Crystal Structure of Bright Fluorescent Protein BrUSLEE with Subnanosecond Fluorescence Lifetime; Electric and Dynamic Properties
Abstract
1. Introduction
2. Results and Discussion
2.1. Overall Structure
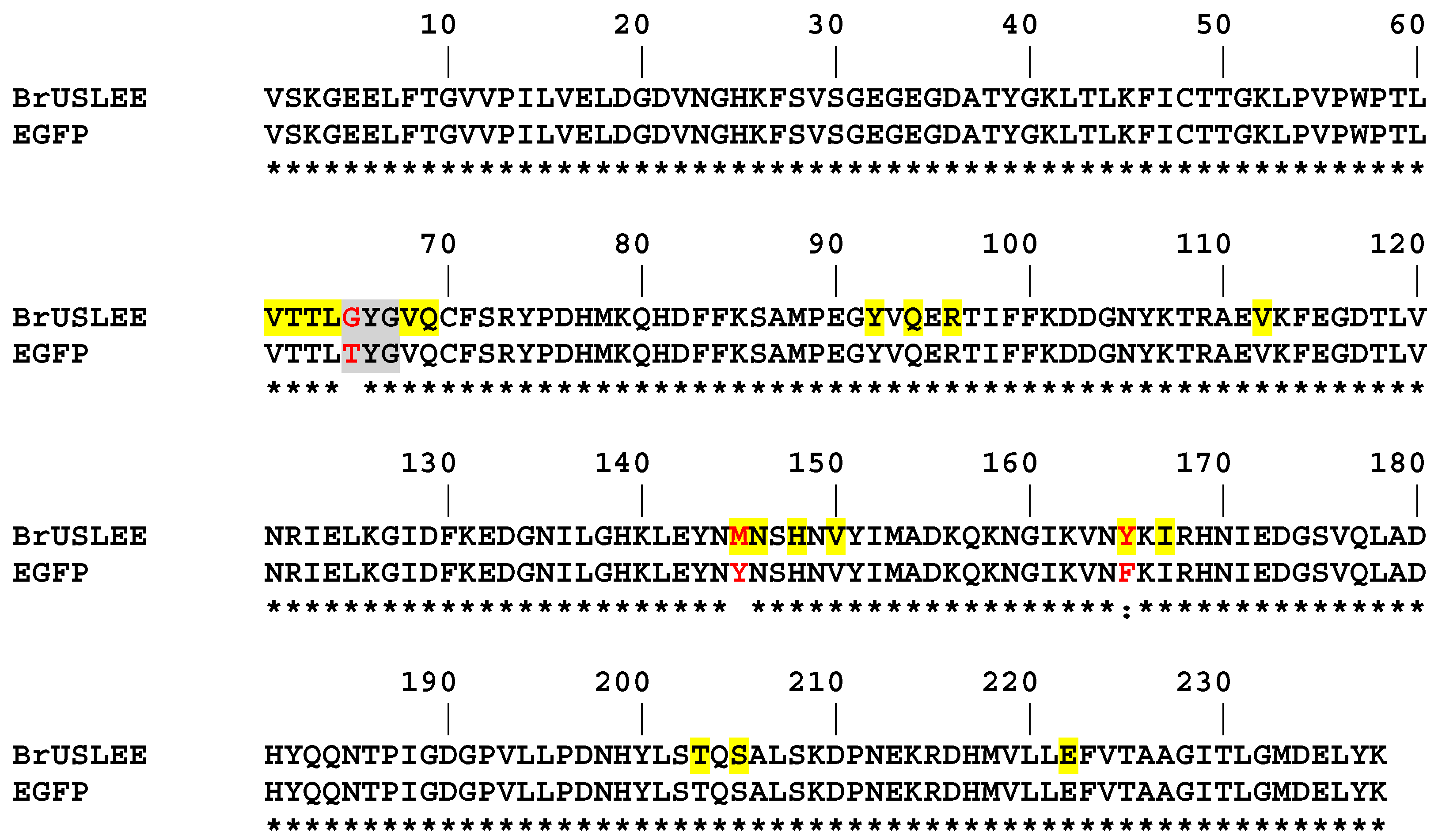
2.2. Structural Features of the Chromophore Area
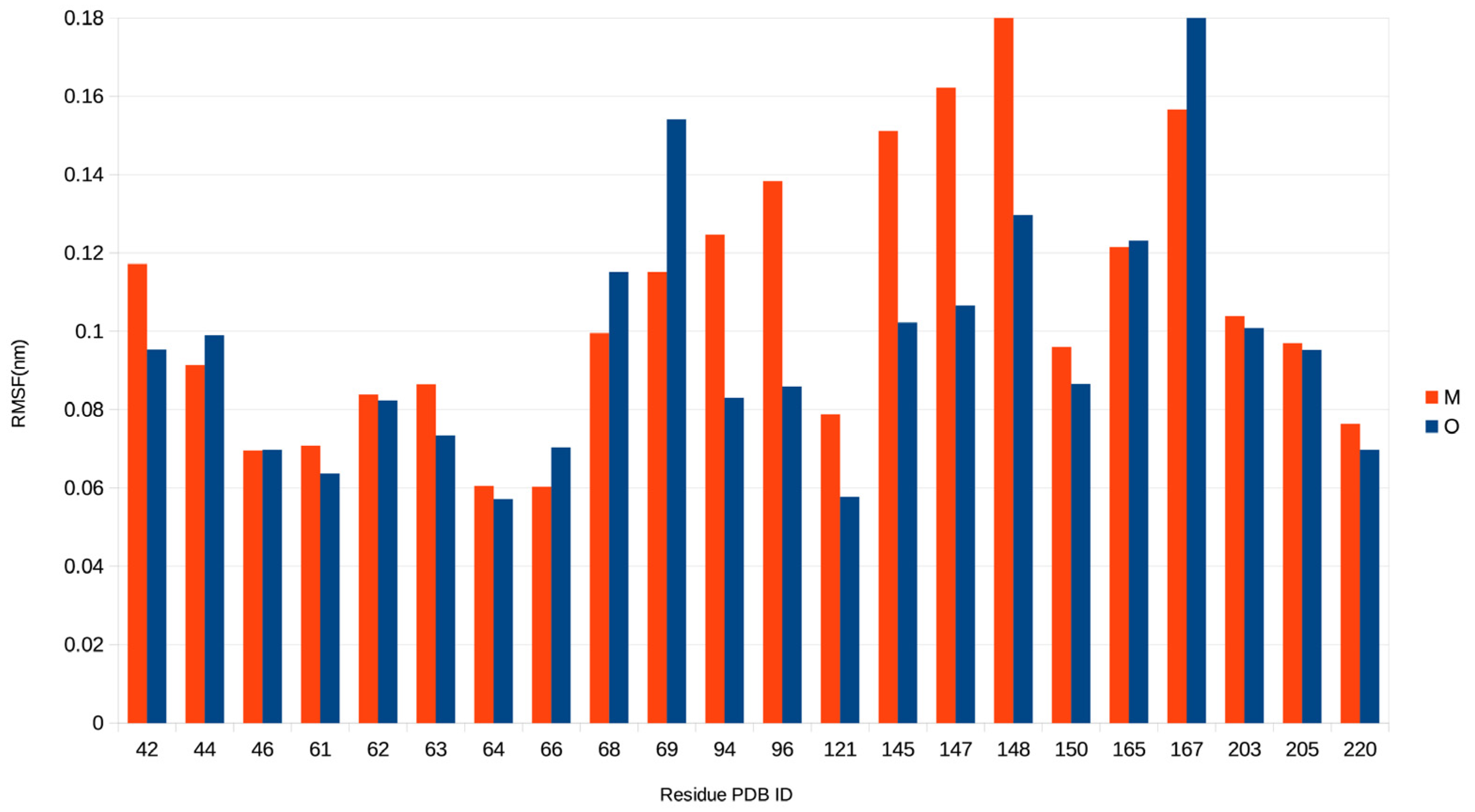
2.3. Electric and Dynamic Properties of the Chromophore-Binding Site
2.4. Conclusive Summary
3. Materials and Methods
3.1. Crystallization
3.2. X-ray Data Collection, Structure Solution, and Crystallographic Refinement
3.3. Molecular Dynamics Simulation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chudakov, D.M.; Matz, M.V.; Lukyanov, S.; Lukyanov, K.A. Fluorescent Proteins and Their Applications in Imaging Living Cells and Tissues. Physiol. Rev. 2010, 90, 1103–1163. [Google Scholar] [CrossRef]
- Wu, B.; Piatkevich, K.D.; Lionnet, T.; Singer, R.H. Modern Fluorescent Proteins and Imaging Technologies to Study Gene Expression, Nuclear Localization, and Dynamics. Curr. Opin. Cell Biol. 2011, 23, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Lam, A.J.; St-Pierre, F.; Gong, Y.; Marshall, J.D.; Cranfill, P.J.; Baird, M.A.; McKeown, M.R.; Wiedenmann, J.; Davidson, M.W.; Schnitzer, M.J.; et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 2012, 9, 1005–1012. [Google Scholar] [CrossRef]
- Mishin, A.S.; Belousov, V.V.; Solntsev, K.M.; Lukyanov, K.A. Novel uses of fluorescent proteins. Curr. Opin. Chem. Biol. 2015, 27, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, F. Back to the Future: Genetically Encoded Fluorescent Proteins as Inert Tracers of the Intracellular Environment. Int. J. Mol. Sci. 2020, 21, 4164. [Google Scholar] [CrossRef]
- Periasamy, A.; Clegg, R.M. FLIM Microscopy in Biology and Medicine, 1st ed.; CRC Press: Boca Raton, FL, USA, 2020; 472p. [Google Scholar]
- Datta, R.; Heaster, T.M.; Sharick, J.T.; Gillette, A.A.; Skala, M.C. Fluorescence lifetime imaging microscopy: Fundamentals and advances in instrumentation, analysis, and applications. J. Biomed Opt. 2020, 25, 071203. [Google Scholar] [CrossRef] [PubMed]
- Margineanu, A.; Chan, J.J.; Kelly, D.J.; Warren, S.C.; Flatters, D.; Kumar, S.; Katan, M.; Dunsby, C.W.; French, P.M.W. Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM). Sci. Rep. 2016, 6, 28186. [Google Scholar] [CrossRef] [PubMed]
- Mamontova, A.V.; Solovyev, I.D.; Savitsky, A.P.; Shakhov, A.M.; Lukyanov, K.A.; Bogdanov, A.M. Bright GFP with subnanosecond fluorescence lifetime. Sci. Rep. 2018, 8, 13224. [Google Scholar] [CrossRef]
- Mamontova, A.V.; Shakhov, A.M.; Lukyanov, K.A.; Bogdanov, A.M. Deciphering the Role of Positions 145 and 165 in Fluorescence Lifetime Shortening in the EGFP Variants. Biomolecules 2020, 10, 1547. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System; Delano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [PubMed]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Cryst. D Biol. Cryst. 2010, D66, 22–25. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Cryst. D Biol. Cryst. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Skubak, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Cryst. D Biol. Cryst. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Cryst. Biol. Cryst. 2010, D66, 486–501. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926. [Google Scholar] [CrossRef]
- Beglov, D.; Roux, B. Finite representation of an infinite bulk system: Solvent boundary potential for computer simulations. J. Chem. Phys. 1994, 100, 9050. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Pall, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputer. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- MacKerell, A.D., Jr.; Bashford, D.; Bellott, M.L.; Dunbrack, R.L., Jr.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowits, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577. [Google Scholar] [CrossRef]
- Reuter, N.; Lin, H.; Thiel, W. Green Fluorescent Proteins: Empirical Force Field for the Neutral and Deprotonated Forms of the Chromophore. Molecular Dynamics Simulations of the Wild Type and S65T Mutant. J. Phys. Chem. B 2002, 106, 6310–6321. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671. [Google Scholar] [CrossRef] [PubMed]



| Fluorescent Protein | λex/λem | EC, M−1·cm−1 | FQY | Relative Brightness *, % | FL, ns | Relative Photostability, % | |
|---|---|---|---|---|---|---|---|
| In Vitro | In Cellulo | ||||||
| EGFP | 489/509 | 55,000 | 0.60 | 100 | 2.83 ± 20 | 100 ± 8 | 100 ± 20 |
| BrUSLEE | 487/509 | 86,000 | 0.30 | 78 | 0.66 ± 35 | 190 ± 4 | 230 ± 45 |
| H-Bonds * | d (Å) |
|---|---|
| Asn146(ND2)∙∙∙∙(OD1)Asn146 | 3.28 |
| Asn(144)(ND2)∙∙∙∙(OG)Ser147 | 3.17 |
| Asn(170)(ND2)∙∙∙∙(OG) Ser147 | 3.59 |
| Leu207(O)∙∙∙∙(NE2)Gln204 | 2.66 |
| Asn212(ND2)∙∙∙∙(OH)Tyr39 | 3.62 |
| Hydrophobic cluster * | |
| Phe123 + Leu221 + Ala206 | |
| Chromophore | H-Bonds | π–π |
|---|---|---|
| EGFP | 3.6 | 0.81 |
| BrUSLEE | 2.4 | 0.37 |
| H-Bonds | |||
|---|---|---|---|
| EGFP | BrUSLEE | ||
| Residue | PrOc | Residue | PrOc |
| VAL61 | 0.98 | VAL61 | 0.92 |
| ARG96 | 0.83 | ARG96 | 0.49 |
| GLN94 | 0.77 | GLN94 | 0.33 |
| HIS148 | 0.5 | SER205 | 0.25 |
| GLN69 | 0.23 | GLN69 | 0.22 |
| TYR145 | 0.09 | SER147 | 0.16 |
| π–π Interactions | |||
| PHE165 | 0.81 | TYR165 | 0.37 |
| Protein | BrUSLEE |
|---|---|
| Crystallographic data | |
| Space group | P212121 |
| Cell dimensions (Å, °) | a = 75.66, b = 122.71 c = 167.40 |
| Z(Z’). | 4(6) |
| Estimated solvent content (%) | 47.0 |
| Temperature (K) | 100 |
| Wavelength (Å) | 1.00 |
| Resolution range (Å) | 29.72–2.38 (2.49–2.38) |
| Total observations | 427,530 |
| Unique reflections observed | 62,234 |
| Redundancy | 6.9 (5.6) |
| I/σ(I) | 20.1 (1.6) |
| Rmerge | 0.094 (0.930) |
| Completeness (%) | 99.9 (99.3) |
| Refinement statistics | |
| Non-H atoms in model | |
| Protein | 1826 [24 × 231 res)] |
| Water | 185 |
| Rwork | 0.203 |
| Rfree | 0.271 |
| Fraction of free reflections (%) | 2.0 |
| RMSD from ideal values: | |
| Bond lengths (Å) | 0.011 |
| Bond angles (grad) | 1.404 |
| Torsion angles (period 3; grad) | 17.2 |
| Chirality (Å3) | 0.074 |
| General planes (Å) | 0.014 |
| Ramachandran statistics (%) | |
| Preferred/Allowed/Outliers | 95.8/4.0/0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goryacheva, E.; Efremov, R.; Krylov, N.; Artemyev, I.; Bogdanov, A.; Mamontova, A.; Pletnev, S.; Pletneva, N.; Pletnev, V. Crystal Structure of Bright Fluorescent Protein BrUSLEE with Subnanosecond Fluorescence Lifetime; Electric and Dynamic Properties. Int. J. Mol. Sci. 2023, 24, 6403. https://doi.org/10.3390/ijms24076403
Goryacheva E, Efremov R, Krylov N, Artemyev I, Bogdanov A, Mamontova A, Pletnev S, Pletneva N, Pletnev V. Crystal Structure of Bright Fluorescent Protein BrUSLEE with Subnanosecond Fluorescence Lifetime; Electric and Dynamic Properties. International Journal of Molecular Sciences. 2023; 24(7):6403. https://doi.org/10.3390/ijms24076403
Chicago/Turabian StyleGoryacheva, Ekaterina, Roman Efremov, Nikolai Krylov, Igor Artemyev, Alexey Bogdanov, Anastasia Mamontova, Sergei Pletnev, Nadya Pletneva, and Vladimir Pletnev. 2023. "Crystal Structure of Bright Fluorescent Protein BrUSLEE with Subnanosecond Fluorescence Lifetime; Electric and Dynamic Properties" International Journal of Molecular Sciences 24, no. 7: 6403. https://doi.org/10.3390/ijms24076403
APA StyleGoryacheva, E., Efremov, R., Krylov, N., Artemyev, I., Bogdanov, A., Mamontova, A., Pletnev, S., Pletneva, N., & Pletnev, V. (2023). Crystal Structure of Bright Fluorescent Protein BrUSLEE with Subnanosecond Fluorescence Lifetime; Electric and Dynamic Properties. International Journal of Molecular Sciences, 24(7), 6403. https://doi.org/10.3390/ijms24076403







