Establishment of an Efficient Agrobacterium-Mediated Genetic Transformation System to Enhance the Tolerance of the Paraquat Stress in Engineering Goosegrass (Eleusine Indica L.)
Abstract
1. Introduction
2. Results and Discussion
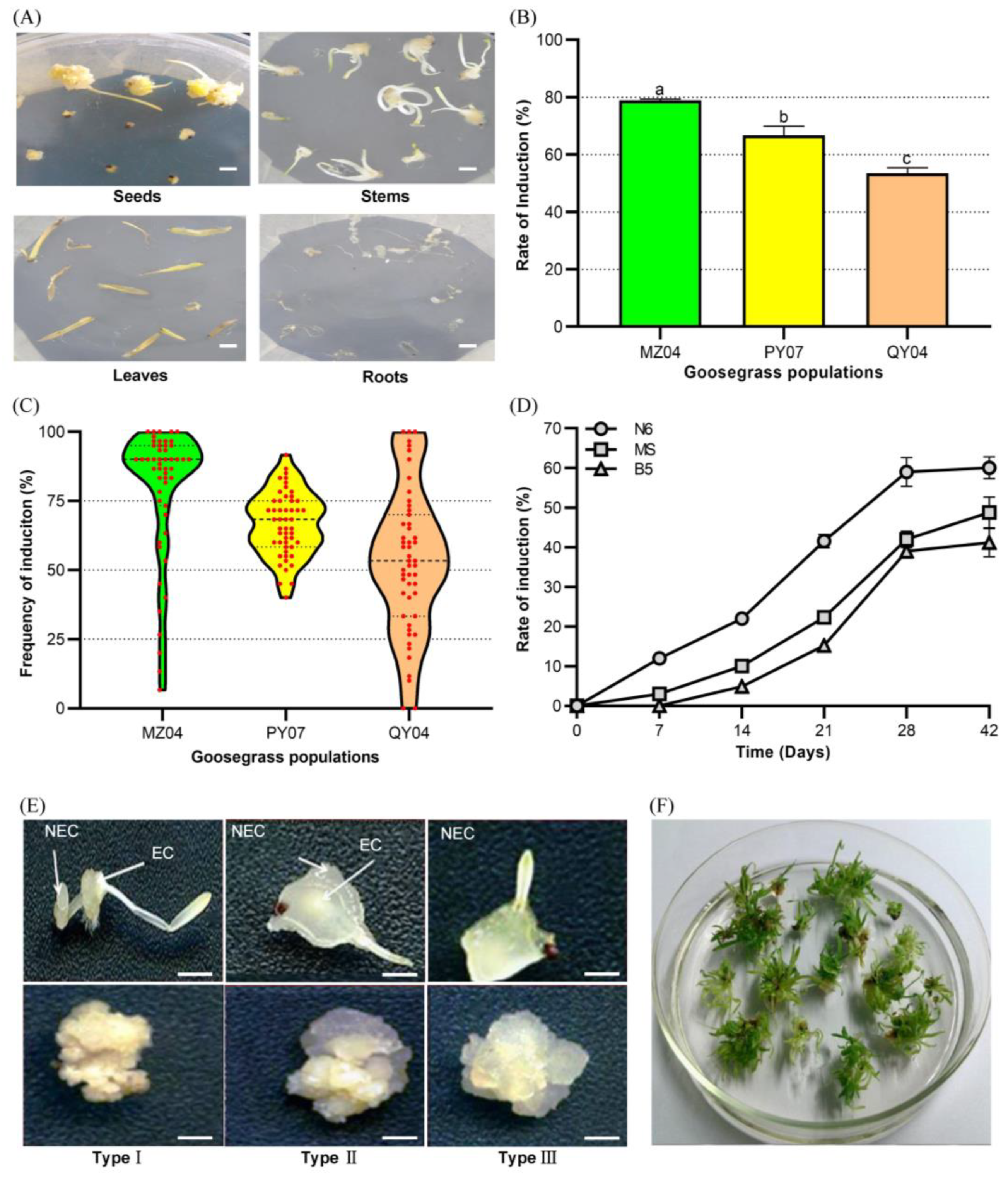
2.1. Effects of the Explant and Genetic Backgrounds in Goosegrass Callus Induction
2.2. Effects of the Developmental Regulators on Goosegrass Calli Induction from Seeds
2.3. Effects of Regulators on Agrobacterium-Mediated Infection and Regeneration of Goosegrass Calli
2.4. Goosegrass Transformed with the Paraquat-Resistant Gene EiKCS
2.5. Characterization of Paraquat-Resistant Transgenic Goosegrass Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. General Improvements of the Transformation System in Goosegrass
4.3. Vector Construction for Overexpressing EiKCS
4.4. Transformation System for Goosegrass
4.5. Regeneration System of Goosegrass Calli
4.6. Molecular Screening of Transgenic EiKCS Goosegrass
4.7. Real-Time Quantitative RT-PCR
4.8. Parallel Reaction Monitoring
4.9. Paraquat-Resistance Analysis of Transgenic EiKCS Goosegrass
4.10. Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2:4-D | 2:4-Dichlorophenoxyacetic acid |
| 6-BA | 6-Benzylaminopurine |
| KT | Kinetin |
| IAA | Indole-3-acetic acid |
| NAA | 1-naphthlcetic acid |
| AS | Asparagine |
References
- Caroline, A.E.; Strömberg, A.; Staver, C. The history and challenge of grassy biomes. Science 2022, 377, 592–593. [Google Scholar]
- McSteen, P.; Kellogg, E.A. Molecular, cellular, and developmental foundations of grass diversity. Science 2022, 377, 599–602. [Google Scholar] [CrossRef]
- Bai, Y.F.; Cotrufo, M.F. Grassland soil carbon sequestration: Current understanding, challenges, and solutions. Science 2022, 377, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Buisson, E.; Archibald, S.; Fidelis, A.; Suding, K.N. Ancient grasslands guide ambitious goals in grassland restoration. Science 2022, 377, 594–598. [Google Scholar] [CrossRef]
- Lopez, B.; Pamela, J.H.; Caroline, A. The unrecognized value of grass. Science 2022, 377, 590–591. [Google Scholar] [CrossRef]
- Unsworth, R.K.F.; Cullen-Unsworth, L.C.; Jones, B.L.H.; Lilley, R.J. The planetary role of seagrass conservation. Science 2022, 377, 609–613. [Google Scholar] [CrossRef]
- Heap, I. The International Herbicide-Resistant Weed Database. 2022. Available online: www.weedscience.org (accessed on 22 August 2022).
- Mcelroy, J.S.; Harris, J.R.; Price, A.; Harkess, A.; Li, X. Identification of a paraquat resistant goosegrass (Eleusine indica) population from a central alabama vegetable production field. Weed Sci. 2021, 69, 648–652. [Google Scholar] [CrossRef]
- Xiao, W.; Li, J.J.; Zhang, Y.X.; Guo, Y.J.; Fang, W.P.; Valverde, B.E.; Yin, J.; Qiang, S.; Chen, S.G. A fungal Bipolaris bicolor strain as a potential bioherbicide for goosegrass (Eleusine indica) control. Pest Manag. Sci. 2022, 78, 1251–1264. [Google Scholar] [CrossRef]
- Vazquez-Garcia, J.G.; Alcántara-de la Cruz, R.; Rojano-Delgado, A.M.; Palma-Bautista, C.; Vasconcelos, J.M.D.; De Prado, R. Multiple herbicide resistance evolution: The case of Eleusine indica in Brazil. J. Agric. Food Chem. 2021, 69, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Plaza, G.; Hoyos, V.; Vazquez-Garcia, J.G.; Alcántara-de la Cruz, R.; De Prado, R. First case of multiple resistance to EPSPS and PSI in Eleusine indica (L.) Gaertn. collected in rice and herbicide-resistant crops in Colombia. Agronomy 2021, 11, 96. [Google Scholar]
- Kumar, S.; Agarwal, K.; Kothari, S.L. In vitro induction and enlargement of apical domes and formation of multiple shoots in millet, Eleusine coracana (L.) Gaertn. and crowfoot grass, Eleusine indica (L.) Gaertn. Curr. Sci. 2001, 81, 1482–1485. [Google Scholar]
- Nyporko, A.Y.; Yemets, A.I.; Klimkina, L.A.; Blume, Y.B. Sensitivity of Eleusine indica callus to trifluralin and amiprophosmethyl in correlation with the binding of these compounds to tubulin. Russ. J. Plant Physiol. 2002, 49, 413–418. [Google Scholar] [CrossRef]
- Yemets, A.I.; Klimkina, L.A.; Tarassenko, L.V.; Blume, Y.B. Efficient callus formation and plant regeneration of goosegrass [Eleusine indica (L.) Gaertn.]. Plant Cell Rep. 2003, 21, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Yemets, A.; Radchuk, V.; Bayer, O.; Bayer, G.; Pakhomov, A.; Baird, W.V.; Blume, Y.B. Development of transformation vectors based upon a modified plant α-tubulin gene as the selectable marker. Cell Biol. Int. 2008, 32, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Upadhyaya, G.; Sen, M.; Roy, A. In vitro callus induction and plant regeneration of rice (Oryza sativa L.) var. ‘Sita’, ‘Rupali’ and ‘Swarna Masuri’. Asian J. Plant Sci. Res. 2015, 5, 24–27. [Google Scholar]
- Liu, Y.R.; Cen, H.F.; Yan, J.P.; Zhang, Y.W.; Zhang, W.J. Inside out: High-efficiency plant regeneration and Agrobacterium-mediated transformation of upland and lowland switchgrass cultivars. Plant Cell Rep. 2015, 34, 1099–1108. [Google Scholar] [CrossRef]
- Nazish, T.; Huang, Y.J.; Zhang, J.; Xia, J.Q.; Alfatih, A.; Luo, C.; Cai, X.T.; Xi, J.; Xu, P.; Xiang, C.B. Understanding paraquat resistance mechanisms in Arabidopsis thaliana to facilitate the development of paraquatresistant crops. Plant Commun. 2022, 3, 100321. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.Y.; Zhao, M.M.; Wang, W.J.; Wang, Q.; Huang, M.Q.; Li, C.Q.; Lian, Q.C.; Xia, J.Q.; Qi, J.; Xiang, C.B.; et al. Changing Gly311 to an acidic amino acid in the mate family protein DTX6 enhances Arabidopsis resistance to the dihydropyridine herbicides. Mol. Plant 2021, 14, 11. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.Q.; Nazish, T.; Javaid, A.; Ali, M.; Liu, Q.Q.; Wang, L.; Zhang, Z.Y.; Zhang, Z.S.; Huang, Y.J.; Wu, J.; et al. A gain-of-function mutation of the MATE family transporter DTX6 confers paraquat resistance in Arabidopsis. Mol. Plant 2021, 14, 2126–2133. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.L. The identification of an accumulation system for diamines and polyamines into the lung and its relevance to paraquat toxicity. Arch. Toxicol. Suppl. 1982, 5, 1–4. [Google Scholar]
- Palmer, A.J.; Wallace, H.M. The polyamine transport system as a target for anticancer drug development. Amino Acids 2010, 38, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.T.; Andérica, R.A.C.; Pedraza, C.J. New insights into antioxidant strategies against paraquat toxicity. Free Radic. Res. 2014, 48, 623–640. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Shinozaki, K. Identification of Polyamine Transporters in Plants: Paraquat Transport Provides Crucial Clues. Plant Cell Physiol. 2014, 55, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.S.; Cao, L.M.; Huang, W.Q.; Liu, J.X.; Lu, H.P. Disruption of three polyamine uptake transporter genes in rice by CRISPR/Cas9 gene editing confers tolerance to herbicide paraquat. Abiotech 2022, 3, 140–145. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Shen, X.F.; Ma, Q.B.; Yang, C.Y.; Liu, S.M.; Chen, Y. Transcriptome profiling to discover putative genes associated with paraquat resistance in goosegrass (Eleusine indica L.). PLoS ONE 2014, 9, e99940. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.Y.; Wei, J.P.; Dong, Z.X.; Shen, X.F.; Chen, Y. Differences of endogenous polyamines and putative genes associated with paraquat resistance in goosegrass (Eleusine indica L.). PLoS ONE 2019, 14, e0216513. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.X.; Li, J.; Liu, S.M.; Zhu, X.F.; Chen, Y.; Shen, X.F. Effects of spermidine and salinity stress on growth and biochemical response of paraquat-susceptibe and -resistant goosegrass (Eleusine indica L.). Weed Biol. Manag. 2019, 19, 75–84. [Google Scholar] [CrossRef]
- Luo, Q.Y.; Chen, S.; Zhu, J.Z.; Ye, L.H.; Nathan, D.H.; Basak, S.; Mcelroy, J.S.; Chen, Y. Overexpression of EiKCS confers paraquat-resistance in rice (Oryza sativa L.) by promoting the polyamine pathway. Pest Manag. Sci. 2021, 78, 246–262. [Google Scholar]
- Chan, M.T.; Chang, H.H.; Ho, S.L.; Tong, W.F.; Yu, S.M. Agrobacterium-mediated production of transgenic rice plants expressing a chimeric alpha-amylase promoter/beta-glucuronidase gene. Plant Mol. Biol. 1993, 22, 491–506. [Google Scholar] [CrossRef]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar]
- Ishida, Y.; Saito, H.; Ohta, S.; Hiei, Y.; Komari, T.; Kumashiro, T. High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 1996, 14, 745–750. [Google Scholar] [CrossRef] [PubMed]
- James, C. Global Status of Commercialized Transgenic Crops (GMO). Int. Serv. Acquis. Agribiotech Appl. 2003, 41, 175–176. [Google Scholar]
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Kumar, V.; Bellinder, R.R.; Gupta, R.K.; Malik, R.K.; Brainard, D.C. Role of herbicide-resistant rice in promoting resource conservation technologies in rice–wheat cropping systems of India: A review. Crop Prot. 2008, 27, 290–301. [Google Scholar] [CrossRef]
- Dong, H.R.; Huang, Y.; Wang, K.J. The development of herbicide resistance crop plants using CRISPR/Cas9-mediated gene editing. Genes 2021, 12, 912. [Google Scholar] [CrossRef]
- Jin, M.; Chen, L.; Deng, X.W.; Tang, X.Y. Development of herbicide resistance genes and their application in rice. Crop J. 2022, 10, 26–35. [Google Scholar] [CrossRef]
- Hu, F.; Dong, H.R.; Seng, X.F.; Liu, J.; Wei, J.P.; Chen, Y. Resistance Level of Eleusine indica populations to paraquat, glyphosate and glufosinate. Southwest China J. Agric. Sci. 2018, 31, 335–340. [Google Scholar]
- Gupta, M.; Duhan, P.; Kajla, S.; Chaudhury, A. Optimization of media for establishment of Glycyrrhiza glabra micropropagation. Ann. Agric. Bio Res. 2014, 19, 197–201. [Google Scholar]
- Khan, S.; Fahim, N.; Singh, P.; Rahman, L.U. Agrobacterium tumefaciens mediated genetic transformation of Ocimum gratissimum: A medicinally important crop. Ind. Crops Prod. 2015, 71, 138–146. [Google Scholar] [CrossRef]
- Zhao, N.; Yan, Y.Y.; Luo, Y.L.; Zou, N.; Liu, W.T.; Wang, J.X. Unravelling mesosulfuron-methyl phytotoxicity and metabolism-based herbicide resistance in Alopecurus aequalis: Insight into regulatory mechanisms using proteomics. Sci. Total Environ. 2019, 20, 486–497. [Google Scholar] [CrossRef]






| Regulators | Concentration (mg L−1) | Regeneration Rate (%) |
|---|---|---|
| Means ± SD | ||
| 6-BA | 0.50 | 0 ± 0 d |
| 6-BA | 1.00 | 0 ± 0 d |
| 6-BA | 2.00 | 0 ± 0 d |
| 6-BA + 2,4-D | 0.50 + 0.50 | 0 ± 0 d |
| 6-BA + 2,4-D | 1.00 + 0.50 | 0 ± 0 d |
| 6-BA + 2,4-D | 2.00 + 0.50 | 0 ± 0 d |
| 6-BA + KT | 0.50 + 0.50 | 71.05 ± 0.18 a |
| 6-BA + KT | 1.00 + 1.00 | 58.03 ± 0.08 b |
| 6-BA + KT | 2.00 + 2.00 | 50.02 ± 0.14 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Q.; Chen, S.; Nian, H.; Ma, Q.; Ding, Y.; Hao, Q.; Wei, J.; Patel, J.D.; McElroy, J.S.; Liu, Y.; et al. Establishment of an Efficient Agrobacterium-Mediated Genetic Transformation System to Enhance the Tolerance of the Paraquat Stress in Engineering Goosegrass (Eleusine Indica L.). Int. J. Mol. Sci. 2023, 24, 6629. https://doi.org/10.3390/ijms24076629
Luo Q, Chen S, Nian H, Ma Q, Ding Y, Hao Q, Wei J, Patel JD, McElroy JS, Liu Y, et al. Establishment of an Efficient Agrobacterium-Mediated Genetic Transformation System to Enhance the Tolerance of the Paraquat Stress in Engineering Goosegrass (Eleusine Indica L.). International Journal of Molecular Sciences. 2023; 24(7):6629. https://doi.org/10.3390/ijms24076629
Chicago/Turabian StyleLuo, Qiyu, Shu Chen, Hai Nian, Qibing Ma, Yuyao Ding, Qinwen Hao, Jiping Wei, Jinesh D. Patel, Joseph Scott McElroy, Yaoguang Liu, and et al. 2023. "Establishment of an Efficient Agrobacterium-Mediated Genetic Transformation System to Enhance the Tolerance of the Paraquat Stress in Engineering Goosegrass (Eleusine Indica L.)" International Journal of Molecular Sciences 24, no. 7: 6629. https://doi.org/10.3390/ijms24076629
APA StyleLuo, Q., Chen, S., Nian, H., Ma, Q., Ding, Y., Hao, Q., Wei, J., Patel, J. D., McElroy, J. S., Liu, Y., & Chen, Y. (2023). Establishment of an Efficient Agrobacterium-Mediated Genetic Transformation System to Enhance the Tolerance of the Paraquat Stress in Engineering Goosegrass (Eleusine Indica L.). International Journal of Molecular Sciences, 24(7), 6629. https://doi.org/10.3390/ijms24076629






