Site-2 Protease Slr1821 Regulates Carbon/Nitrogen Homeostasis during Ammonium Stress Acclimation in Cyanobacterium Synechocystis sp. PCC 6803
Abstract
1. Introduction
2. Results and Discussion
2.1. Functional Slr1821 Is Essential to the Effective Acclimation of Synechocystis 6803 to High Concentration of NH4+
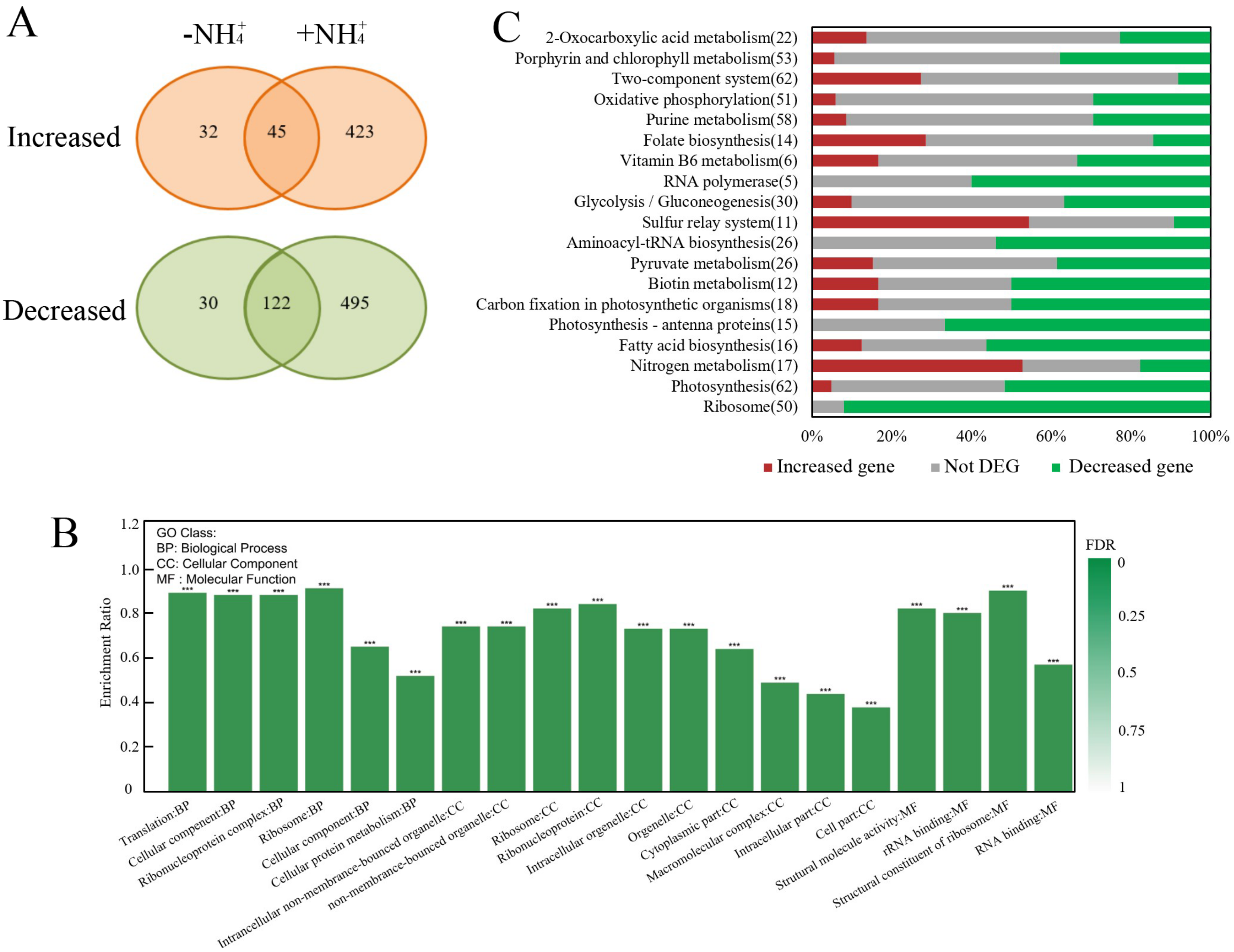
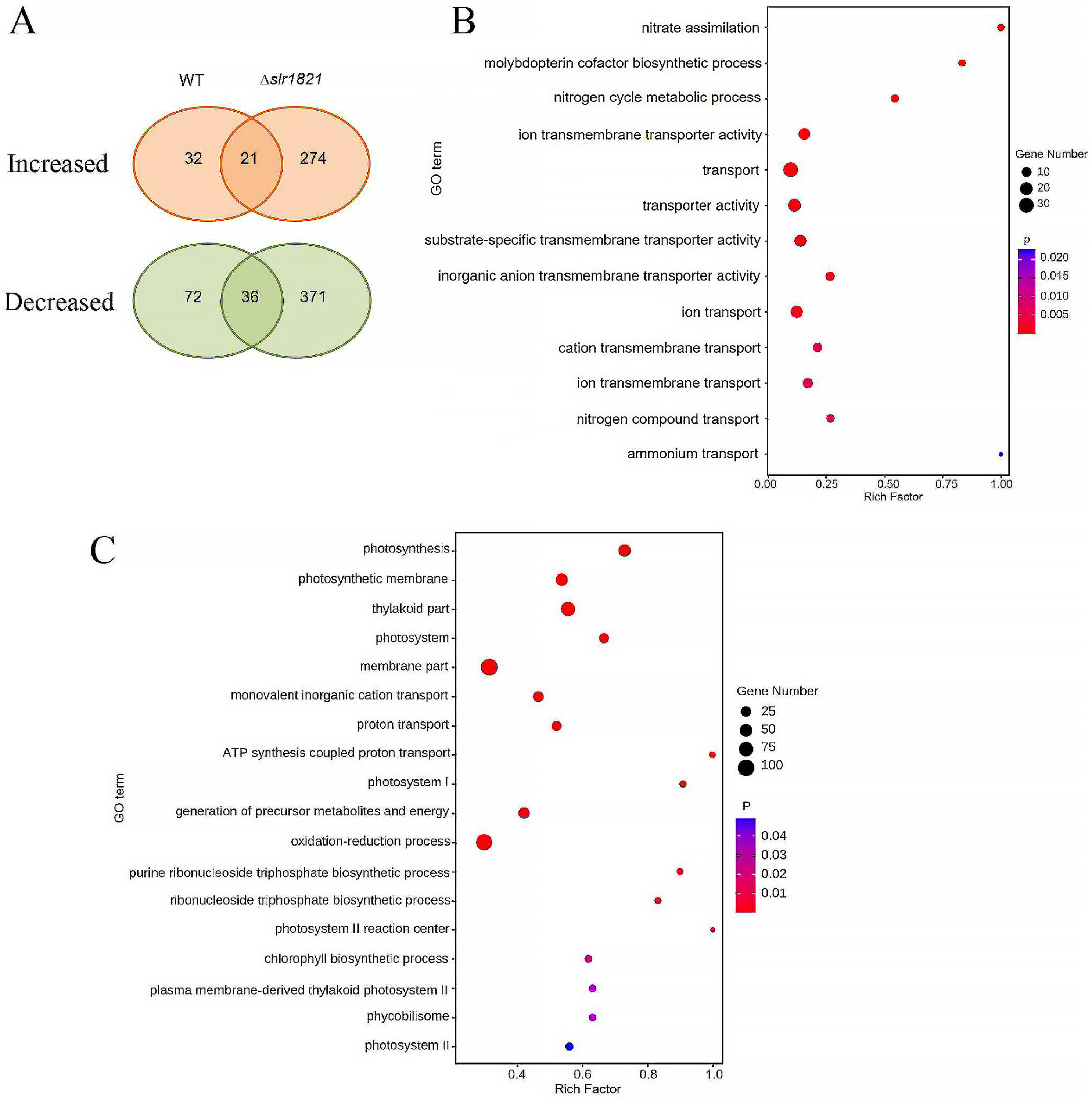
2.2. Global Transcriptomic Comparison of WT and ∆slr1821 Knockout Mutant upon NH4+ Stress
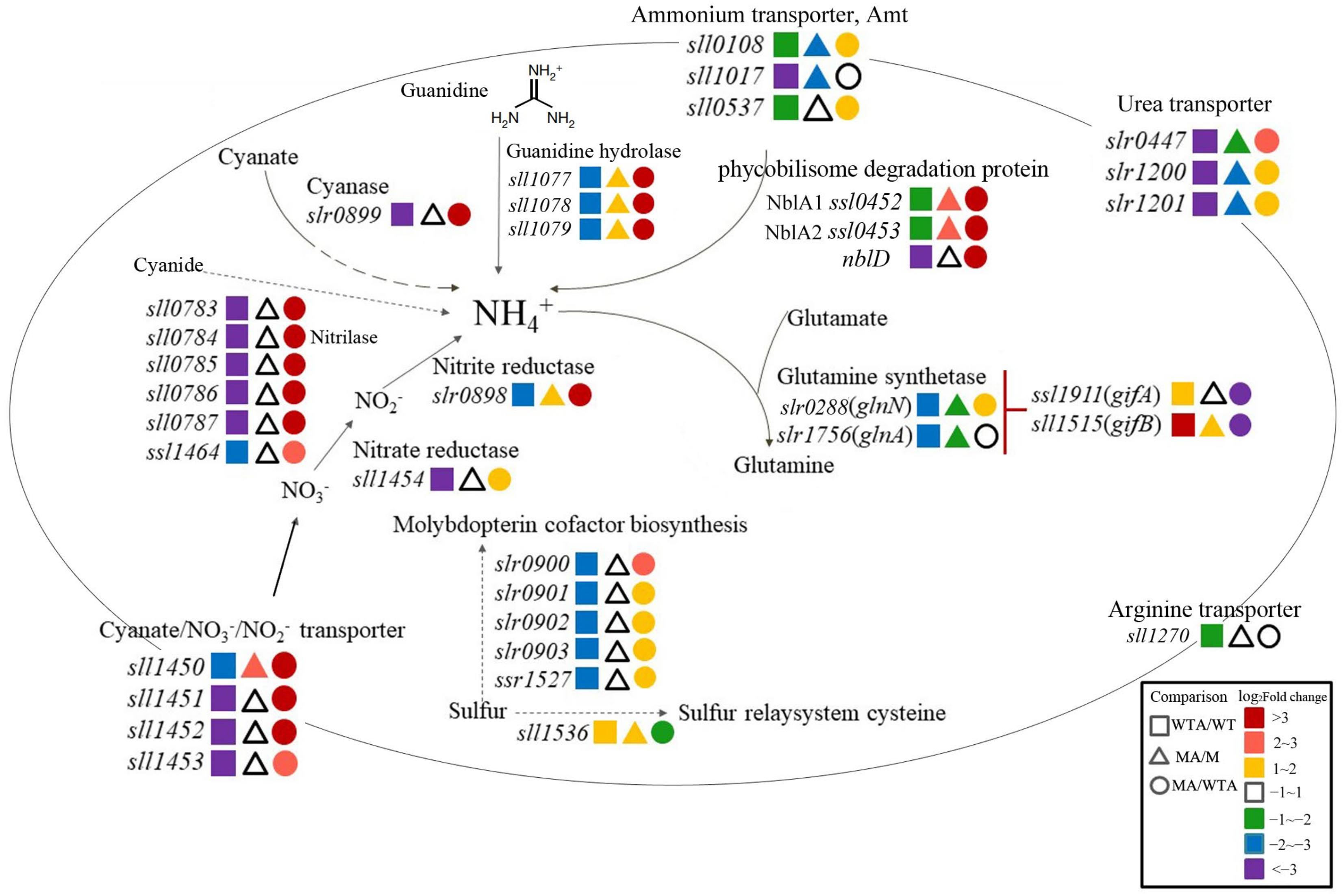
2.3. Slr1821 Is Indispensable for the High Concentration NH4+-Induced Suppression of Nitrogen Uptake, Assimilation and Mobilization
2.3.1. Nitrogen Uptake and Assimilation
2.3.2. Ammonium Integration and Nitrogen Mobilization
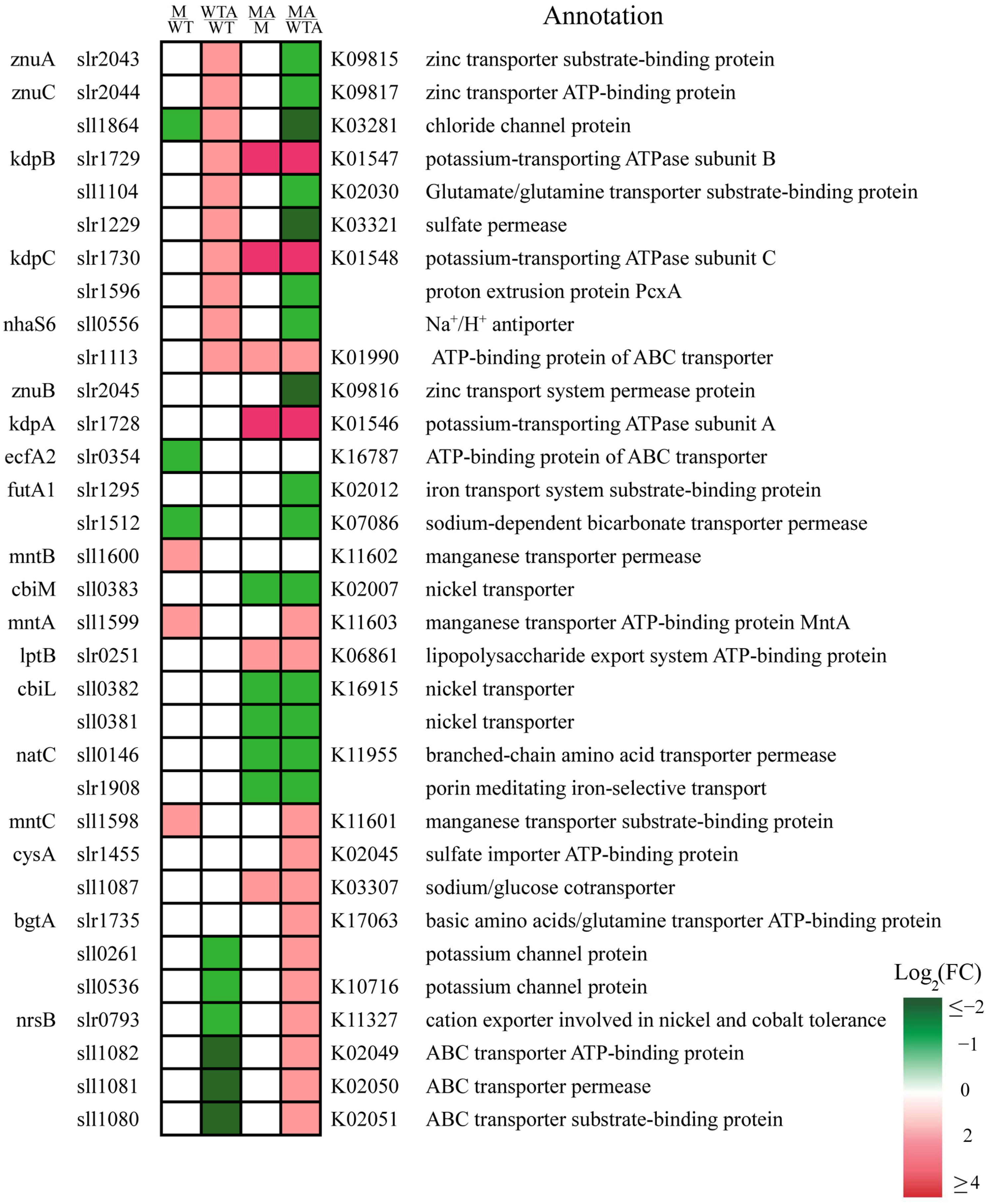
2.4. Slr1821 Knockout Disrupts the Proper Response of Transporters and Channels in the Ammonium-Specific Stimulon
2.5. Slr1821 Knockout Interferes with the Proper Response of Regulators upon Ammonium Stress
2.6. Knockout of slr1821 Resulted in Defective Photosynthesis, Compromised Translational and Transcriptional Machinery and Redirection of Carbon Flux
2.6.1. Defective Photosystem and Carbon Fixation
2.6.2. Disrupted Oxidative Phosphorylation and Central Carbon Metabolism
2.6.3. Compromised Translational and Transcriptional Machinery
2.6.4. Interfered Direction of Carbon Flux among PHB and Other Carbon Reservoirs
2.6.5. Extensive Stress Response
3. Materials and Methods
3.1. Strains and Cultivation
3.2. Construction of ∆slr1821 Strain
3.3. RNA Extraction, Library Construction, Sequencing and Bioinformatics Analysis
3.4. Physiological Characterization
3.5. RNA Isolation and Quantitative RT-PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Srisawat, P.; Higuchi-Takeuchi, M.; Numata, K. Microbial autotrophic biorefineries: Perspectives for biopolymer production. Polym. J. 2022, 54, 1139–1151. [Google Scholar] [CrossRef]
- Esteves-Ferreira, A.A.; Inaba, M.; Fort, A.; Araujo, W.L.; Sulpice, R. Nitrogen metabolism in cyanobacteria: Metabolic and molecular control, growth consequences and biotechnological applications. Crit. Rev. Microbiol. 2018, 44, 541–560. [Google Scholar] [CrossRef] [PubMed]
- Cassier-Chauvat, C.; Blanc-Garin, V.; Chauvat, F. Genetic, Genomics, and Responses to Stresses in Cyanobacteria: Biotechnological Implications. Genes 2021, 12, 500. [Google Scholar] [CrossRef]
- Rachedi, R.; Foglino, M.; Latifi, A. Stress Signaling in Cyanobacteria: A Mechanistic Overview. Life 2020, 10, 312. [Google Scholar] [CrossRef] [PubMed]
- Perin, G.; Fletcher, T.; Sagi-Kiss, V.; Gaboriau, D.C.A.; Carey, M.R.; Bundy, J.G.; Jones, P.R. Calm on the surface, dynamic on the inside. Molecular homeostasis of Anabaena sp. PCC 7120 nitrogen metabolism. Plant Cell Environ. 2021, 44, 1885–1907. [Google Scholar] [CrossRef] [PubMed]
- Forchhammer, K.; Selim, K.A. Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol. Rev. 2020, 44, 33–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.C.; Zhou, C.Z.; Burnap, R.L.; Peng, L. Carbon/Nitrogen Metabolic Balance: Lessons from Cyanobacteria. Trends Plant Sci. 2018, 23, 1116–1130. [Google Scholar] [CrossRef] [PubMed]
- Diez, J.; Lopez-Lozano, A.; Dominguez-Martin, M.A.; Gomez-Baena, G.; Munoz-Marin, M.C.; Melero-Rubio, Y.; Garcia-Fernandez, J.M. Regulatory and metabolic adaptations in the nitrogen assimilation of marine picocyanobacteria. FEMS Microbiol. Rev. 2023, 47, fuac043. [Google Scholar] [CrossRef]
- Orthwein, T.; Scholl, J.; Spat, P.; Lucius, S.; Koch, M.; Macek, B.; Hagemann, M.; Forchhammer, K. The novel PII-interactor PirC identifies phosphoglycerate mutase as key control point of carbon storage metabolism in cyanobacteria. Proc. Natl. Acad. Sci. USA 2021, 118, e2019988118. [Google Scholar] [CrossRef] [PubMed]
- Forchhammer, K.; Selim, K.A.; Huergo, L.F. New views on PII signaling: From nitrogen sensing to global metabolic control. Trends Microbiol. 2022, 30, 722–735. [Google Scholar] [CrossRef]
- Dai, G.-Z.; Qiu, B.-S.; Forchhammer, K. Ammonium tolerance in the cyanobacterium Synechocystis sp. strain PCC 6803 and the role of the psbA multigene family. Plant Cell Environ. 2014, 37, 840–851. [Google Scholar] [CrossRef]
- Erisman, J.W.; Galloway, J.N.; Seitzinger, S.; Bleeker, A.; Dise, N.B.; Petrescu, A.M.R.; Leach, A.M.; de Vries, W. Consequences of human modification of the global nitrogen cycle. Philos. Trans. R. Soc. B-Biol. Sci. 2013, 368, 20130116. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Zhang, X.; Lam, S.K.; Yu, Y.; van Grinsven, H.J.M.; Zhang, S.; Wang, X.; Bodirsky, B.L.; Wang, S.; Duan, J.; et al. Cost-effective mitigation of nitrogen pollution from global croplands. Nature 2023, 613, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Varjani, S.; Rakholiya, P.; Shindhal, T.; Shah, A.V.; Huu Hao, N. Trends in dye industry effluent treatment and recovery of value added products. J. Water Process Eng. 2021, 39, 101734. [Google Scholar] [CrossRef]
- Erisman, J.W. How ammonia feeds and pollutes the world. Science 2021, 374, 685–686. [Google Scholar] [CrossRef]
- Bhatia, S.K.; Mehariya, S.; Bhatia, R.K.; Kumar, M.; Pugazhendhi, A.; Awasthi, M.K.; Atabani, A.E.; Kumar, G.; Kim, W.; Seo, S.-O.; et al. Wastewater based microalgal biorefinery for bioenergy production: Progress and challenges. Sci. Total Environ. 2021, 751, 141599. [Google Scholar] [CrossRef]
- Priyadharshini, S.D.; Babu, P.S.; Manikandan, S.; Subbaiya, R.; Govarthanan, M.; Karmegam, N. Phycoremediation of wastewater for pollutant removal: A green approach to environmental protection and long-term remediation. Environ. Pollut. 2021, 290, 117989. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Mishra, A.; Pant, D.; Malaviya, P. Recent advances in microalgae-based remediation of industrial and non-industrial wastewaters with simultaneous recovery of value-added products. Bioresour. Technol. 2022, 344, 126129. [Google Scholar] [CrossRef] [PubMed]
- Jian, S.F.; Liao, Q.; Song, H.X.; Liu, Q.; Lepo, J.E.; Guan, C.Y.; Zhang, J.H.; Ismail, A.M.; Zhang, Z.H. NRT1.1-Related NH4+ Toxicity Is Associated with a Disturbed Balance between NH4+ Uptake and Assimilation. Plant Physiol. 2018, 178, 1473–1488. [Google Scholar] [CrossRef]
- Sun, L.; Di, D.W.; Li, G.J.; Kronzucker, H.J.; Wu, X.Y.; Shi, W.M. Endogenous ABA alleviates rice ammonium toxicity by reducing ROS and free ammonium via regulation of the SAPK9-bZIP20 pathway. J. Exp. Bot. 2020, 71, 4562–4577. [Google Scholar] [CrossRef]
- Yang, S.Y.; Hao, D.L.; Jin, M.; Li, Y.; Liu, Z.T.; Huang, Y.N.; Chen, T.X.; Su, Y.H. Internal ammonium excess induces ROS-mediated reactions and causes carbon scarcity in rice. BMC Plant Biol. 2020, 20, 143. [Google Scholar] [CrossRef]
- Hachiya, T.; Inaba, J.; Wakazaki, M.; Sato, M.; Toyooka, K.; Miyagi, A.; Kawai-Yamada, M.; Sugiura, D.; Nakagawa, T.; Kiba, T.; et al. Excessive ammonium assimilation by plastidic glutamine synthetase causes ammonium toxicity in Arabidopsis thaliana. Nat. Commun. 2021, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; von Wiren, N. Ammonium as a signal for physiological and morphological responses in plants. J. Exp. Bot. 2017, 68, 2581–2592. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, Q.; Xiong, L.; Kronzucker, H.J.; Kraemer, U.; Shi, W. Arabidopsis Plastid AMOS1/EGY1 Integrates Abscisic Acid Signaling to Regulate Global Gene Expression Response to Ammonium Stress. Plant Physiol. 2012, 160, 2040–2051. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Bi, Y.R.; Li, N. EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J. 2005, 41, 364–375. [Google Scholar] [CrossRef]
- Guo, D.; Gao, X.R.; Li, H.; Zhang, T.; Chen, G.; Huang, P.B.; An, L.J.; Li, N. EGY1 plays a role in regulation of endodermal plastid size and number that are involved in ethylene-dependent gravitropism of light-grown Arabidopsis hypocotyls. Plant Mol. Biol. 2008, 66, 345–360. [Google Scholar] [CrossRef]
- Brown, M.S.; Ye, J.; Rawson, R.B.; Goldstein, J.L. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell 2000, 100, 391–398. [Google Scholar] [CrossRef]
- Kuehnle, N.; Dederer, V.; Lemberg, M.K. Intramembrane proteolysis at a glance: From signalling to protein degradation. J. Cell Sci. 2019, 132, jcs217745. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, X. New insights into S2P signaling cascades: Regulation, variation, and conservation. Protein Sci. 2010, 19, 2015–2030. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, G.; Qin, C.; Wang, Y.; Wei, D. Slr0643, an S2P homologue, is essential for acid acclimation in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 2012, 158, 2765–2780. [Google Scholar] [CrossRef]
- Lei, H.; Chen, G.; Wang, Y.; Ding, Q.; Wei, D. Sll0528, a Site-2-Protease, Is Critically Involved in Cold, Salt and Hyperosmotic Stress Acclimation of Cyanobacterium Synechocystis sp. PCC 6803. Int. J. Mol. Sci. 2014, 15, 22678–22693. [Google Scholar] [CrossRef] [PubMed]
- Giner-Lamia, J.; Robles-Rengel, R.; Hernandez-Prieto, M.A.; Muro-Pastor, M.I.; Florencio, F.J.; Futschik, M.E. Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 2017, 45, 11800–11820. [Google Scholar] [CrossRef]
- Harano, Y.; Suzuki, I.; Maeda, S.I.; Kaneko, T.; Tabata, S.; Omata, T. Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942. J. Bacteriol. 1997, 179, 5744–5750. [Google Scholar] [CrossRef]
- Kopf, M.; Klahn, S.; Scholz, I.; Matthiessen, J.K.F.; Hess, W.R.; Voss, B. Comparative Analysis of the Primary Transcriptome of Synechocystis sp. PCC 6803. DNA Res. 2014, 21, 527–539. [Google Scholar] [CrossRef]
- Krauspe, V.; Fahrner, M.; Spat, P.; Steglich, C.; Frankenberg-Dinkel, N.; Macek, B.; Schilling, O.; Hess, W.R. Discovery of a small protein factor involved in the coordinated degradation of phycobilisomes in cyanobacteria. Proc. Natl. Acad. Sci. USA 2021, 118, e2012277118. [Google Scholar] [CrossRef]
- Baier, K.; Nicklisch, S.; Grundner, C.; Reinecke, J.; Lockau, W. Expression of two nblA-homologous genes is required for phycobilisome degradation in nitrogen-starved Synechocystis sp. PCC6803. FEMS Microbiol. Lett. 2001, 195, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Krauspe, V.; Timm, S.; Hagemann, M.; Hess, W.R. Phycobilisome Breakdown Effector NbID Is Required to Maintain Cellular Amino Acid Composition during Nitrogen Starvation. J. Bacteriol. 2022, 204, JB0015821. [Google Scholar] [CrossRef] [PubMed]
- Hauf, W.; Schlebusch, M.; Huge, J.; Kopka, J.; Hagemann, M.; Forchhammer, K. Metabolic Changes in Synechocystis PCC6803 upon Nitrogen-Starvation: Excess NADPH Sustains Polyhydroxybutyrate Accumulation. Metabolites 2013, 3, 101–118. [Google Scholar] [CrossRef]
- Jones, L.B.; Ghosh, P.; Lee, J.H.; Chou, C.N.; Kunz, D.A. Linkage of the Nit1C gene cluster to bacterial cyanide assimilation as a nitrogen source. Microbiology 2018, 164, 956–968. [Google Scholar] [CrossRef]
- Wang, B.; Xu, Y.; Wang, X.; Yuan, J.S.; Johnson, C.H.; Young, J.D.; Yu, J. A guanidine-degrading enzyme controls genomic stability of ethylene-producing cyanobacteria. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Funck, D.; Sinn, M.; Fleming, J.R.; Stanoppi, M.; Dietrich, J.; Lopez-Igual, R.; Mayans, O.; Hartig, J.S. Discovery of a Ni+-dependent guanidine hydrolase in bacteria. Nature 2022, 603, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, L.; Sun, T.; Jiang, J.; Tian, L.; Cui, J.; Zhang, W. A transporter Slr1512 involved in bicarbonate and pH-dependent acclimation mechanism to high light stress in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta-Bioenerg. 2021, 1862, 148336. [Google Scholar] [CrossRef] [PubMed]
- Gutekunst, K.; Hoffmann, D.; Westernstroeer, U.; Schulz, R.; Garbe-Schoenberg, D.; Appel, J. In-vivo turnover frequency of the cyanobacterial NiFe-hydrogenase during photohydrogen production outperforms in-vitro systems. Sci. Rep. 2018, 8, 6083. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.W.; Jiang, H.B.; Lis, H.; Li, Z.K.; Deng, B.; Shang, J.L.; Sun, C.Y.; Keren, N.; Qiu, B.S. A unique porin meditates iron-selective transport through cyanobacterial outer membranes. Environ. Microbiol. 2021, 23, 376–390. [Google Scholar] [CrossRef]
- Huang, F.; Fulda, S.; Hagemann, M.; Norling, B. Proteomic screening of salt-stress-induced changes in plasma membranes of Synechocystis sp. strain PCC 6803. Proteomics 2006, 6, 910–920. [Google Scholar] [CrossRef]
- Katoh, H.; Hagino, N.; Ogawa, T. Iron-binding activity of FutA1 subunit of an ABC-type iron transporter in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 2001, 42, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Bolay, P.; Schluter, S.; Grimm, S.; Riediger, M.; Hess, W.R.; Klaehn, S. The transcriptional regulator RbcR controls ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) genes in the cyanobacterium Synechocystis sp. PCC 6803. New Phytol. 2022, 235, 432–445. [Google Scholar] [CrossRef]
- Tabei, Y.; Okada, K.; Tsuzuki, M. Sll1330 controls the expression of glycolytic genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 2007, 355, 1045–1050. [Google Scholar] [CrossRef]
- Joseph, A.; Aikawa, S.; Sasaki, K.; Teramura, H.; Hasunuma, T.; Matsuda, F.; Osanai, T.; Hirai, M.Y.; Kondo, A. Rre37 stimulates accumulation of 2-oxoglutarate and glycogen under nitrogen starvation in Synechocystis sp. PCC 6803. FEBS Lett. 2014, 588, 466–471. [Google Scholar] [CrossRef]
- Khan, R.I.; Wang, Y.; Afrin, S.; Wang, B.; Liu, Y.; Zhang, X.; Chen, L.; Zhang, W.; He, L.; Ma, G. Transcriptional regulator PrqR plays a negative role in glucose metabolism and oxidative stress acclimation in Synechocystis sp. PCC 6803. Sci. Rep. 2016, 6, 32507. [Google Scholar] [CrossRef]
- Uchiyama, J.; Asakura, R.; Moriyama, A.; Kubo, Y.; Shibata, Y.; Yoshino, Y.; Tahara, H.; Matsuhashi, A.; Sato, S.; Nakamura, Y.; et al. Sll0939 is induced by Slr0967 in the cyanobacterium Synechocystis sp. PCC6803 and is essential for growth under various stress conditions. Plant Physiol. Biochem. 2014, 81, 36–43. [Google Scholar] [CrossRef]
- Wang, H.L.; Postier, B.L.; Burnap, R.L. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 2004, 279, 5739–5751. [Google Scholar] [CrossRef]
- Song, J.-Y.; Cho, H.S.; Cho, J.-I.; Jeon, J.-S.; Lagarias, J.C.; Park, Y.-I. Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 2011, 108, 10780–10785. [Google Scholar] [CrossRef] [PubMed]
- Huckauf, J.; Nomura, C.; Forchhammer, K.; Hagemann, M. Stress responses of Synechocystis sp. strain PCC 6803 mutants impaired in genes encoding putative alternative sigma factors. Microbiology 2000, 146, 2877–2889. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, J.; Kanesaki, Y.; Iwata, N.; Asakura, R.; Funamizu, K.; Tasaki, R.; Agatsuma, M.; Tahara, H.; Matsuhashi, A.; Yoshikawa, H.; et al. Genomic analysis of parallel-evolved cyanobacterium Synechocystis sp. PCC 6803 under acid stress. Photosynth. Res. 2015, 125, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Klaehn, S.; Mikkat, S.; Riediger, M.; Georg, J.; Hess, W.R.; Hagemann, M. Integrative analysis of the salt stress response in cyanobacteria. Biol. Direct. 2021, 16, 1–23. [Google Scholar] [CrossRef]
- Kobayashi, M.; Ishizuka, T.; Katayama, M.; Kanehisa, M.; Bhattacharyya-Pakrasi, M.; Pakrasi, H.B.; Ikeuchi, M. Response to oxidative stress involves a novel peroxiredoxin gene in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2004, 45, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Houot, L.; Floutier, M.; Marteyn, B.; Michaut, M.; Picciocchi, A.; Legrain, P.; Aude, J.-C.; Cassier-Chauvat, C.; Chauvat, F. Cadmium triggers an integrated reprogramming of the metabolism of Synechocystis PCC6803, under the control of the Slr1738 regulator. BMC Genom. 2007, 8, 350. [Google Scholar] [CrossRef]
- Hakkila, K.; Valev, D.; Antal, T.; Tyystjarvi, E.; Tyystjarvi, T. Group 2 Sigma Factors are Central Regulators of Oxidative Stress Acclimation in Cyanobacteria. Plant Cell Physiol. 2019, 60, 436–447. [Google Scholar] [CrossRef]
- Turunen, O.; Koskinen, S.; Kurkela, J.; Karhuvaara, O.; Hakkila, K.; Tyystjaervi, T. Roles of Close Homologues SigB and SigD in Heat and High Light Acclimation of the Cyanobacterium Synechocystis sp. PCC 6803. Life 2022, 12, 162. [Google Scholar] [CrossRef]
- Antal, T.; Kurkela, J.; Parikainen, M.; Karlund, A.; Hakkila, K.; Tyystjarvi, E.; Tyystjarvi, T. Roles of Group 2 Sigma Factors in Acclimation of the Cyanobacterium Synechocystis sp. PCC 6803 to Nitrogen Deficiency. Plant Cell Physiol. 2016, 57, 1309–1318. [Google Scholar] [CrossRef]
- Ishii, A.; Hihara, Y. An AbrB-like transcriptional regulator, Sll0822, is essential for the activation of nitrogen-regulated genes in Synechocystis sp. PCC 6803. Plant Physiol. 2008, 148, 660–670. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Kaniya, Y.; Kaneko, Y.; Hihara, Y. Physiological Roles of the cyAbrB Transcriptional Regulator Pair Sll0822 and Sll0359 in Synechocystis sp. strain PCC 6803. J. Bacteriol. 2011, 193, 3702–3709. [Google Scholar] [CrossRef] [PubMed]
- Kaniya, Y.; Kizawa, A.; Miyagi, A.; Kawai-Yamada, M.; Uchimiya, H.; Kaneko, Y.; Nishiyama, Y.; Hihara, Y. Deletion of the Transcriptional Regulator cyAbrB2 Deregulates Primary Carbon Metabolism in Synechocystis sp. PCC 6803. Plant Physiol. 2013, 162, 1153–1163. [Google Scholar] [CrossRef]
- Chen, X.; Schreiber, K.; Appel, J.; Makowka, A.; Fahnrich, B.; Roettger, M.; Hajirezaei, M.R.; Sonnichsen, F.D.; Schonheit, P.; Martin, W.F.; et al. The Entner-Doudoroff pathway is an overlooked glycolytic route in cyanobacteria and plants. Proc. Natl. Acad. Sci. USA 2016, 113, 5441–5446. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Brune, D.; Vermaas, W.F.J. The gamma-aminobutyric acid shunt contributes to closing the tricarboxylic acid cycle in Synechocystis sp. PCC 6803. Mol. Microbiol. 2014, 93, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lei, G.; Wu, X.; Wang, F.; Lai, C.; Li, Z. Expression, purification and characterization of sll1981 protein from cyanobacterium Synechocystis sp. PCC6803. Protein Expr. Purif. 2017, 139, 21–28. [Google Scholar] [CrossRef]
- Balaji, S.; Gopi, K.; Muthuvelan, B. A review on production of poly beta hydroxybutyrates from cyanobacteria for the production of bio plastics. Algal Res.-Biomass Biofuels Bioprod. 2013, 2, 278–285. [Google Scholar] [CrossRef]
- Hauf, W.; Watzer, B.; Roos, N.; Klotz, A.; Forchhammer, K. Photoautotrophic Polyhydroxybutyrate Granule Formation Is Regulated by Cyanobacterial Phasin PhaP in Synechocystis sp. Strain PCC 6803. Appl. Environ. Microbiol. 2015, 81, 4411–4422. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.; Orthwein, T.; Alford, J.T.; Forchhammer, K. The Slr0058 Protein from Synechocystis sp. PCC 6803 Is a Novel Regulatory Protein Involved in PHB Granule Formation. Front. Microbiol. 2020, 11, 809. [Google Scholar] [CrossRef]
- Schlebusch, M.; Forchhammer, K. Requirement of the Nitrogen Starvation-Induced Protein Sll0783 for Polyhydroxybutyrate Accumulation in Synechocystis sp. Strain PCC 6803. Appl. Environ. Microbiol. 2010, 76, 6101–6107. [Google Scholar] [CrossRef] [PubMed]
- Madsen, M.A.; Hamilton, G.; Herzyk, P.; Amtmann, A. Environmental Regulation of PndbA600, an Auto-Inducible Promoter for Two-Stage Industrial Biotechnology in Cyanobacteria. Front. Bioeng. Biotechnol. 2021, 8, 619055. [Google Scholar] [CrossRef] [PubMed]
- Kammerscheit, X.; Chauvat, F.; Cassier-Chauvat, C. First in vivo Evidence That Glutathione-S-Transferase Operates in Photo-Oxidative Stress in Cyanobacteria. Front. Microbiol. 2019, 10, 1899. [Google Scholar] [CrossRef]
- Karlsen, J.; Asplund-Samuelsson, J.; Thomas, Q.; Jahn, M.; Hudson, E.P. Ribosome Profiling of Synechocystis Reveals Altered Ribosome Allocation at Carbon Starvation. Msystems 2018, 3, e00126-18. [Google Scholar] [CrossRef]
- Vachiranuvathin, P.; Tharasirivat, V.; Hemnusornnanon, T.; Jantaro, S. Native SodB Overexpression of Synechocystis sp. PCC 6803 Improves Cell Growth Under Alcohol Stresses Whereas Its Gpx2 Overexpression Impacts on Growth Recovery from Alcohol Stressors. Appl. Biochem. Biotechnol. 2022, 194, 5748–5766. [Google Scholar] [CrossRef]


| Gene | Annotation of Gene Product | Log2FC | |||
|---|---|---|---|---|---|
| M WT | WTA WT | MA M | MA WTA | ||
| ncr1071 | non-coding RNA, Ncr1071 | −2.25 | 1.76 | 0.52 | −3.48 |
| sll0998 | LysR-type transcriptional regulator, RbcR | −0.89 | 1.4 | −0.87 | −3.16 |
| slr1731 | sensor histidine kinase, KdpD | 0.67 | 1.1 | 5.36 | 4.93 |
| sll0528 | zinc metalloprotease | 0.43 | 0.74 | 3.73 | 3.42 |
| sll1592 | two-component response regulator | 0.35 | 0.33 | 2.17 | 2.19 |
| sll0822 | transcriptional regulator cyAbrB2 | −0.61 | 0.17 | −2.64 | −3.42 |
| sll1590 | two-component sensor histidine kinase | 0.16 | 0.14 | 3.11 | 3.13 |
| sll1423 | global nitrogen regulator, NtcA | −0.57 | 0.01 | −1.29 | −1.86 |
| slr1738 | transcription regulator PerR | 1.21 | −0.08 | 1.13 | 2.43 |
| slr0473 | cyanobacterial phytochrome 1, two-component sensor histidine kinase | −0.16 | −0.33 | −1.39 | −1.22 |
| sll2012 | group 2 RNA polymerase sigma factor, SigD | −0.08 | −0.49 | 1.44 | 1.84 |
| slr0895 | transcriptional regulator, PrqR | −0.37 | −1.07 | −2.04 | −1.35 |
| sll1293 | chemotaxis signal transducer | 0.06 | −1.19 | −0.2 | 1.05 |
| slr1545 | group 3 RNA polymerase sigma factor, SigG | 0.63 | −1.34 | −1.7 | 0.27 |
| sll1330 | two-component system response regulator, Rre37 | 0.38 | −1.44 | 0.2 | 2.02 |
| sll0782 | protein kinase, transcriptional regulator | 0.77 | −1.58 | 0.8 | 3.16 |
| slr1214 | two-component response regulator PatA subfamily | 0.55 | −1.66 | −1.03 | 1.17 |
| sll1294 | methyl-accepting chemotaxis protein | −0.3 | −1.88 | −0.6 | 0.98 |
| sll1296 | two-component hybrid sensor and regulator | −0.76 | −2.15 | −1.29 | 0.11 |
| sll0944 | PII interactive protein, PirC | 0.67 | −2.28 | −0.61 | 2.34 |
| ssl0707 | nitrogen regulatory protein PII | −0.19 | −2.64 | −2.56 | −0.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.; Li, S.; Ouyang, T.; Chen, G. Site-2 Protease Slr1821 Regulates Carbon/Nitrogen Homeostasis during Ammonium Stress Acclimation in Cyanobacterium Synechocystis sp. PCC 6803. Int. J. Mol. Sci. 2023, 24, 6606. https://doi.org/10.3390/ijms24076606
Lin S, Li S, Ouyang T, Chen G. Site-2 Protease Slr1821 Regulates Carbon/Nitrogen Homeostasis during Ammonium Stress Acclimation in Cyanobacterium Synechocystis sp. PCC 6803. International Journal of Molecular Sciences. 2023; 24(7):6606. https://doi.org/10.3390/ijms24076606
Chicago/Turabian StyleLin, Shiqi, Shiliang Li, Tong Ouyang, and Gu Chen. 2023. "Site-2 Protease Slr1821 Regulates Carbon/Nitrogen Homeostasis during Ammonium Stress Acclimation in Cyanobacterium Synechocystis sp. PCC 6803" International Journal of Molecular Sciences 24, no. 7: 6606. https://doi.org/10.3390/ijms24076606
APA StyleLin, S., Li, S., Ouyang, T., & Chen, G. (2023). Site-2 Protease Slr1821 Regulates Carbon/Nitrogen Homeostasis during Ammonium Stress Acclimation in Cyanobacterium Synechocystis sp. PCC 6803. International Journal of Molecular Sciences, 24(7), 6606. https://doi.org/10.3390/ijms24076606






