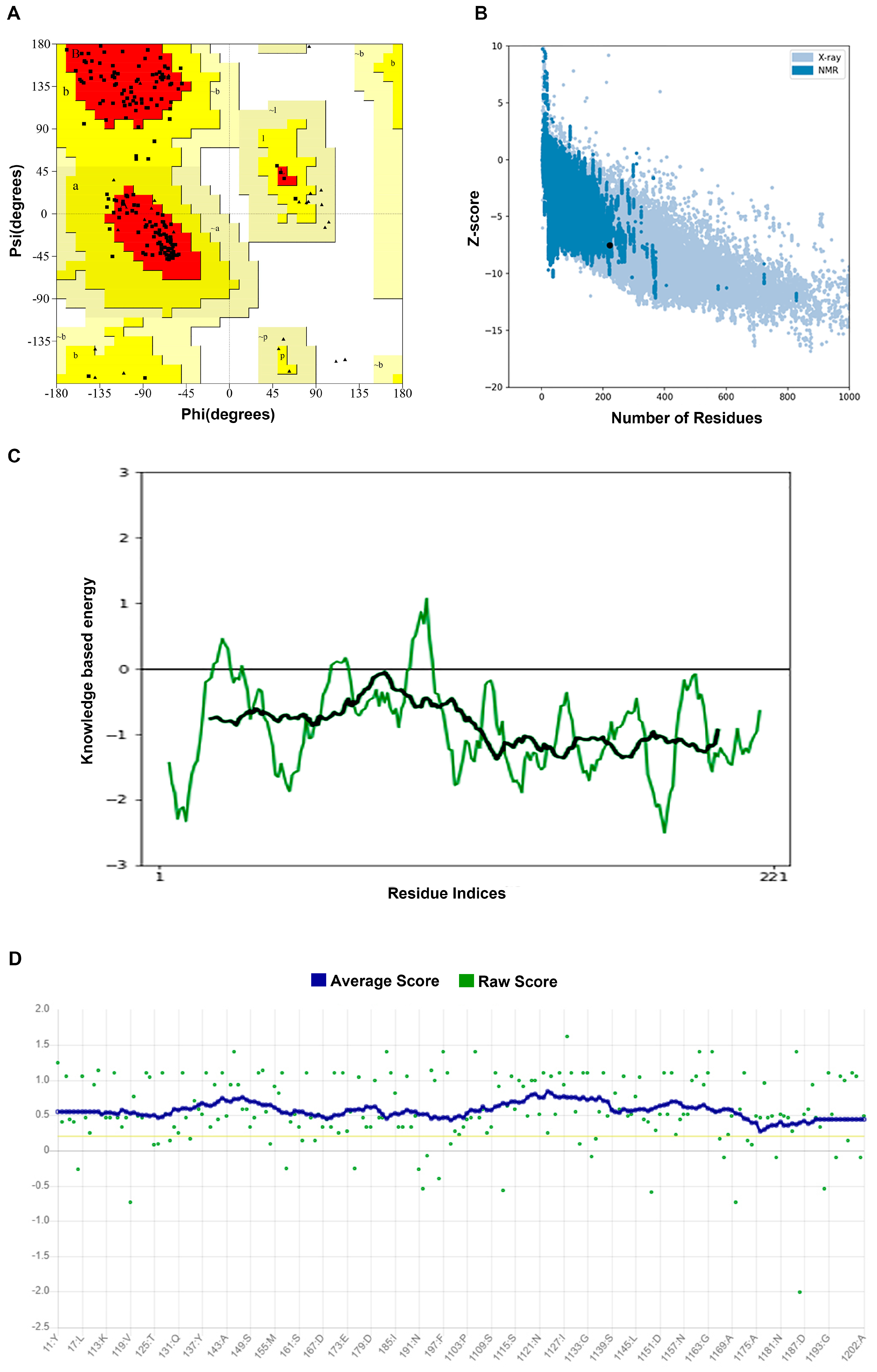
2.1. Structure Prediction and Evaluation
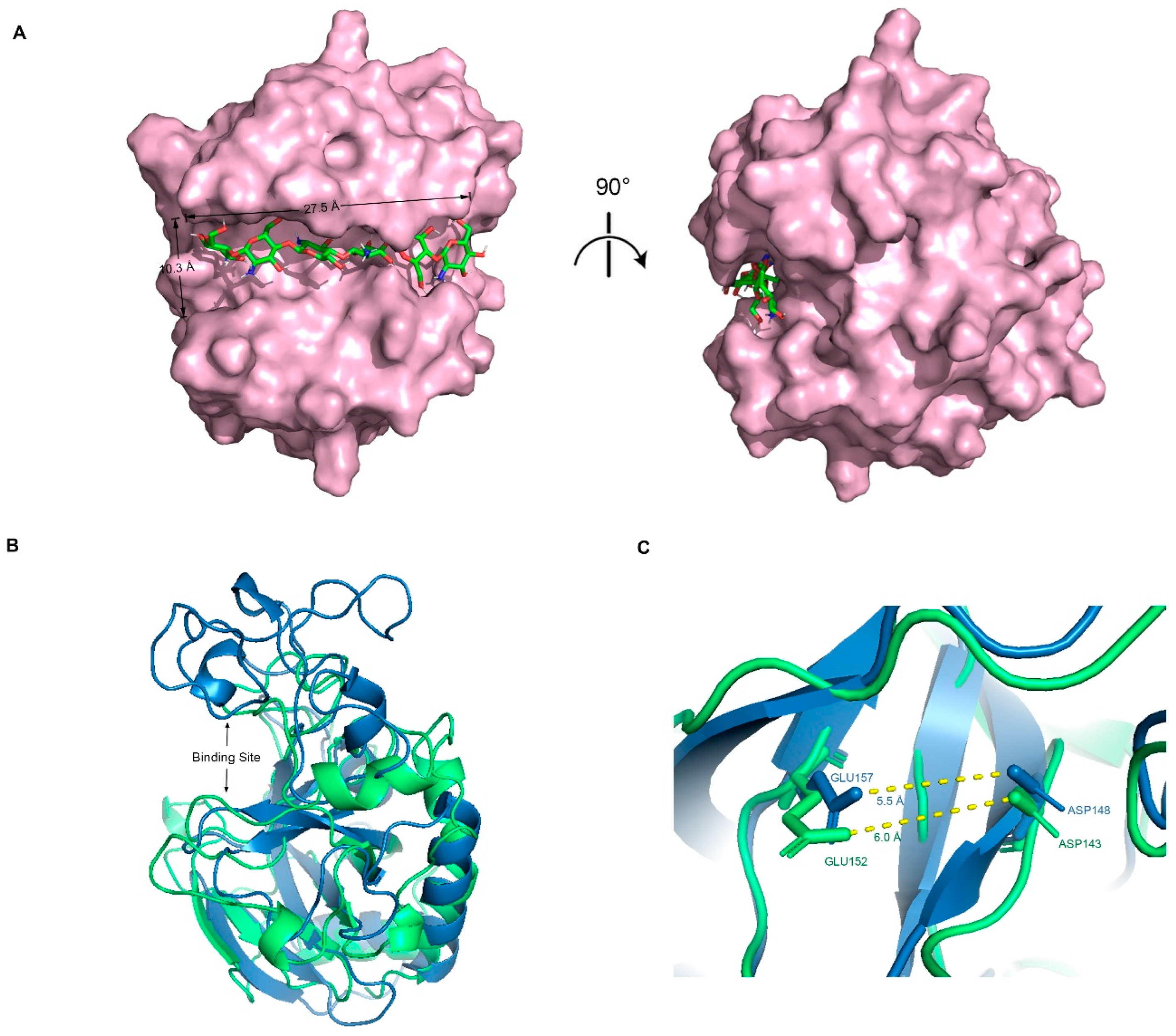
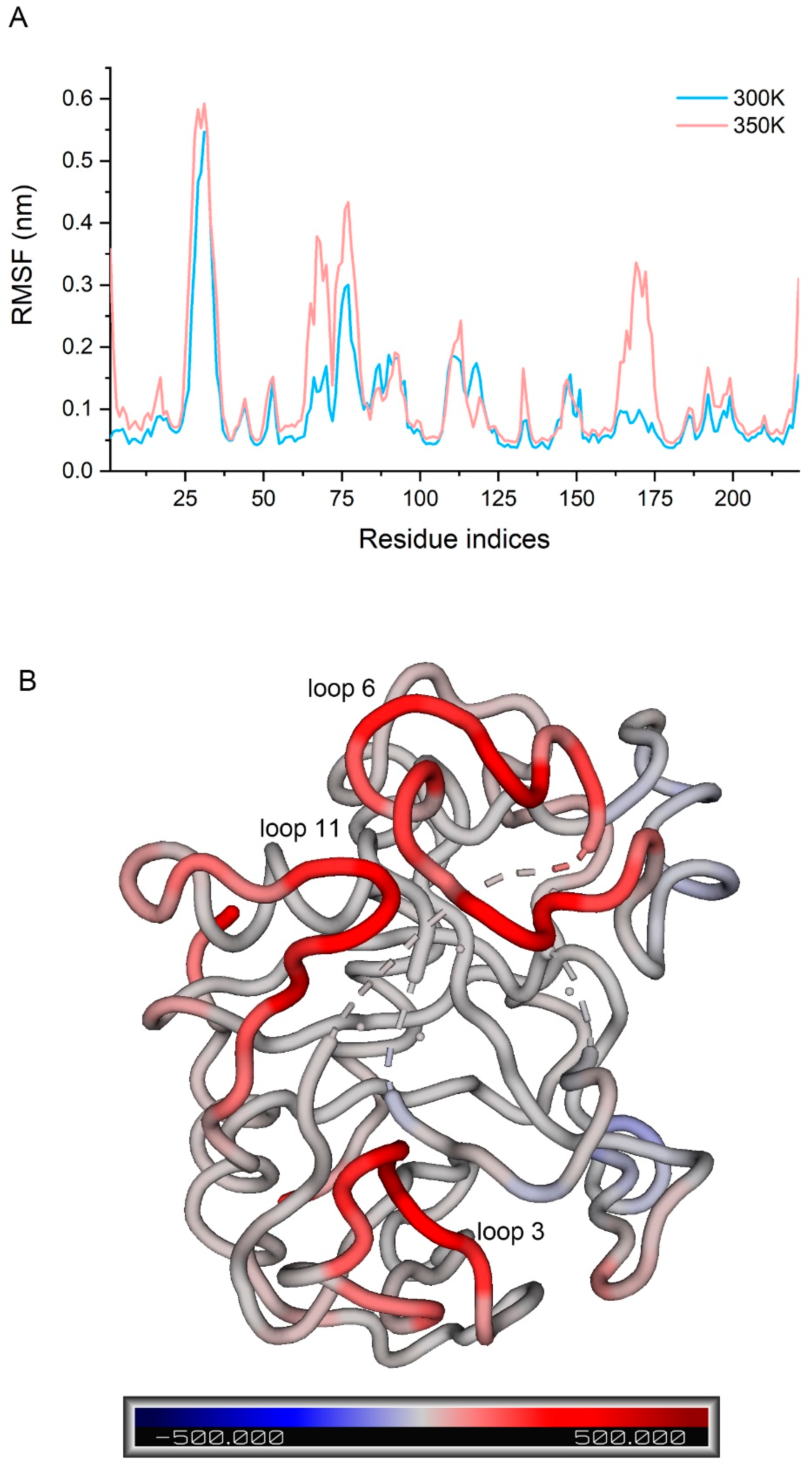
2.2. Structural Analysis and Catalytic Residues
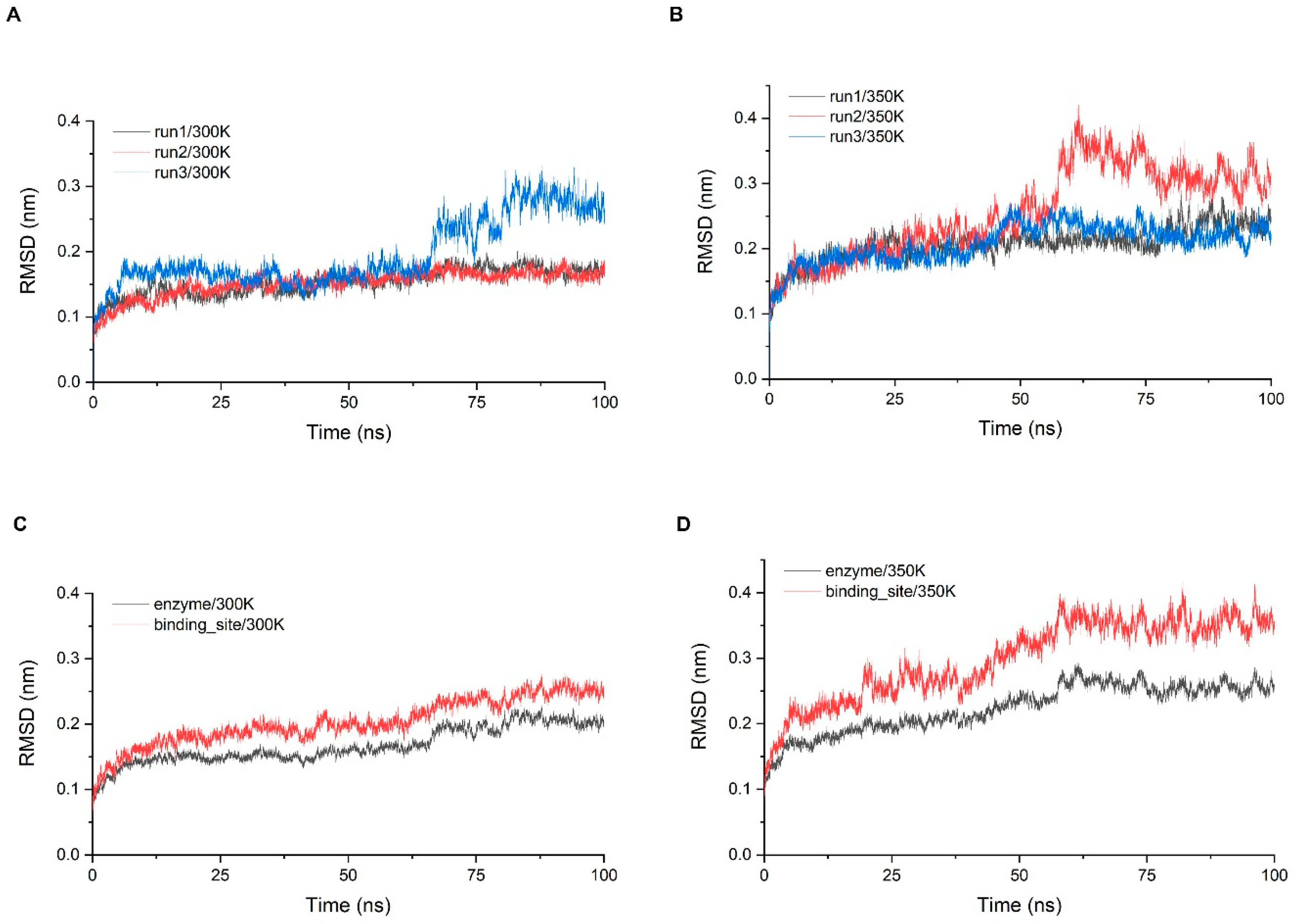
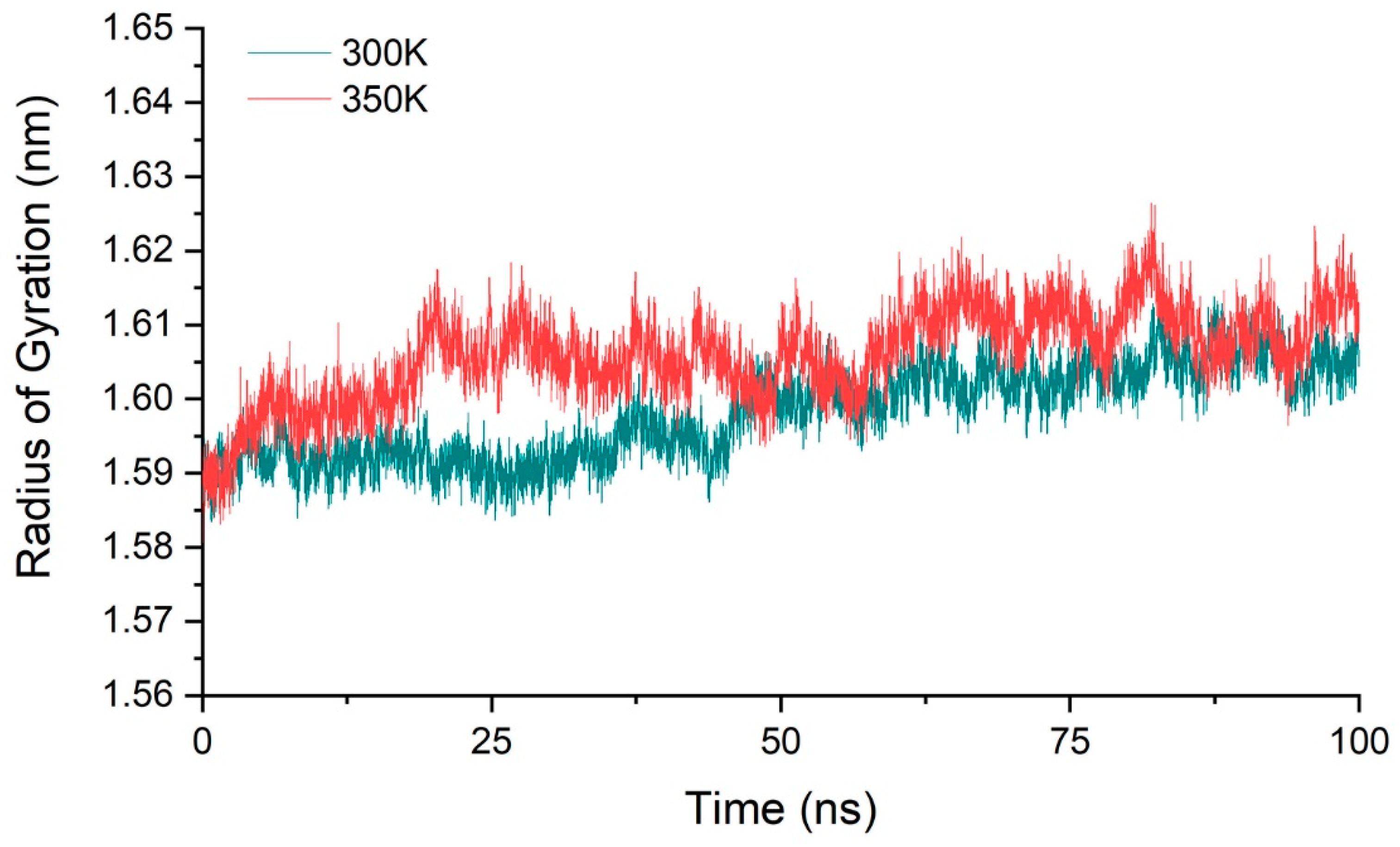
2.3. Stability of Simulation System
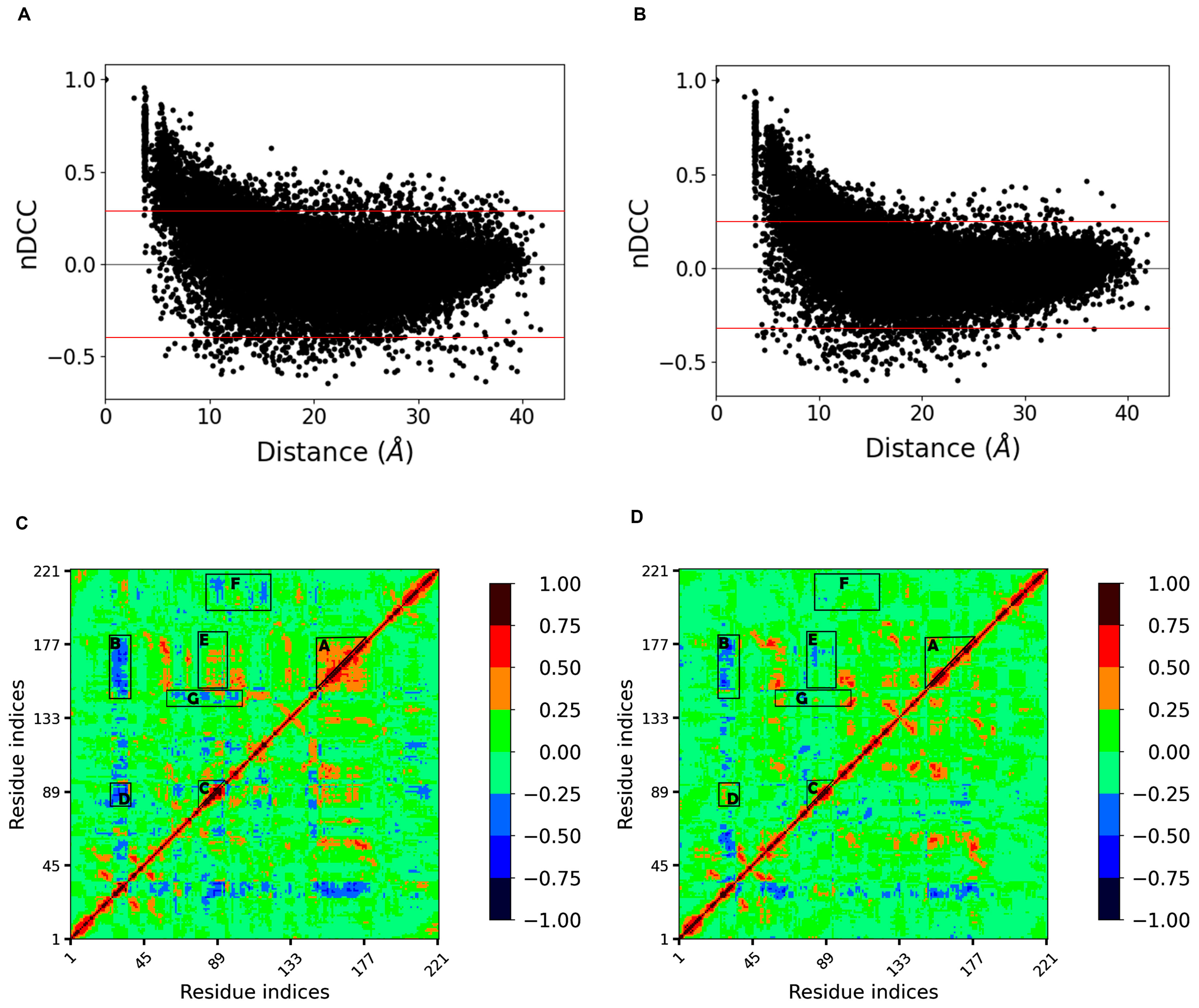
2.4. Dynamic Cross Correlation Analysis
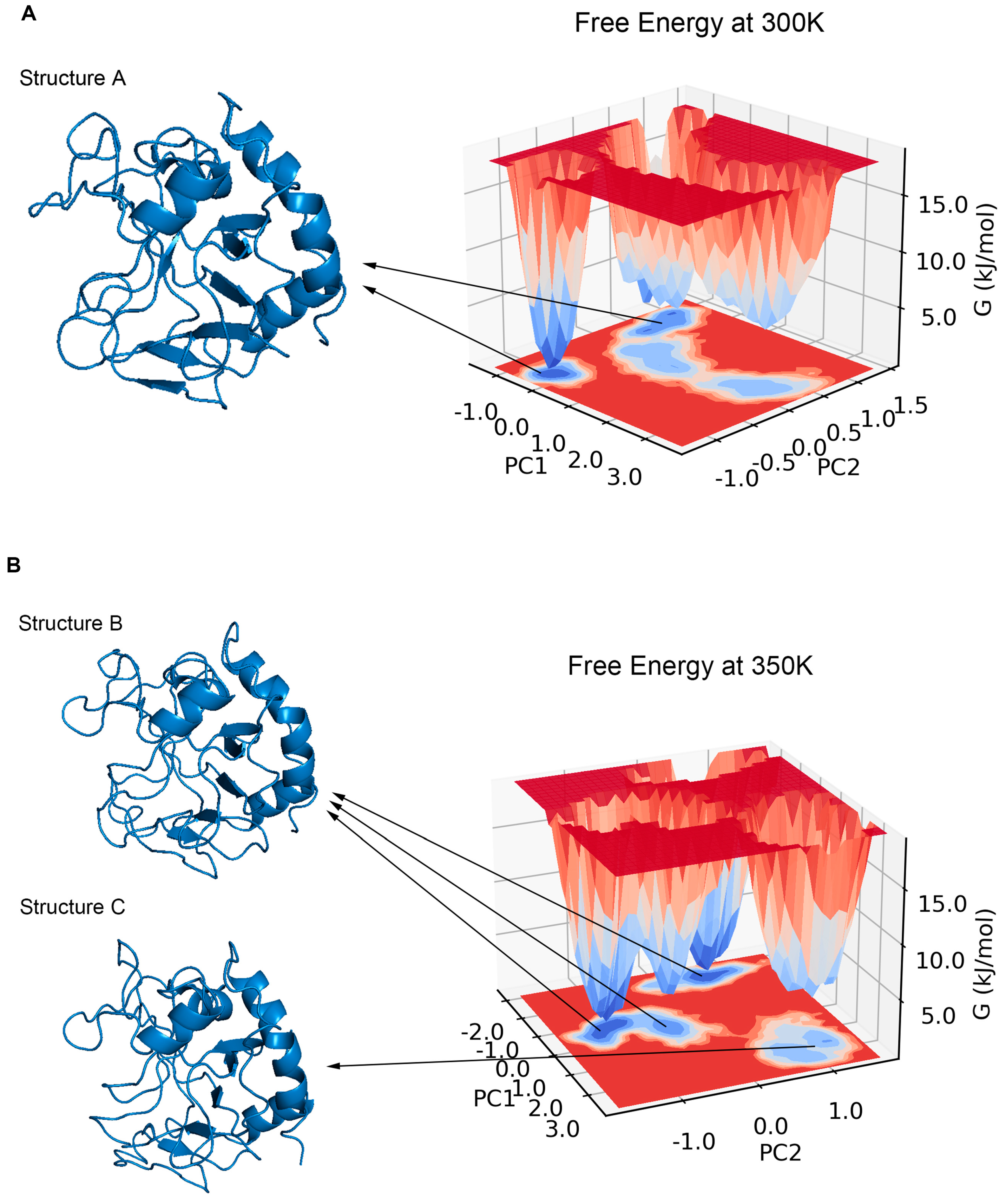
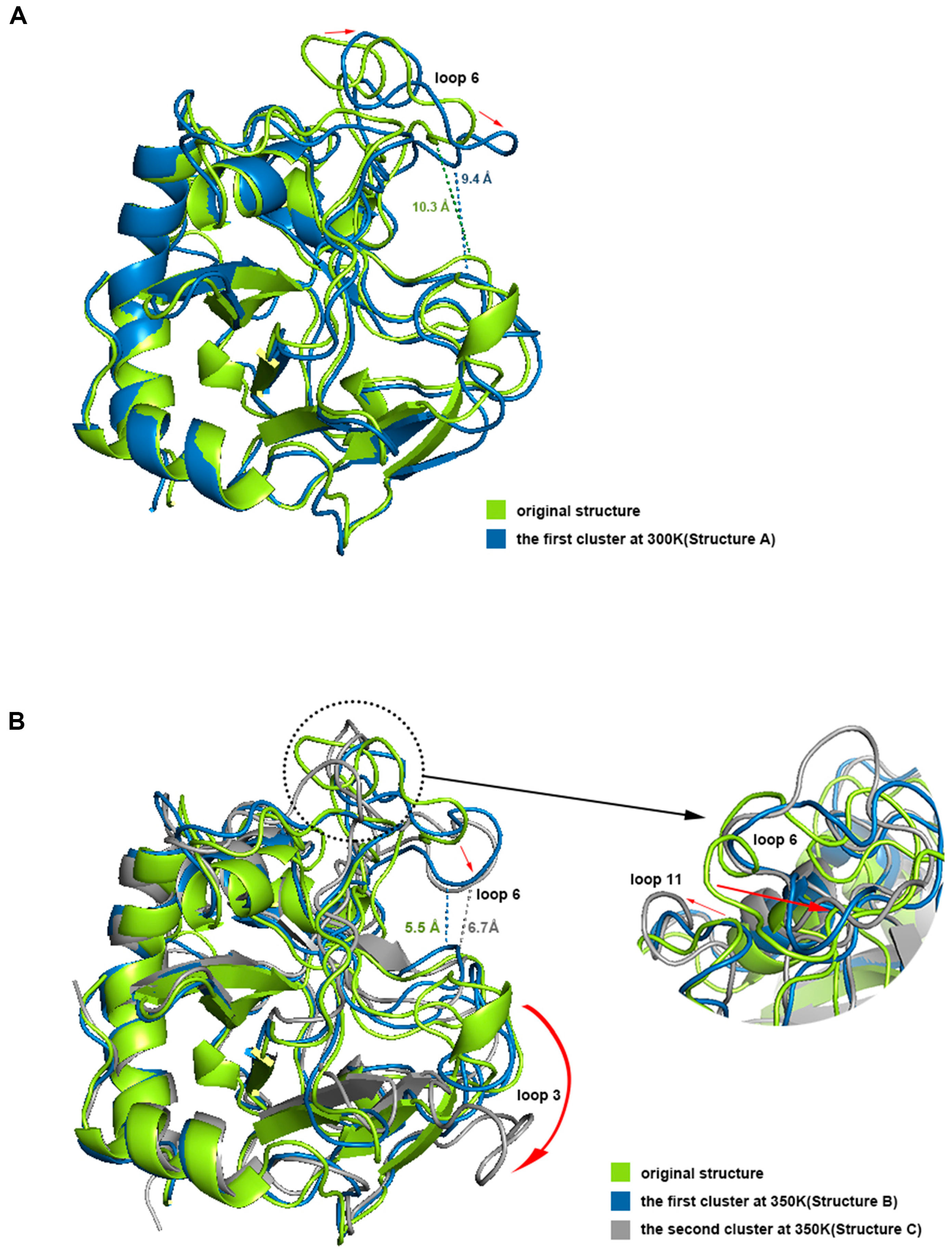
2.5. Major Conformation Analysis
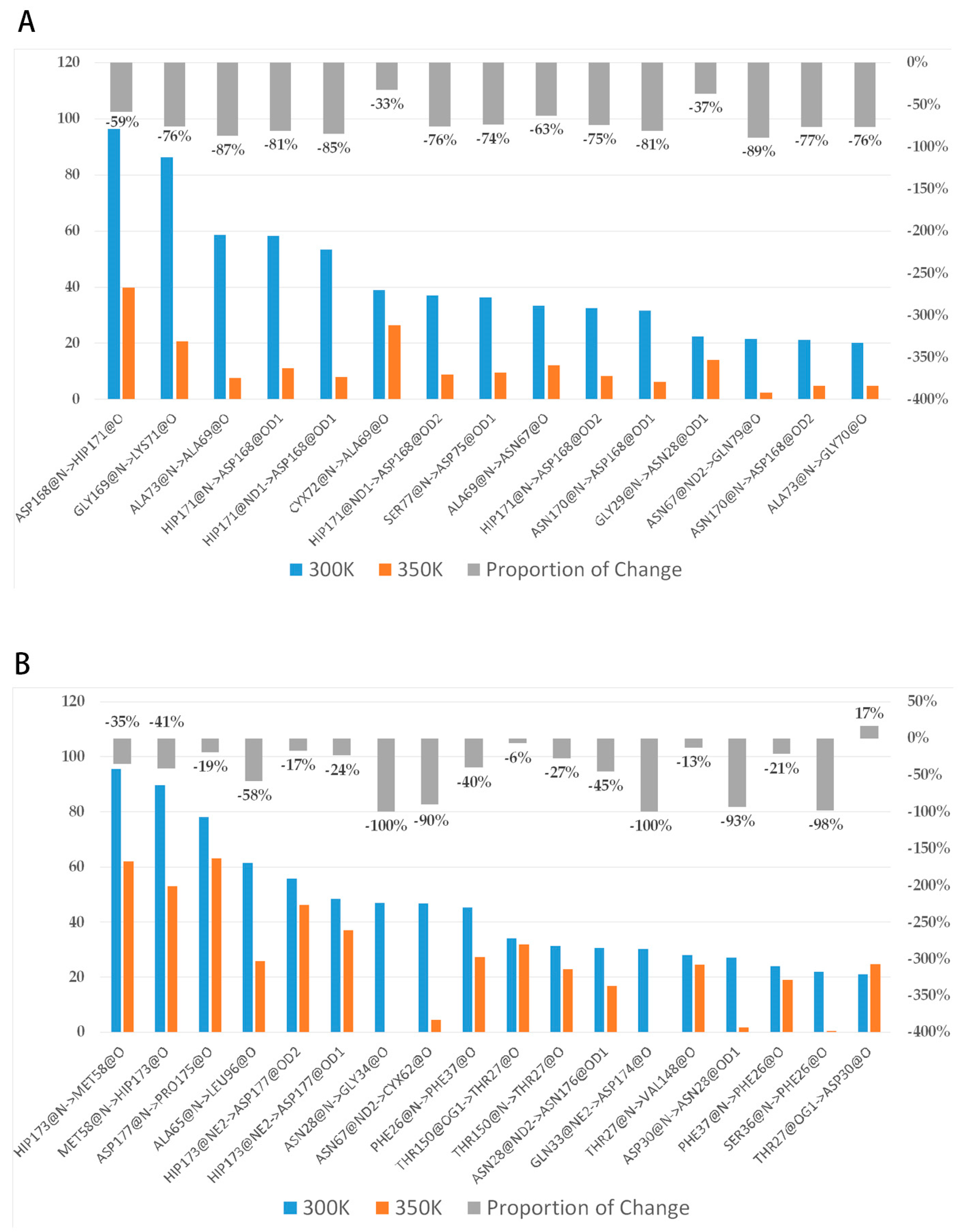
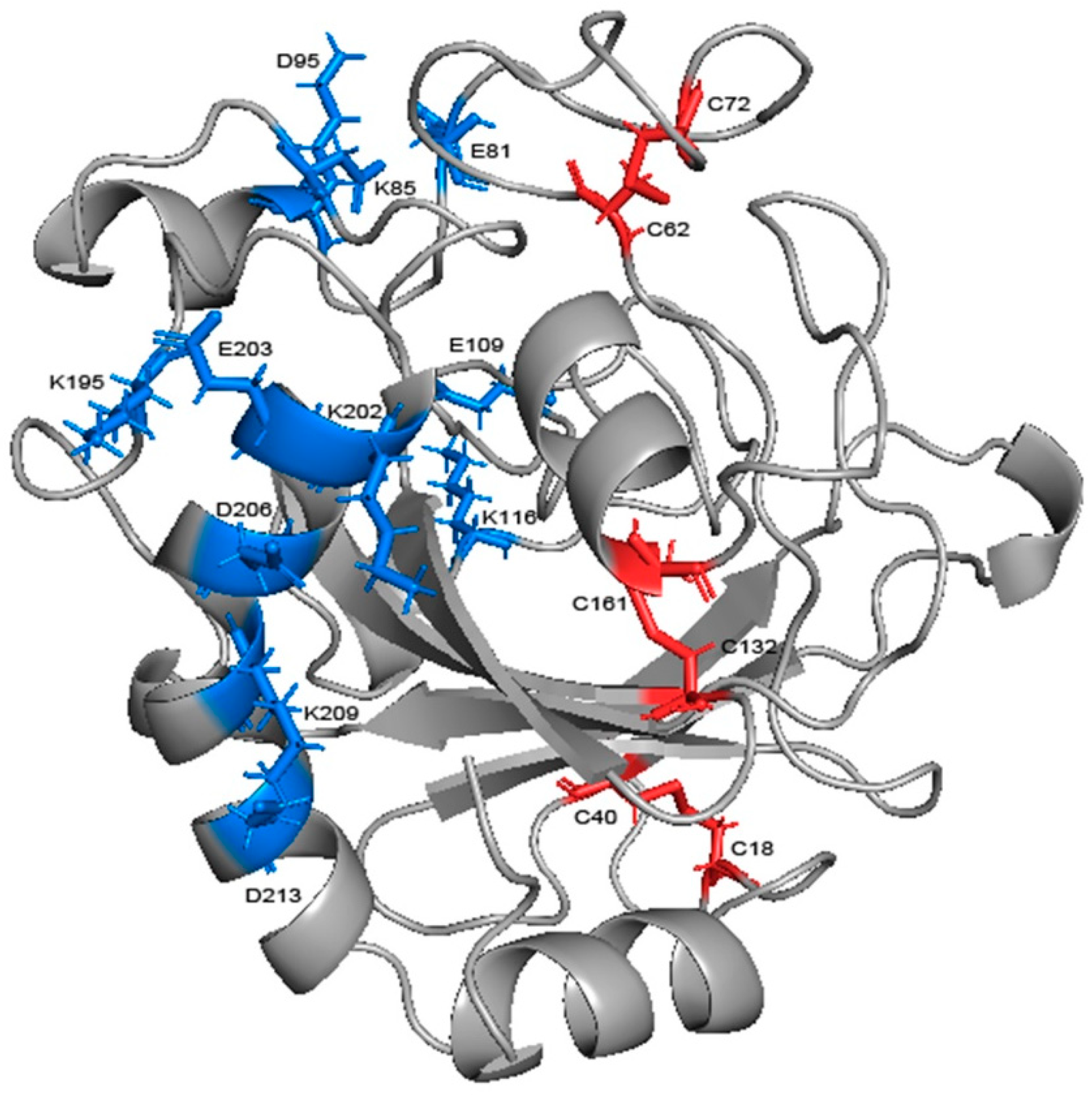
2.6. Residues Interaction Analysis
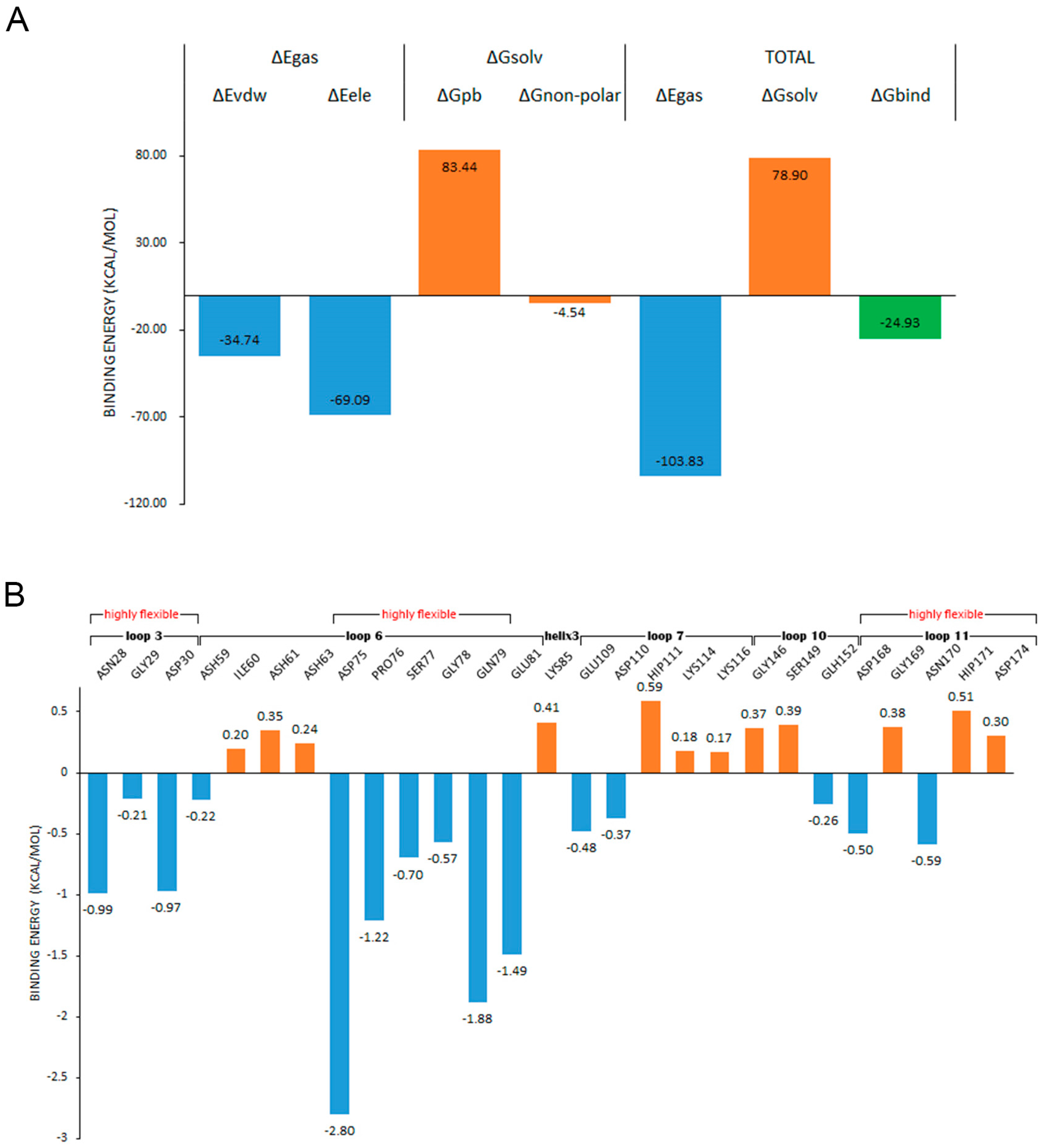
2.7. Binding Free Energy Analysis
3. Materials and Methods
3.1. Structural Prediction and Model Evaluation
3.2. Protonation, Binding Site Search, and Molecular Docking
3.3. Molecular Modeling and Molecular Dynamics Simulation
3.4. Analytical Methods
3.4.1. Conformational Fluctuation Analyses
3.4.2. Dynamic Cross Correlation
3.4.3. Principal Component Analysis, Free Energy Landscape and Cluster Analysis
3.4.4. Interaction Analysis
3.4.5. Binding Free Energy Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix B
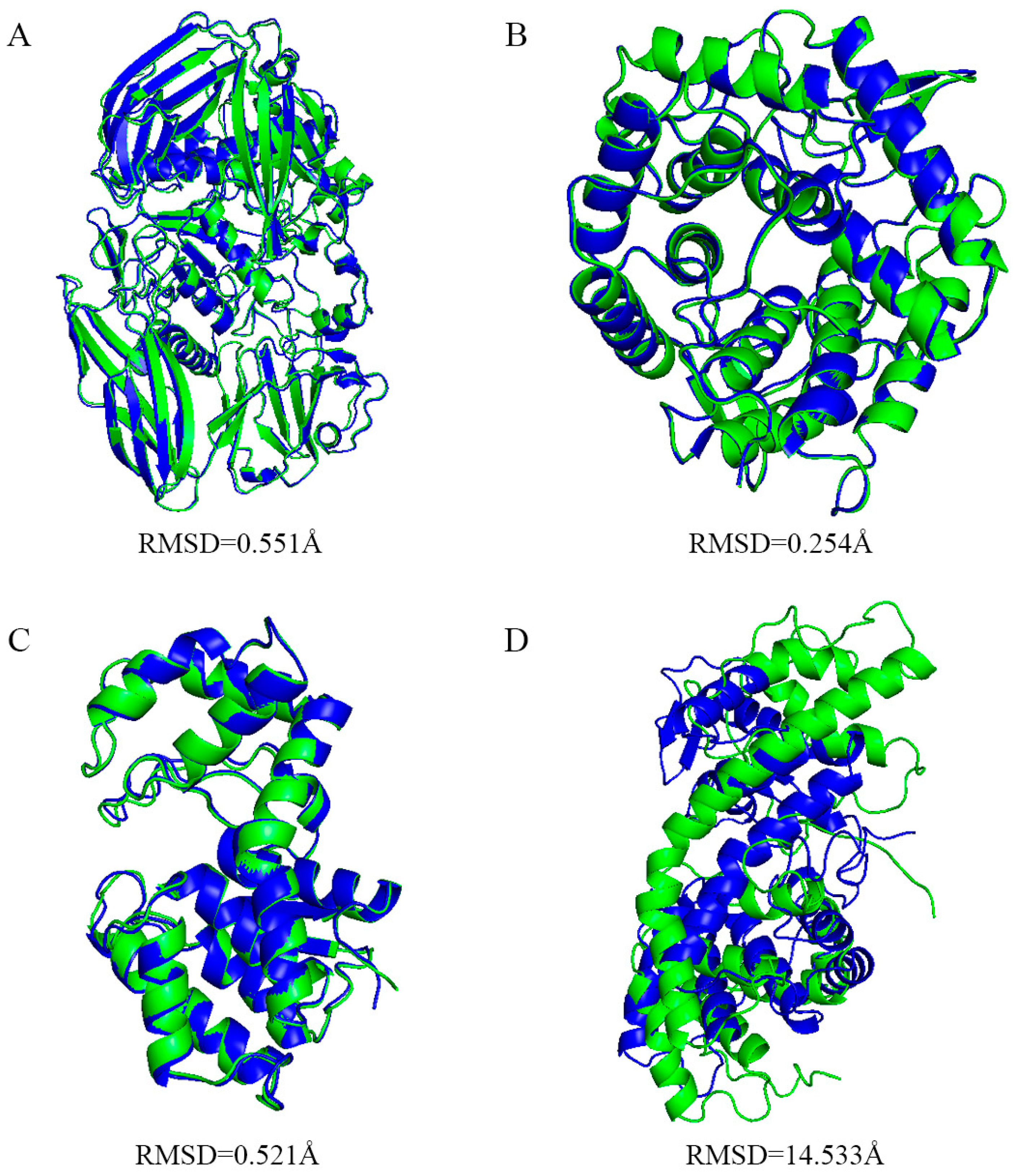


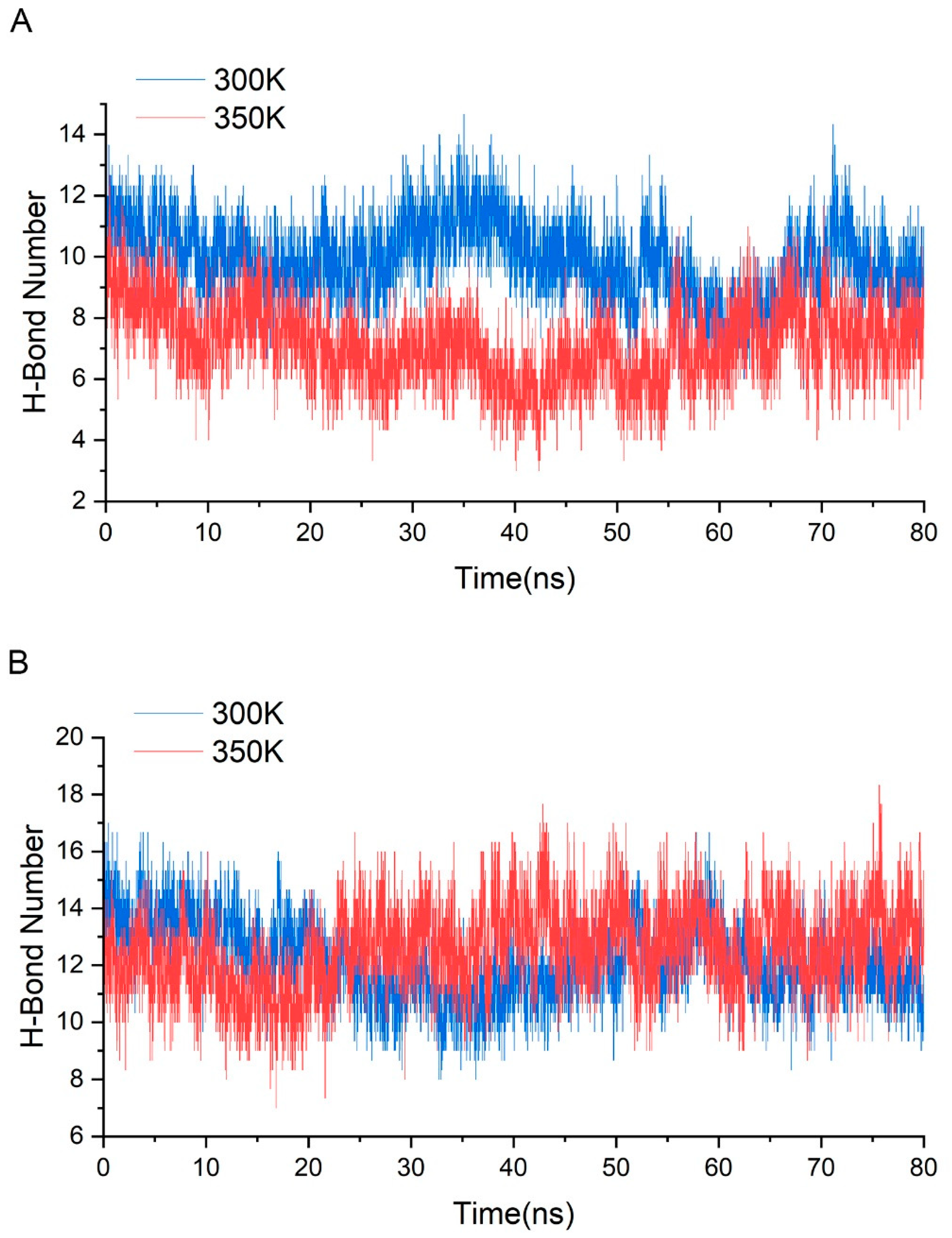

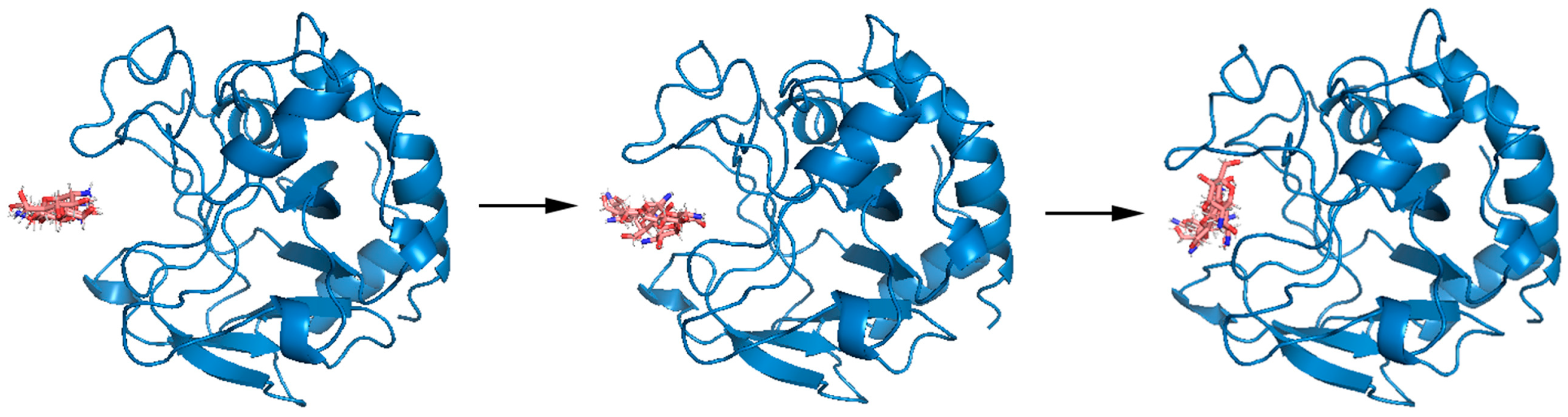
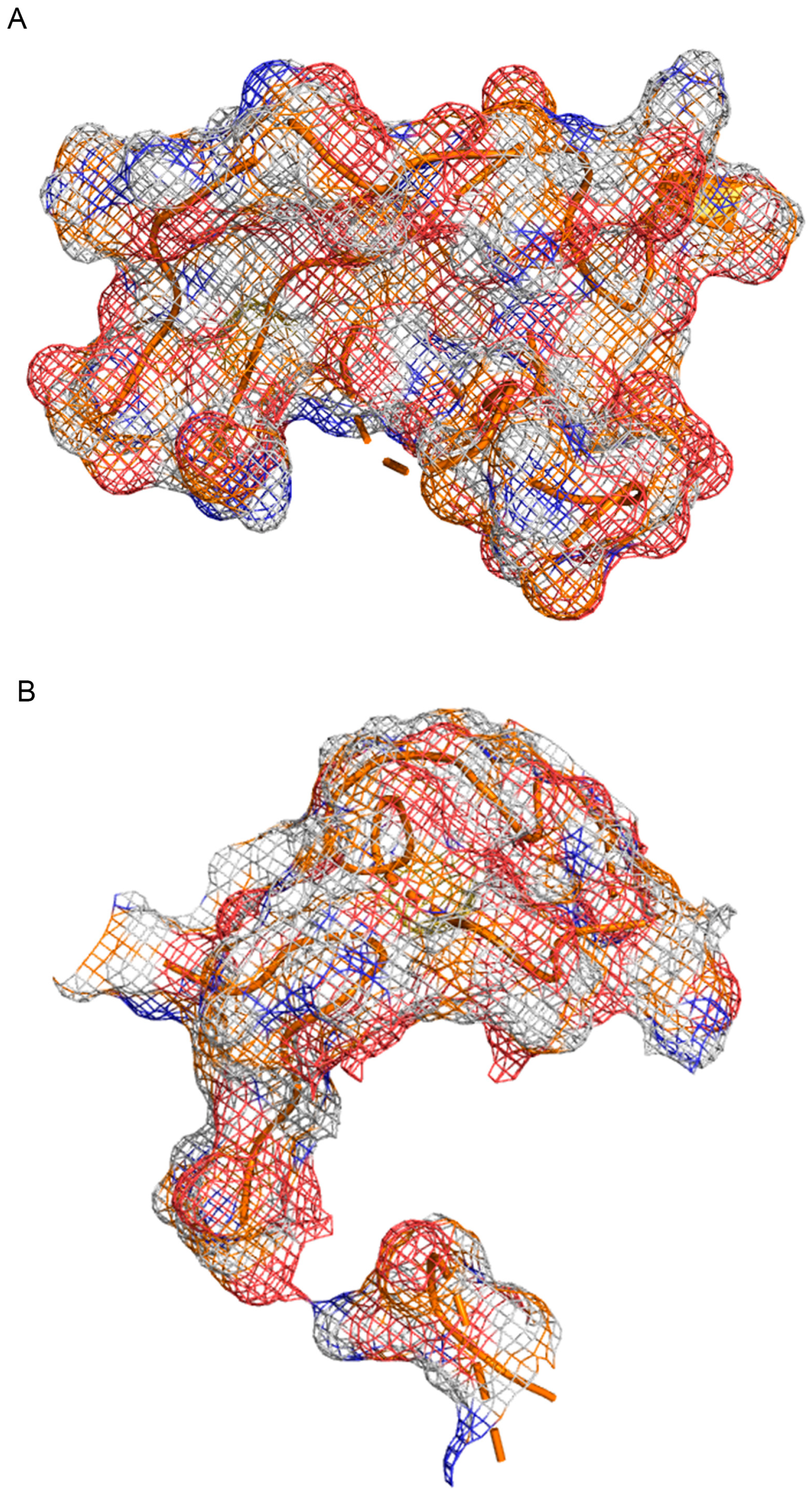
Appendix C
| Classification | Residue Composition |
|---|---|
| helix1-7 | ASN5-HID14, ALA31-GLN33, LYS85-PHE91, PRO117-HIE120, ILE155-CYS161, LYS186-ALA188, ALA201-GLY218 |
| sheet1-6 | LEU22-PHE26, PHE37-CYS40, ALA46-SER50, LEU125-CYS132,GLN135-TRP141, LEU179-PHE183 |
| loop1-14 | TYR1-PRO4, LYS15-VAL21, THR27-ASP30, GLY34-SER36, GLY41-GLY45, SER51-PHE84, GLY92-LYS116, GLY121-PRO124, ASN133-GLY134, GLY142-SER154, PHE162-VAL178, THR184-SER185, VAL189-ASN200, LEU219-ALA221 |
| Original | Protonated | Residue Number |
|---|---|---|
| ASP | ASH | 59, 61, 63, 143 |
| GLU | GLH | 152 |
| HIS | HID | 14 |
| HIS | HIE | 101, 120 |
| HIS | HIP | 111, 137, 166, 171, 173 |
| Residue | ASP61 | ASP63 | TYR103 | ASP143 | GLU152 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neighbours | 103 | 63 | 61 | 61 | 103 | 143 | 61 | 63 | 143 | 61 | 103 | 63 | 61 | 59 | 103 |
| Value | 4.09 | 2.36 | 2.18 | 2.36 | 2.02 | 1.73 | 4.09 | 2.02 | 1.92 | 2.18 | 1.92 | 1.73 | 1.58 | 1.19 | 0.98 |
| Binding Energy (kcal/mol) | Residue | RMSF Difference(nm) | Average of Differences(nm) |
|---|---|---|---|
| −2.80 | ASP75 | 0.07 | 0.04 |
| −1.88 | GLN79 | 0.12 | |
| −1.49 | GLU81 | 0.03 | |
| −1.22 | PRO76 | 0.13 | |
| −0.99 | ASN28 | 0.2 | |
| −0.97 | ASP30 | 0.07 | |
| −0.70 | SER77 | 0.13 | |
| −0.59 | ASN170 | 0.22 | |
| −0.57 | GLY78 | 0.15 | |
| −0.50 | ASP168 | 0.21 |
References
- Zhang, E.; Xing, R.; Liu, S.; Qin, Y.; Li, K.; Li, P. Advances in Chitosan-Based Nanoparticles for Oncotherapy. Carbohydr. Polym. 2019, 222, 115004. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Park, P.J.; Kim, S.K. Antimicrobial Effect of Chitooligosaccharides Produced by Bioreactor. Carbohydr. Polym. 2001, 44, 71–76. [Google Scholar] [CrossRef]
- Jeon, Y.J.; Kim, S.K. Antitumor Activity of Chitosan Oligosaccharides Produced in Ultrafiltration Membrance Reactor System. J. Microbiol. Biotechnol. 2002, 12, 503–507. [Google Scholar]
- Zeng, D.; Wu, J.; Kennedy, J.F. Application of a Chitosan Flocculant to Water Treatment. Carbohydr. Polym. 2008, 71, 135–139. [Google Scholar] [CrossRef]
- El Hadrami, A.; Adam, L.R.; El Hadrami, I.; Daayf, F. Chitosan in Plant Protection. Mar. Drugs 2010, 8, 968–987. [Google Scholar] [CrossRef]
- Dutta, P.K.; Tripathi, S.; Mehrotra, G.K.; Dutta, J. Perspectives for Chitosan Based Antimicrobial Films in Food Applications. Food Chem. 2009, 114, 1173–1182. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, H.; Liu, C.; Jiang, Y.; Yu, G.; Mu, X.; Wang, X. Magnetic Cellulose-Chitosan Hydrogels Prepared from Ionic Liquids as Reusable Adsorbent for Removal of Heavy Metal Ions. Chem. Commun. 2012, 48, 7350–7352. [Google Scholar] [CrossRef]
- Thadathil, N.; Velappan, S.P. Recent Developments in Chitosanase Research and Its Biotechnological Applications: A Review. Food Chem. 2014, 150, 392–399. [Google Scholar] [CrossRef]
- Lodhi, G.; Kim, Y.S.; Hwang, J.W.; Kim, S.K.; Jeon, Y.J.; Je, J.Y.; Ahn, C.B.; Moon, S.H.; Jeon, B.T.; Park, P.J. Chitooligosaccharide and Its Derivatives: Preparation and Biological Applications. Biomed Res. Int. 2014, 2014, 654913. [Google Scholar] [CrossRef]
- Zhu, X.F.; Tan, H.Q.; Zhu, C.; Liao, L.; Zhang, X.Q.; Wu, M. Cloning and Overexpression of a New Chitosanase Gene from Penicillium Sp. D-1. AMB Express 2012, 2, 13. [Google Scholar] [CrossRef]
- Saito, J.I.; Kita, A.; Higuchi, Y.; Nagata, Y.; Ando, A.; Miki, K. Crystal Structure of Chitosanase from Bacillus Circulans MH-K1 at 1.6-Å Resolution and Its Substrate Recognition Mechanism. J. Biol. Chem. 1999, 274, 30818–30825. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Chang, C.H.; Wu, Y.J.; Li, Y.K. Exploration of Glycosyl Hydrolase Family 75, a Chitosanase from Aspergillus fumigatus. J. Biol. Chem. 2006, 281, 3137–3144. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, Z.; Hong, Y.; Ni, J.; Sheng, D.; Shen, Y. Cloning, Expression and Characterization of a Thermostable Exo-β-D-Glucosaminidase from the Hyperthermophilic Archaeon Pyrococcus Horikoshii. Biotechnol. Lett. 2006, 28, 1655–1660. [Google Scholar] [CrossRef]
- van Bueren, A.L.; Ghinet, M.G.; Gregg, K.; Fleury, A.; Brzezinski, R.; Boraston, A.B. The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis. J. Mol. Biol. 2009, 385, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Adachi, W.; Sakihama, Y.; Shimizu, S.; Sunami, T.; Fukazawa, T.; Suzuki, M.; Yatsunami, R.; Nakamura, S.; Takénaka, A. Crystal Structure of Family GH-8 Chitosanase with Subclass II Specificity from Bacillus Sp. K17. J. Mol. Biol. 2004, 343, 785–795. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Li, Y.-K. An Aspergillus Chitosanase with Potential for Large-Scale Preparation of Chitosan Oligosaccharides. Biotechnol. Appl. Biochem. 2000, 32, 197–203. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The Carbohydrate-Active Enzyme Database: Functions and Literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Shimosaka, M.; Sato, K.; Nishiwaki, N.; Miyazawa, T.; Okazaki, M. Analysis of Essential Carboxylic Amino Acid Residues for Catalytic Activity of Fungal Chitosanases by Site-Directed Mutagenesis. J. Biosci. Bioeng. 2005, 100, 545–550. [Google Scholar] [CrossRef]
- Wu, R.; Smith, C.A.; Buchko, G.W.; Blaby, I.K.; Paez-Espino, D.; Kyrpides, N.C.; Yoshikuni, Y.; McDermott, J.E.; Hofmockel, K.S.; Cort, J.R.; et al. Structural Characterization of a Soil Viral Auxiliary Metabolic Gene Product—A Functional Chitosanase. Nat. Commun. 2022, 13, 5485. [Google Scholar] [CrossRef]
- Fukamizo, T.; Honda, Y.; Goto, S.; Boucher, I.; Brzezinski, R. Reaction Mechanism of Chitosanase from Streptomyces Sp. N174. Biochem. J. 1995, 311, 377–383. [Google Scholar] [CrossRef]
- Liu, Y.L.; Jiang, S.; Ke, Z.M.; Wu, H.S.; Chi, C.W.; Guo, Z.Y. Recombinant Expression of a Chitosanase and Its Application in Chitosan Oligosaccharide Production. Carbohydr. Res. 2009, 344, 815–819. [Google Scholar] [CrossRef]
- Singh, R.; Weikert, T.; Basa, S.; Moerschbacher, B.M. Structural and Biochemical Insight into Mode of Action and Subsite Specificity of a Chitosan Degrading Enzyme from Bacillus Spec. MN. Sci. Rep. 2019, 9, 1132. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Dong, Y.W.; Liao, M.L.; Meng, X.L.; Somero, G.N. Structural Flexibility and Protein Adaptation to Temperature: Molecular Dynamics Analysis of Malate Dehydrogenases of Marine Molluscs. Proc. Natl. Acad. Sci. USA 2018, 115, 1274–1279. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef] [PubMed]
- Montefiori, M.; Pilotto, S.; Marabelli, C.; Moroni, E.; Ferraro, M.; Serapian, S.A.; Mattevi, A.; Colombo, G. Impact of Mutations on NPAC Structural Dynamics: Mechanistic Insights from MD Simulations. J. Chem. Inf. Model. 2019, 59, 3927–3937. [Google Scholar] [CrossRef]
- Serapian, S.A.; Marchetti, F.; Triveri, A.; Morra, G.; Meli, M.; Moroni, E.; Sautto, G.A.; Rasola, A.; Colombo, G. The Answer Lies in the Energy: How Simple Atomistic Molecular Dynamics Simulations May Hold the Key to Epitope Prediction on the Fully Glycosylated SARS-CoV-2 Spike Protein. J. Phys. Chem. Lett. 2020, 11, 8084–8093. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L. The {PyMOL} Molecular Graphics System, Version~1.8; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Jeffryes, J.; Strutz, J.; Henry, C.; Tyo, K.E.J. Metabolic in Silico Network Expansions to Predict and Exploit Enzyme Promiscuity. Methods Mol. Biol. 2019, 1927, 11–21. [Google Scholar]
- Madan, B.; Lee, S.G. Sequence and Structural Features of Subsite Residues in GH10 and GH11 Xylanases. Biotechnol. Bioprocess Eng. 2018, 23, 311–318. [Google Scholar] [CrossRef]
- Tekpinar, M.; Neron, B.; Delarue, M. Extracting Dynamical Correlations and Identifying Key Residues for Allosteric Communication in Proteins by Correlationplus. J. Chem. Inf. Model. 2021, 61, 4832–4838. [Google Scholar] [CrossRef]
- Clarkson, M.W.; Gilmore, S.A.; Edgell, M.H.; Lee, A.L. Dynamic Coupling and Allosteric Behavior in a Nonallosteric Protein. Biochemistry 2006, 45, 7693–7699. [Google Scholar] [CrossRef]
- Zhuravleva, A.; Korzhnev, D.M.; Nolde, S.B.; Kay, L.E.; Arseniev, A.S.; Billeter, M.; Orekhov, V.Y. Propagation of Dynamic Changes in Barnase Upon Binding of Barstar: An NMR and Computational Study. J. Mol. Biol. 2007, 367, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.A.M.; Loccisano, A.E.; Firestine, S.M.; Evanseck, J.D. Chapter 13 Principal Components Analysis: A Review of Its Application on Molecular Dynamics Data. Annu. Rep. Comput. Chem. 2006, 2, 233–261. [Google Scholar]
- Vucinic, J.; Novikov, G.; Montanier, C.Y.; Dumon, C.; Schiex, T.; Barbe, S. A Comparative Study to Decipher the Structural and Dynamics Determinants Underlying the Activity and Thermal Stability of Gh-11 Xylanases. Int. J. Mol. Sci. 2021, 22, 5961. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Li, F.; Ding, M.; Zhang, R.; Shi, T.; Jiang, W. Molecular Dynamic Simulation: Structural Insights of Multi-Stranded Curdlan in Aqueous Solution. Carbohydr. Polym. 2021, 261, 117844. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.T.; Hung, H.M.; Tran, K.N.; Nguyen, M.T. Replica Exchange Molecular Dynamics Study of the Amyloid Beta (11-40) Trimer Penetrating a Membrane. RSC Adv. 2017, 7, 7346–7357. [Google Scholar] [CrossRef]
- Pace, C.N.; Shirley, B.A.; McNutt, M.; Gajiwala, K. Forces Contributing to the Conformational Stability of Proteins. FASEB J. 1996, 10, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Fu, H.; Fryar, K.L.; Landua, J.; Trevino, S.R.; Schell, D.; Thurlkill, R.L.; Imura, S.; Scholtz, J.M.; Gajiwala, K.; et al. Contribution of Hydrogen Bonds to Protein Stability. Protein Sci. 2014, 23, 652–661. [Google Scholar] [CrossRef]
- de Souza, A.R.; de Araújo, G.C.; Zanphorlin, L.M.; Ruller, R.; Franco, F.C.; Torres, F.A.G.; Mertens, J.A.; Bowman, M.J.; Gomes, E.; Da Silva, R. Engineering Increased Thermostability in the GH-10 Endo-1, 4-β-Xylanase from Thermoascus Aurantiacus CBMAI 756. Int. J. Biol. Macromol. 2016, 93, 20–26. [Google Scholar] [CrossRef]
- Gruber, K.; Klintschar, G.; Hayn, M.; Schlacher, A.; Steiner, W.; Kratky, C. Thermophilic Xylanase from Thermomyces Lanuginosus: High-Resolution X- Ray Structure and Modeling Studies. Biochemistry 1998, 37, 13475–13485. [Google Scholar] [CrossRef]
- Vieira, D.S.; Degrève, L. An Insight into the Thermostability of a Pair of Xylanases: The Role of Hydrogen Bonds. Mol. Phys. 2009, 107, 59–69. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, Y.; Li, X.; Bai, Y.; Xia, W.; Ma, R.; Luo, H.; Shi, P.; Yao, B. Impact of Disulfide Bonds on the Folding and Refolding Capability of a Novel Thermostable GH45 Cellulase. Appl. Microbiol. Biotechnol. 2018, 102, 9183–9192. [Google Scholar] [CrossRef]
- Wang, C.; Greene, D.; Xiao, L.; Qi, R.; Luo, R. Recent Developments and Applications of the MMPBSA Method. Front. Mol. Biosci. 2018, 4, 87. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022 196 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Rullmann, J.A.C.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for Checking the Quality of Protein Structures Solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Bowie, J.U.; Lüthy, R.; Eisenberg, D. A Method to Identify Protein Sequences That Fold into a Known Three-Dimensional Structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of Protein Models with Three-Dimensional Profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Sippl, M.J. Recognition of Errors in Three-dimensional Structures of Proteins. Proteins Struct. Funct. Bioinforma. 1993, 17, 355–362. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Gordon, J.C.; Myers, J.B.; Folta, T.; Shoja, V.; Heath, L.S.; Onufriev, A. H++: A Server for Estimating PKas and Adding Missing Hydrogens to Macromolecules. Nucleic Acids Res. 2005, 33, W368–W371. [Google Scholar] [CrossRef]
- Myers, J.; Grothaus, G.; Narayanan, S.; Onufriev, A. A Simple Clustering Algorithm Can Be Accurate Enough for Use in Calculations of PKs in Macromolecules. Proteins Struct. Funct. Genet. 2006, 63, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Anandakrishnan, R.; Aguilar, B.; Onufriev, A.V. H++ 3.0: Automating PK Prediction and the Preparation of Biomolecular Structures for Atomistic Molecular Modeling and Simulations. Nucleic Acids Res. 2012, 40, W537–W541. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, J.; Doerr, S.; Martínez-Rosell, G.; Rose, A.S.; De Fabritiis, G. DeepSite: Protein-Binding Site Predictor Using 3D-Convolutional Neural Networks. Bioinformatics 2017, 33, 3036–3042. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An Overview of the Amber Biomolecular Simulation Package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber Biomolecular Simulation Programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. Ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 26, 114. [Google Scholar] [CrossRef]
- Izadi, S.; Anandakrishnan, R.; Onufriev, A.V. Building Water Models: A Different Approach. J. Phys. Chem. Lett. 2014, 5, 3863–3871. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle: An Analytical Version of the SHAKE and RATTLE Algorithm for Rigid Water Models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N⋅log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Lemak, A.S.; Balabaev, N.K. On the Berendsen Thermostat. Mol. Simul. 1994, 13, 177–187. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; Van Gunsteren, W.F.; Mark, A.E. Peptide Folding: When Simulation Meets Experiment. Angew. Chemie-Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Miller, D.W.; Agard, D.A. Enzyme Specificity under Dynamic Control: A Normal Mode Analysis of α-Lytic Protease. J. Mol. Biol. 1999, 286, 267–278. [Google Scholar] [CrossRef] [PubMed]

| Type | Hydrogen Bond (Donor->Acceptor) | 300 K (%) | 350 K (%) | Difference (%) | Percentage (%) |
|---|---|---|---|---|---|
| Intra-Group | GLY64@N->GLN79@O | 3.18 | 31.77 | 28.59 | 899 |
| HIP171@N->ASP168@O | 0.06 | 22.54 | 22.48 | 37,400 | |
| CYX72@N->SER68@O | 4.5 | 25.69 | 21.19 | 471 | |
| Inter-Group | GLN79@NE2->GLU81@O | 0.64 | 34.1 | 33.46 | 5228 |
| ALA65@N->GLY80@O | 0 | 30.79 | 30.79 | brand new | |
| LYS35@N->ALA31@O | 0.03 | 30.39 | 30.36 | 101,200 | |
| ILE60@N->ASN170@O | 0 | 24.33 | 24.33 | brand new | |
| PHE26@N->GLY34@O | 0 | 22.82 | 22.82 | brand new | |
| ASN57@ND2->HIP173@O | 6.01 | 26.49 | 20.48 | 341 |
| Salt Bridge | 300 K (%) | 350 K (%) | Difference (%) | Percentage (%) |
|---|---|---|---|---|
| GLU81-LYS85 | 36.60 | 30.64 | −5.95 | −16.27 |
| LYS85-ASP95 | 19.73 | 30.30 | 10.57 | 53.60 |
| GLU109-LYS116 | 34.29 | 43.00 | 8.70 | 25.38 |
| LYS195-GLU203 | 50.74 | 45.75 | −4.99 | −9.83 |
| LYS202-ASP206 | 4.01 | 20.23 | 16.22 | 404.25 |
| LYS209-ASP213 | 87.28 | 73.61 | −13.66 | −15.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Liu, S.; Li, K.; Xing, R.; Chen, X.; Li, P. A Computational Biology Study on the Structure and Dynamics Determinants of Thermal Stability of the Chitosanase from Aspergillus fumigatus. Int. J. Mol. Sci. 2023, 24, 6671. https://doi.org/10.3390/ijms24076671
Wang Q, Liu S, Li K, Xing R, Chen X, Li P. A Computational Biology Study on the Structure and Dynamics Determinants of Thermal Stability of the Chitosanase from Aspergillus fumigatus. International Journal of Molecular Sciences. 2023; 24(7):6671. https://doi.org/10.3390/ijms24076671
Chicago/Turabian StyleWang, Qian, Song Liu, Kecheng Li, Ronge Xing, Xiaolin Chen, and Pengcheng Li. 2023. "A Computational Biology Study on the Structure and Dynamics Determinants of Thermal Stability of the Chitosanase from Aspergillus fumigatus" International Journal of Molecular Sciences 24, no. 7: 6671. https://doi.org/10.3390/ijms24076671
APA StyleWang, Q., Liu, S., Li, K., Xing, R., Chen, X., & Li, P. (2023). A Computational Biology Study on the Structure and Dynamics Determinants of Thermal Stability of the Chitosanase from Aspergillus fumigatus. International Journal of Molecular Sciences, 24(7), 6671. https://doi.org/10.3390/ijms24076671







