Curcumin and Plumbagin Synergistically Target the PI3K/Akt/mTOR Pathway: A Prospective Role in Cancer Treatment
Abstract
1. Introduction
2. Results and Discussion
2.1. Sequential Molecular Docking Determines the Synergy of Curcumin and Plumbagin
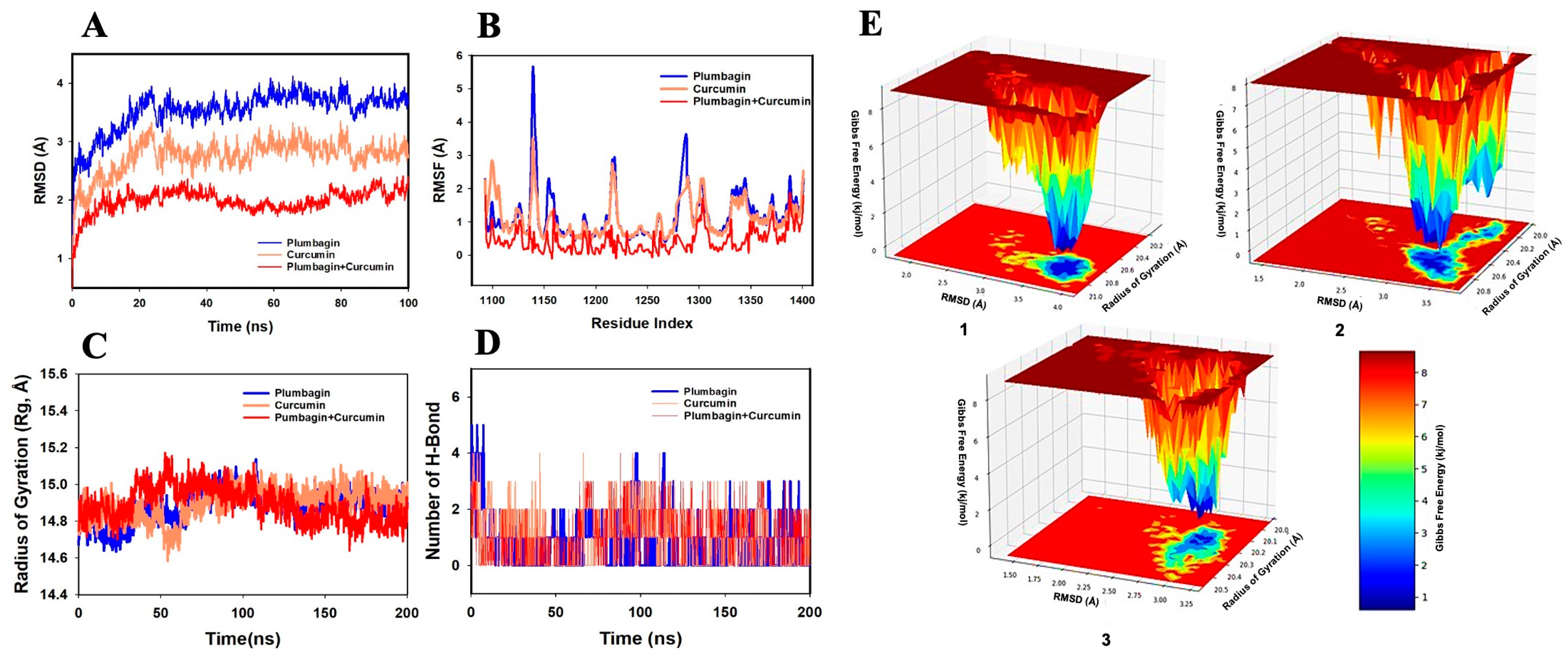
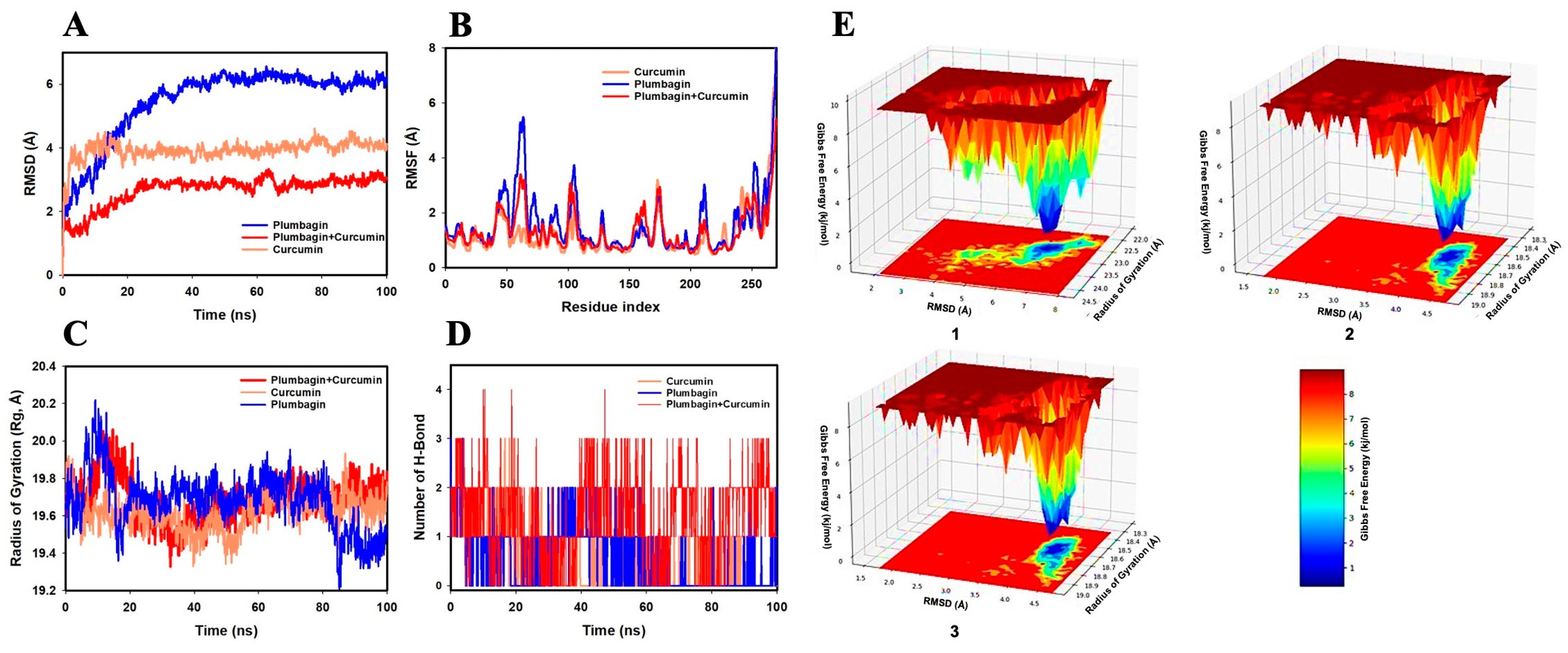
2.2. MD Simulation Analysis of Curcumin and Plumbagin Targeting PI3K/Akt/mTOR
2.3. MM-GBSA Analysis of Curcumin and Plumbagin Synergistic Targeting of PI3K/Akt/mTOR
3. Materials and Methods
3.1. Preparation of Target Proteins
3.2. Preparation of Ligands
3.3. Molecular Docking
3.4. MD Simulation
3.5. Binding Free Energy Analysis
- ΔGbind denotes the binding free energy;
- ΔGMM denotes the difference between the free energies of ligand–protein complexes and the total energies of protein and ligand in isolation;
- ΔGSolv denotes the difference in the GSA solvation energies of the ligand–protein complex and the sum of the solvation energies of the protein and the ligand in the unbound state;
- ΔGSA denotes the difference in the surface area energies for the protein and the ligand.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The Ever-increasing Importance of Cancer as a Leading Cause of Premature Death Worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef]
- Sever, R.; Brugge, J.S. Signal Transduction in Cancer. Cold Spring Harb. Perspect. Med. 2015, 5, a006098. [Google Scholar] [CrossRef]
- Islam, B.U.; Khan, M.S.; Husain, F.M.; Rehman, M.T.; Zughaibi, T.A.; Abuzenadah, A.M.; Urooj, M.; Kamal, M.A.; Tabrez, S. MTOR Targeted Cancer Chemoprevention by Flavonoids. Curr. Med. Chem. 2021, 28, 8068–8082. [Google Scholar] [CrossRef]
- Muhammad, N.; Usmani, D.; Tarique, M.; Naz, H.; Ashraf, M.; Raliya, R.; Tabrez, S.; Zughaibi, T.A.; Alsaieedi, A.; Hakeem, I.J.; et al. The Role of Natural Products and Their Multitargeted Approach to Treat Solid Cancer. Cells 2022, 11, 2209. [Google Scholar] [CrossRef]
- Zughaibi, T.A.; Suhail, M.; Tarique, M.; Tabrez, S. Targeting PI3K/Akt/MTOR Pathway by Different Flavonoids: A Cancer Chemopreventive Approach. Int. J. Mol. Sci. 2021, 22, 12455. [Google Scholar] [CrossRef]
- Suhail, M.; Khan, M.S.; Ahmad, A.; Zughaibi, T.A.; Husain, F.M.; Rehman, M.T.; Tabrez, S. Flavonoids and PI3K/Akt/MTOR Signaling Cascade: A Potential Crosstalk in Anticancer Treatment. Curr. Med. Chem. 2021, 28, 8083–8097. [Google Scholar] [CrossRef]
- Tan, A.C. Targeting the PI3K/Akt/MTOR Pathway in Non-small Cell Lung Cancer (NSCLC). Thorac. Cancer 2020, 11, 511–518. [Google Scholar] [CrossRef]
- Alam, S.S.M.; Uddin, F.; Khan, F.B.; Kamal, M.A.; Hoque, M. Therapeutic and Pharmacological Potential of Tanshinones against Lung Cancer: A Systematic Review. Phytomed. Plus 2022, 2, 100202. [Google Scholar] [CrossRef]
- Uddin, F.; Hoque, M. Non-Flavonoids Targeting Cancer Stem Cells: A Promising Therapeutic Avenue for Cancer Treatment. In Polyphenols-Based Nanotherapeutics for Cancer Management; Tabrez, S., Imran Khan, M., Eds.; Springer: Singapore, 2021; pp. 289–334. ISBN 9789811649349. [Google Scholar]
- Farghadani, R.; Naidu, R. Curcumin: Modulator of Key Molecular Signaling Pathways in Hormone-Independent Breast Cancer. Cancers 2021, 13, 3427. [Google Scholar] [CrossRef]
- Hamzehzadeh, L.; Atkin, S.L.; Majeed, M.; Butler, A.E.; Sahebkar, A. The Versatile Role of Curcumin in Cancer Prevention and Treatment: A Focus on PI3K/AKT Pathway. J. Cell Physiol. 2018, 233, 6530–6537. [Google Scholar] [CrossRef]
- Mansouri, K.; Rasoulpoor, S.; Daneshkhah, A.; Abolfathi, S.; Salari, N.; Mohammadi, M.; Rasoulpoor, S.; Shabani, S. Clinical Effects of Curcumin in Enhancing Cancer Therapy: A Systematic Review. BMC Cancer 2020, 20, 791. [Google Scholar] [CrossRef]
- Zoi, V.; Galani, V.; Lianos, G.D.; Voulgaris, S.; Kyritsis, A.P.; Alexiou, G.A. The Role of Curcumin in Cancer Treatment. Biomedicines 2021, 9, 1086. [Google Scholar] [CrossRef]
- Roy, A. Plumbagin: A Potential Anti-Cancer Compound. Mini Rev. Med. Chem. 2021, 21, 731–737. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Z.; You, W.; Zhou, F.; Guo, Z.; Qian, K.; Xiao, Y.; Wang, X. Suppressive Effects of Plumbagin on the Growth of Human Bladder Cancer Cells via PI3K/AKT/MTOR Signaling Pathways and EMT. Cancer Cell Int. 2020, 20, 520. [Google Scholar] [CrossRef]
- Zhang, X.; Kan, H.; Liu, Y.; Ding, W. Plumbagin Induces Ishikawa Cell Cycle Arrest, Autophagy, and Apoptosis via the PI3K/Akt Signaling Pathway in Endometrial Cancer. Food Chem. Toxicol. 2021, 148, 111957. [Google Scholar] [CrossRef]
- Mokhtari, R.B.; Homayouni, T.S.; Baluch, N.; Morgatskaya, E.; Kumar, S.; Das, B.; Yeger, H. Combination Therapy in Combating Cancer. Oncotarget 2017, 8, 38022–38043. [Google Scholar] [CrossRef]
- Tan, B.L.; Norhaizan, M.E. Curcumin Combination Chemotherapy: The Implication and Efficacy in Cancer. Molecules 2019, 24, 2527. [Google Scholar] [CrossRef]
- Cai, Z.; He, S.; Li, T.; Zhao, L.; Zhang, K. Plumbagin Inhibits Proliferation and Promotes Apoptosis of Ovarian Granulosa Cells in Polycystic Ovary Syndrome by Inactivating PI3K/Akt/MTOR Pathway. Anim. Cells Systems 2020, 24, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Q.; Jiang, Y.; Li, G. Inhibition of the PI3K/AKT-NF-ΚB Pathway With Curcumin Enhanced Radiation-Induced Apoptosis in Human Burkitt’s Lymphoma. J. Pharmacol. Sci. 2013, 121, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Shen, G.; Khor, T.O.; Kim, J.-H.; Kong, A.-N.T. Curcumin Inhibits Akt/Mammalian Target of Rapamycin Signaling through Protein Phosphatase-Dependent Mechanism. Mol. Cancer Ther. 2008, 7, 2609–2620. [Google Scholar] [CrossRef]
- Borges, G.A.; Elias, S.T.; Amorim, B.; Lima, C.L.; Coletta, R.D.; Castilho, R.M.; Squarize, C.H.; Guerra, E.N.S. Curcumin Downregulates the PI3K–AKT–mTOR Pathway and Inhibits Growth and Progression in Head and Neck Cancer Cells. Phytother. Res. 2020, 34, 3311–3324. [Google Scholar] [CrossRef] [PubMed]
- Mukerjee, N.; Das, A.; Jawarkar, R.D.; Maitra, S.; Das, P.; Castrosanto, M.A.; Paul, S.; Samad, A.; Zaki, M.E.A.; Al-Hussain, S.A.; et al. Repurposing Food Molecules as a Potential BACE1 Inhibitor for Alzheimer’s Disease. Front. Aging Neurosci. 2022, 14, 878276. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, I.; Sen, A. In Silico Screening Predicts Common Cold Drug Dextromethorphan along with Prednisolone and Dexamethasone Can Be Effective against Novel Coronavirus Disease (COVID-19). Biomol. Struct. Dyn. 2020, 40, 3706–3710. [Google Scholar] [CrossRef]
- Aier, I.; Varadwaj, P.K.; Raj, U. Structural Insights into Conformational Stability of Both Wild-Type and Mutant EZH2 Receptor. Sci. Rep. 2016, 6, 34984. [Google Scholar] [CrossRef] [PubMed]
- Castrosanto, M.A.; Mukerjee, N.; Ramos, A.R.; Maitra, S.; Manuben, J.J.P.; Das, P.; Malik, S.; Hasan, M.M.; Alexiou, A.; Dey, A.; et al. Abetting Host Immune Response by Inhibiting Rhipicephalus Sanguineus Evasin-1: An in Silico Approach. PLoS ONE 2022, 17, e0271401. [Google Scholar] [CrossRef] [PubMed]
- Martínez, L. Automatic Identification of Mobile and Rigid Substructures in Molecular Dynamics Simulations and Fractional Structural Fluctuation Analysis. PLoS ONE 2015, 10, e0119264. [Google Scholar] [CrossRef] [PubMed]
- Abuzenadah, A.M.; Al-Sayes, F.; Alam, S.S.M.; Hoque, M.; Karim, S.; Hussain, I.M.R.; Tabrez, S. Elucidating Antiangiogenic Potential of Rauwolfia Serpentina: VEGFR-2 Targeting-Based Molecular Docking Study. Evid. Based Complement. Alternat. Med. 2022, 2022, 6224666. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Muralidharan, N.; Sakthivel, R.; Velmurugan, D.; Gromiha, M.M. Computational Studies of Drug Repurposing and Synergism of Lopinavir, Oseltamivir and Ritonavir Binding with SARS-CoV-2 Protease against COVID-19. J. Biomol. Struct. Dyn. 2021, 39, 2673–2678. [Google Scholar] [CrossRef]
- Abuzenadah, A.M.; Al-Sayes, F.; Alam, S.S.M.; Hoque, M.; Karim, S.; Hussain, I.M.R.; Tabrez, S. Identification of Potential Poly (ADP-Ribose) Polymerase-1 Inhibitors Derived from Rauwolfia Serpentina: Possible Implication in Cancer Therapy. Evid. Based Complement. Alternat. Med. 2022, 2022, 3787162. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the ACM/IEEE SC 2006 Conference (SC’06), Tampa, FL, USA, 11–17 November 2006; IEEE: Tampa, FL, USA, 2006; p. 43. [Google Scholar]
- Chow, E.; Rendleman, C.A.; Bowers, K.J.; Dror, R.O.; Gullingsrud, J.; Sacerdoti, F.D.; Shaw, D.E. Desmond Performance on a Cluster of Multicore Processors; DE Shaw Research Technical Report DESRES/TR--2008-01; DE Shaw Research: New York, NY, USA, 2008; p. 14. [Google Scholar]
- Shivakumar, D.; Williams, J.; Wu, Y.; Damm, W.; Shelley, J.; Sherman, W. Prediction of Absolute Solvation Free Energies Using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. J. Chem. Theory Comput. 2010, 6, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant Pressure Molecular Dynamics Algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Martyna, G.J.; Klein, M.L.; Tuckerman, M. Nosé–Hoover Chains: The Canonical Ensemble via Continuous Dynamics. J. Chem. Phys. 1992, 97, 2635–2643. [Google Scholar] [CrossRef]
- Toukmaji, A.Y.; Board, J.A. Ewald Summation Techniques in Perspective: A Survey. Comput. Phys. Commun. 1996, 95, 73–92. [Google Scholar] [CrossRef]
- Kagami, L.P.; das Neves, G.M.; Timmers, L.F.S.M.; Caceres, R.A.; Eifler-Lima, V.L. Geo-Measures: A PyMOL Plugin for Protein Structure Ensembles Analysis. Comput. Biol. Chem. 2020, 87, 107322. [Google Scholar] [CrossRef] [PubMed]
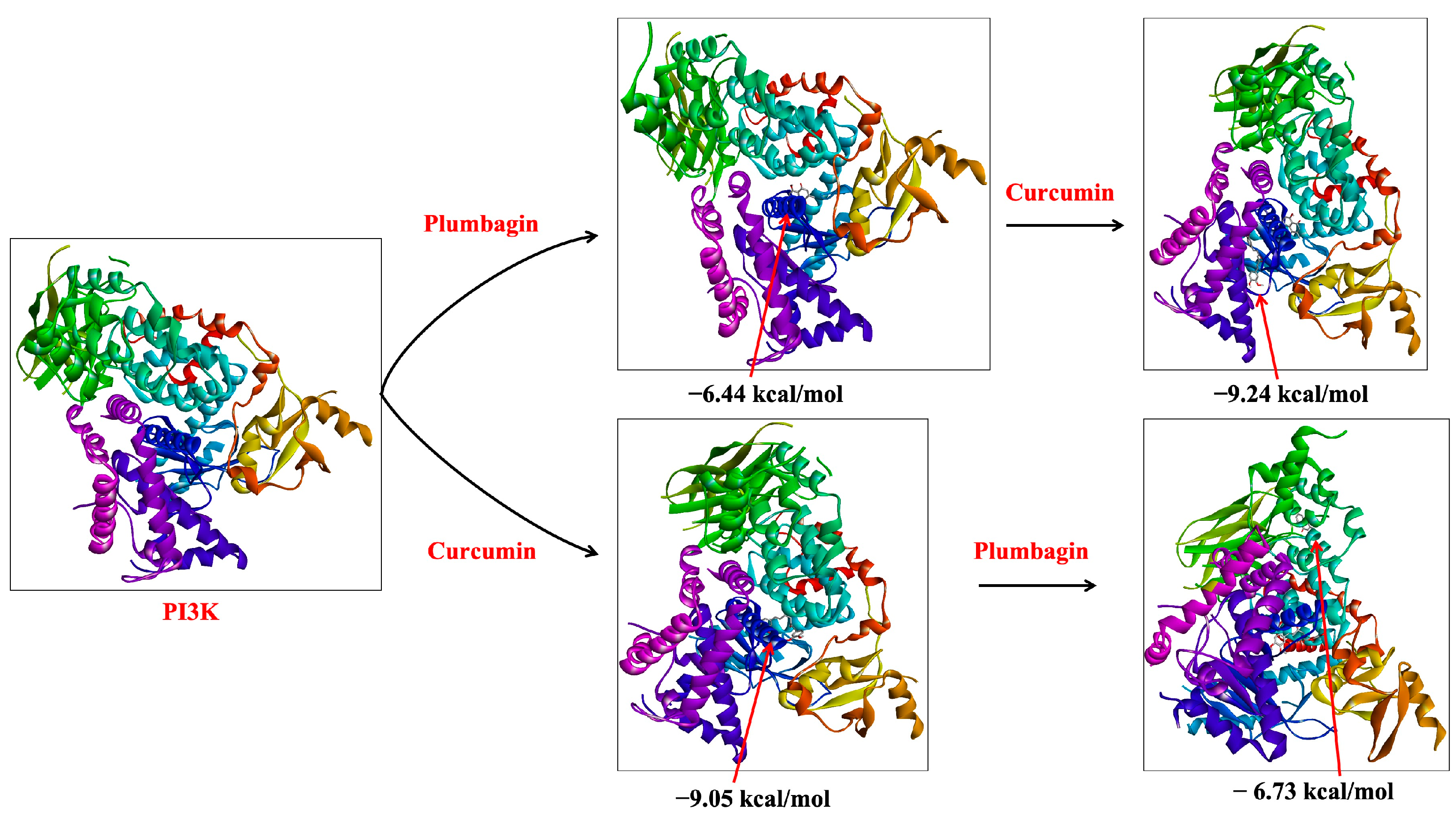

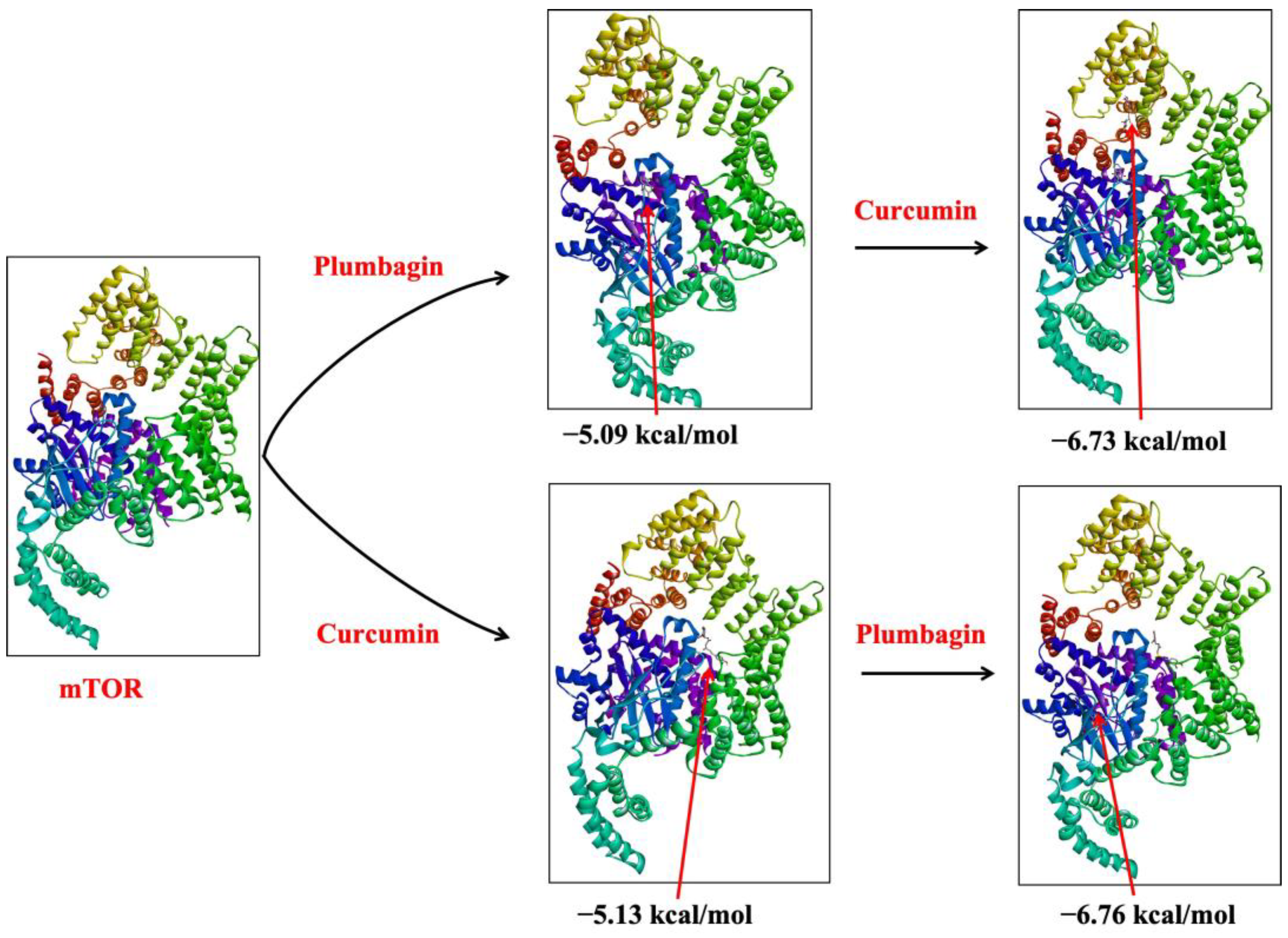

| Sl. No. | Target | Ligand | Docking Complex | ΔG (kcal/mol) | Ki (μM) | Interacting Residues |
|---|---|---|---|---|---|---|
| 1 | PI3K | Plumbagin | PI3K-P | −6.44 | 19.06 | Leu661, Phe694, Phe698, Met842, Leu845, Gln846, Arg849, Gly868, Cys869, Tyr787 |
| 2 | PI3K | Curcumin | PI3K-C | −9.05 | 0.23 | Gln291, Trp201, Asp654, Leu657, His658, Leu660, Leu661, Phe694, Phe698, Arg690, Tyr787, Met842, Leu845, Gln846, Arg849, Tyr867, Gly868, Cys869 |
| 3 | PI3K-Plumbagin complex (PI3K-P) | Curcumin | PI3K-P-C | −9.24 | 0.17 | Pro810, Trp812, Ile831, Lys833, Asp836, Leu838, Asp841, Ile879, Glu880, Ile881, Val882, Lys883, Asp884, Ala885, Thr886, Tyr867, Met953, Phe961, Ile963, Asp964 |
| 4 | PI3K-Curcumin complex (PI3K-C) | Plumbagin | PI3K-C-P | −6.73 | 11.63 | His389, Pro424, Lys425, Trp598, Val604, Tyr608, Ser636, Asp637, Glu638, Asn639 |
| 5 | Akt | Plumbagin | Akt-P | −6.06 | 35.98 | Tyr272, Arg273, Asp274, Leu275, Cys310, Gly311, Thr312, Tyr315, Leu316, Ala317, Val320, Val330, Asp331, Gly334 |
| 6 | Akt | Curcumin | Akt-C | −8.31 | 0.80 | Leu156, Gly157, Lys158, Gly159, Thr160, Phe161, Gly162, Lys163, Val164, Lys179, Met227, Gly233, Glu234, Phe237, Met281, Thr291, Asp292, Tyr437, Phe438, Asp439, Phe442 |
| 7 | Akt-Plumbagin complex (Akt-P) | Curcumin | Akt-P-C | −9.24 | 0.17 | Leu156, Gly157, Val164, Ala177, Lys179, Glu198, Leu202, Thr211, Met227, Glu228, Tyr229, Ala230, Glu234, Phe237, Met281, Thr291, Asp292, Phe293, Tyr437, Phe438, Asp439, Phe442 |
| 8 | Akt-Curcumin complex (Akt-C) | Plumbagin | Akt-C-P | −5.32 | 126.77 | Gly157, Lys158, Gly159, Val164, Glu234, Lys276, Glu278, Asn279, Asp292 |
| 9 | mTOR | Plumbagin | mTOR-P | −5.09 | 187.26 | Asn2206, Leu2209, Ala2210, Ser2215, Leu2216, Asn2219, Leu2220, Ser2221 |
| 10 | mTOR | Curcumin | mTOR-C | −5.13 | 174.96 | Tyr1787, His1791, Asn1898, Asn1899, Gln1901, Asp1902, Thr2207, Leu2208, Asn2211, Asp2212, Val2406, Glu2409, Pro2213, His2410, Ser2413 |
| 11 | mTOR-Plumbagin complex (mTOR-P) | Curcumin | mTOR-P-C | −6.73 | 11.74 | His1454, Trp1456, Glu1485, Ala1486, Ser1584, Tyr1587, Val1591, Gln1627, Arg1628, Ile1629, Glu1631, Asp1632, Lys1635 |
| 12 | mTOR-Curcumin complex (mTOR-C) | Plumbagin | mTOR-C-P | −6.76 | 11.01 | Leu2185, Lys2187, Tyr2225, Ile2237, Gly2238, Trp2239, Val2240, Met2345, Ile2356, Asp2357 |
| Energies (kcal/mol) | PI3K-P | PI3K-C | PI3K-C-P | Akt-P | Akt-C | Akt-C-P | mTOR-P | mTOR-C | mTOR-C-P |
|---|---|---|---|---|---|---|---|---|---|
| ΔGbind | −42.07 ± 2.4 | −38.35 ± 8.40 | −76.93 ± 5.15 | −61.42 ± 4.1 | −66.04 ± 2.63 | −85.26 ± 2.99 | −42.58 ± 6.35 | −41.04 ± 1.13 | −56.81 ± 6.79 |
| ΔGbindLipo | −8.68 ± 2.44 | −11.65 ± 2.08 | −35.33 ± 2.61 | −19.83 ± 2.3 | −23.96 ± 1.03 | −31.50 ± 3.1 | −12.24 ± 1.23 | −13.43 ± 1.6 | −18.08 ± 1.04 |
| ΔGbindvdW | −35.35 ± 6.44 | −29.78 ± 6.17 | −72.26 ± 5.40 | −52.68 ± 2.17 | −51.10 ± 2.0 | −70.63 ± 2.63 | −36.13 ± 2.17 | −34.160 ± 3.0 | −48.49 ± 2.18 |
| ΔGbindCoulomb | −12.4 ± 2.21 | −13.31 ± 8.69 | −27.18 ± 3.11 | −2.14 ± 1.01 | −8.12 ± 1.99 | −43.66 ± 2.88 | −13.83 ± 5.54 | −6.22 ± 0.99 | −25.47 ± 6.20 |
| ΔGbindHbond | −2.03 ± 0.77 | −0.87 ± 0.6 | −0.17 ± 0.1 | −0.06 ± 0.01 | −0.41 ± 0.22 | −1.87 ± 0.5 | −0.37 ± 0.2 | −0.62 ± 0.16 | −1.73 ± 0.34 |
| ΔGbindSolvGB | 8.15 ± 0.09 | 16.30 ± 7.42 | 58.35 ± 6.76 | 13.65 ± 2.27 | 16.5 ± 1.09 | 60.54 ± 2.8 | 19.73 ± 2.78 | 21.2 ± 1.7 | 31.74 ± 3.34 |
| ΔGbindCovalent | 2.01 ± 0.72 | 1.83 ± 1.28 | 3.27 ± 3.02 | 0.85 ± 0.5 | 1.56 ± 1.2 | 4.22 ± 1.07 | 2.97 ± 1.90 | 2.66 ± 1.12 | 5.23 ± 4.41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, I.; Hoque, M.; Alam, S.S.M.; Zughaibi, T.A.; Tabrez, S. Curcumin and Plumbagin Synergistically Target the PI3K/Akt/mTOR Pathway: A Prospective Role in Cancer Treatment. Int. J. Mol. Sci. 2023, 24, 6651. https://doi.org/10.3390/ijms24076651
Ahmad I, Hoque M, Alam SSM, Zughaibi TA, Tabrez S. Curcumin and Plumbagin Synergistically Target the PI3K/Akt/mTOR Pathway: A Prospective Role in Cancer Treatment. International Journal of Molecular Sciences. 2023; 24(7):6651. https://doi.org/10.3390/ijms24076651
Chicago/Turabian StyleAhmad, Iftikhar, Mehboob Hoque, Syed Sahajada Mahafujul Alam, Torki A. Zughaibi, and Shams Tabrez. 2023. "Curcumin and Plumbagin Synergistically Target the PI3K/Akt/mTOR Pathway: A Prospective Role in Cancer Treatment" International Journal of Molecular Sciences 24, no. 7: 6651. https://doi.org/10.3390/ijms24076651
APA StyleAhmad, I., Hoque, M., Alam, S. S. M., Zughaibi, T. A., & Tabrez, S. (2023). Curcumin and Plumbagin Synergistically Target the PI3K/Akt/mTOR Pathway: A Prospective Role in Cancer Treatment. International Journal of Molecular Sciences, 24(7), 6651. https://doi.org/10.3390/ijms24076651








