A Novel D-Psicose 3-Epimerase from Halophilic, Anaerobic Iocasia fonsfrigidae and Its Application in Coconut Water
Abstract
1. Introduction
2. Results
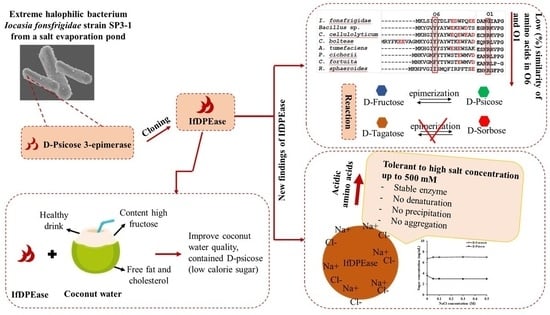
2.1. Amino Acid Sequence Analysis
2.2. Protein Expression and Purification and Allergen Analysis
2.3. pI Values of IfDPEase and Other Ketose 3-Epimerases
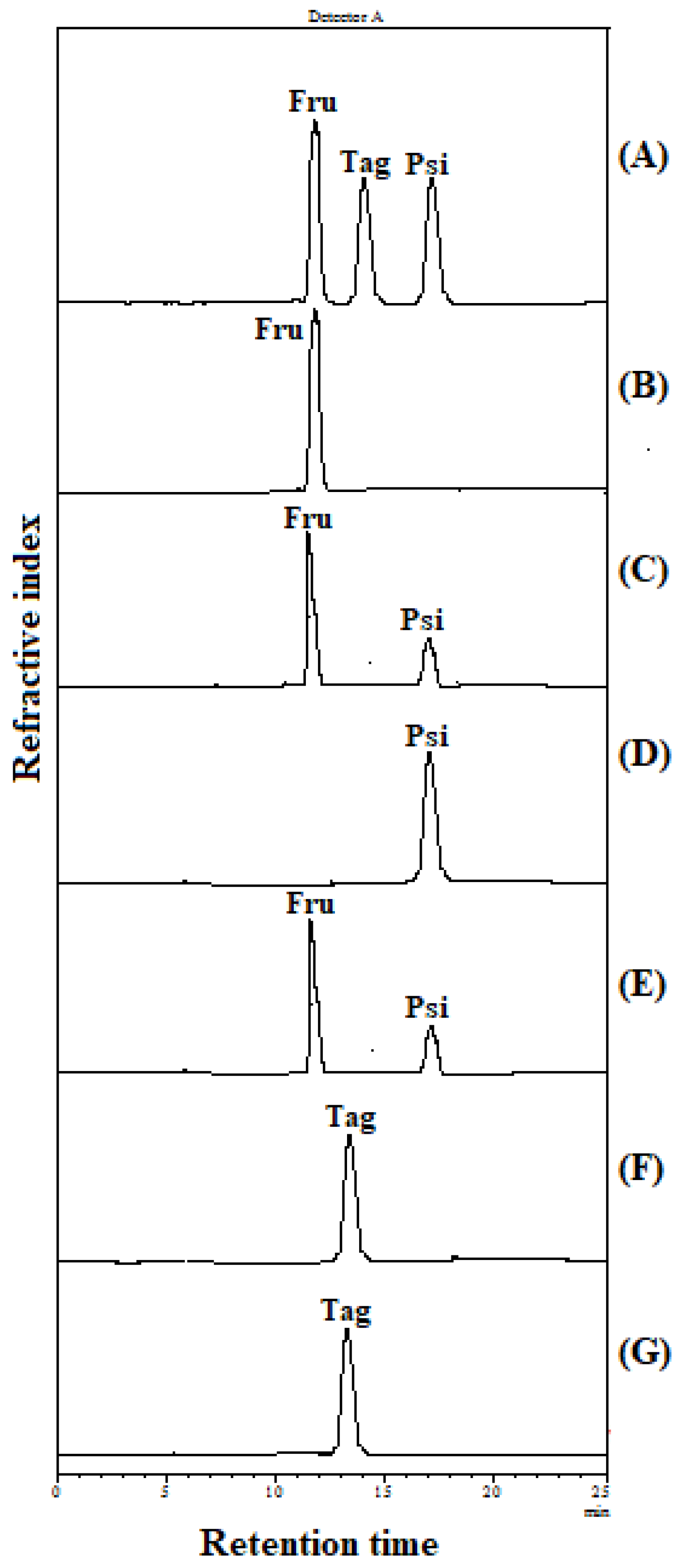
2.4. Substrate Specificity
2.5. Enzyme Kinetics
2.6. Effects of pH, Temperature, and Metal Ions on IfDPEase Activity
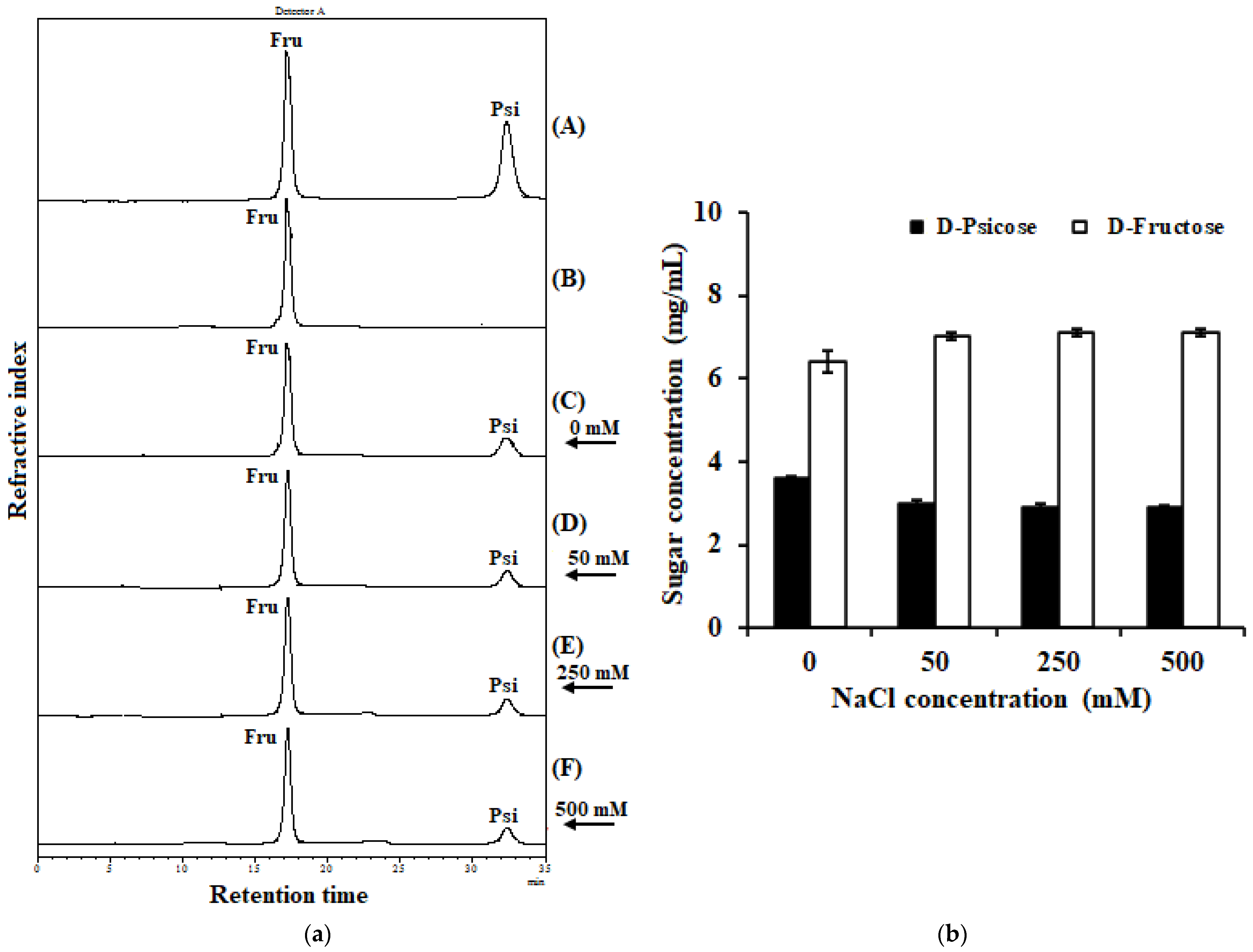
2.7. Effect of Sodium Chloride on IfDPEase Activity
2.8. Conversion of D-Fructose in Coconut Water into D-Psicose by IfDPEase
3. Discussion
4. Materials and Methods
4.1. Strains and Plasmid
4.2. Chemicals and Reagents
4.3. Alignment and Allergenic Protein Sequence Search
4.4. Phylogenetic Tree of IfDPEase with Other Characterized Members
4.5. Gene Manipulation, Expression, and Purification of Recombinant IfDPEase
4.6. Enzyme Assay
4.7. Substrate Specificity of IfDPEase
4.8. Effects of pH, Temperature, and Metal Ions on IfDPEase Activity
4.9. Effect of Sodium Chloride on IfDPEase Activity
4.10. Prediction of the Isoelectric Point (pI) and Acidic Amino Acids on the Surface of Ketose 3-Epimerase Structures
4.11. Determination of Enzyme Kinetics
4.12. Application of IfDPEase to Coconut Water
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xia, Y.; Cheng, Q.; Mu, W.; Hu, X.; Sun, Z.; Qiu, Y.; Liu, X.; Wang, Z. Research advances of D-allulose: An overview of physiological functions, enzymatic biotransformation technologies, and production processes. Foods 2021, 10, 2186. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.N.; Kaushal, G.; Singh, S.P. D-Allulose 3-epimerase of Bacillus sp. origin manifests profuse heat-stability and noteworthy potential of D-fructose epimerization. Microb. Cell Fact. 2021, 20, 60. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, Z.; Zhang, W.; Guang, C.; Mu, W. Characterization of a D-tagatose 3-epimerase from Caballeronia fortuita and its application in rare sugar production. Int. J. Biol. Macromol. 2019, 138, 536–545. [Google Scholar] [CrossRef]
- Tang, X.; An, Y.; Iqbal, M.W.; Cong, H.; Zhang, G.; Zhang, Y.; Ravikumar, Y.; Zabed, H.M.; Zhao, M.; Zhou, H.; et al. The characterization of a novel D-allulose 3-epimerase from Blautia produca and its application in D-allulose production. Foods 2022, 11, 3225. [Google Scholar] [CrossRef]
- Mu, W.; Zhang, W.; Feng, Y.; Jiang, B.; Zhou, L. Recent advances on applications and biotechnological production of D-psicose. Appl. Microbiol. Biotechnol. 2012, 94, 1461–1467. [Google Scholar] [CrossRef]
- DasSarma, S.; DasSarma, P. Halophiles and their enzymes: Negativity put to good use. Curr. Opin. Microbiol. 2015, 25, 120–126. [Google Scholar] [CrossRef]
- Martínez, G.M.; Pire, C.; Martínez-Espinosa, R.M. Hypersaline environments as natural sources of microbes with potential applications in biotechnology: The case of solar evaporation systems to produce salt in Alicante county (Spain). Curr. Res. Microbial. Sci. 2022, 3, 100136. [Google Scholar] [CrossRef]
- Prades, A.; Dornier, M.; Diop, N.; Pain, J.-P. Coconut water uses, composition and properties: A review. Fruits 2014, 67, 87–107. [Google Scholar] [CrossRef]
- Sunil, L.; Appaiah, P.; Prasanth Kumar, P.K.; Gopala Krishna, A.G. Coconut water—A nature’s miracle health drink: Chemistry, health benefits, packaging, storage, and technologies: A review. Indian Coconut J. 2020, 63, 17–25. [Google Scholar]
- Heng, S.; Sutheeworapong, S.; Champreda, V.; Uke, A.; Kosugi, A.; Pason, P.; Waeonukul, R.; Ceballos, R.M.; Ratanakhanokchai, K.; Tachaapaikoon, C. Genomics and cellulolytic, hemicellulolytic and amylolytic potential of Iocasia fonsfrigidae strain SP3-1 for polysaccharide degradation. PeerJ 2022, 10, 14211. [Google Scholar] [CrossRef]
- Chan, H.-C.; Zhu, Y.; Hu, Y.; Ko, T.-P.; Huang, C.-H.; Ren, F.; Chen, C.-C.; Ma, Y.; Guo, R.-T.; Sun, Y. Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars. Protein Cell 2012, 3, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Yong, J.W.H.; Ge, L.; Ng, Y.F.; Tan, S.N. The chemical composition and biological properties of coconut (Cocos nucifera L.) water. Molecules 2009, 14, 5144–5164. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Mu, W.; Chu, F.; Zhang, X.; Jiang, B.; Zhou, L.; Zhang, T. A D-psicose 3-epimerase with neutral pH optimum from Clostridium bolteae for D-psicose production: Cloning, expression, purification, and characterization. Appl. Microbiol. Biotechnol. 2014, 98, 717–725. [Google Scholar] [CrossRef]
- Kim, H.J.; Hyun, E.K.; Kim, Y.S.; Lee, Y.J.; Oh, D.K. Characterization of an Agrobacterium tumefaciens D-psicose 3-epimerase that converts D-fructose to D-psicose. Appl. Environ. Microbiol. 2006, 72, 981–985. [Google Scholar] [CrossRef]
- Yoshida, H.; Yamada, M.; Nishitani, T.; Takada, G.; Izumori, K.; Kamitori, S. Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose. J. Mol. Biol. 2007, 374, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Mu, W.; Jiang, B.; Zhang, T. Characterization of D-tagatose-3-epimerase from Rhodobacter sphaeroides that converts D-fructose into D-psicose. Biotechnol. Lett. 2009, 31, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, H.; Zhang, T.; Jiang, B.; Zhou, L.; Mu, W. Characterization of a D-psicose 3-epimerase from Dorea sp. CAG317 with an acidic pH optimum and a high specific activity. J. Mol. Catal. B Enzym. 2015, 120, 68–74. [Google Scholar] [CrossRef]
- Zhu, Y.; Men, Y.; Bai, W.; Li, X.; Zhang, L.; Sun, Y.; Ma, Y. Overexpression of D-psicose 3-epimerase from Ruminococcus sp. in Escherichia coli and its potential application in D-psicose production. Biotechnol. Lett. 2012, 34, 1901–1906. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Chu, F.; Xing, Q.; Yu, S.; Zhou, L.; Jiang, B. Cloning, expression, and characterization of a D-psicose 3-epimerase from Clostridium cellulolyticum H10. J. Agric. Food Chem. 2011, 59, 7785–7792. [Google Scholar] [CrossRef]
- Itoh, H.; Okaya, H.; Khan, A.R.; Tajima, S.; Hayakawa, S.; Izumori, K. Purification and characterization of D-tagatose 3-epimerase from Pseudomonas sp. ST-24. Biosci. Biotechnol. Biochem. 1994, 58, 2168–2171. [Google Scholar] [CrossRef]
- Santoso, U.; Kubo, K.; Ota, T.; Tadokoro, T.; Maekawa, A. Nutrient composition of kopyor coconuts (Cocos nucifera L.). Food Chem. 1996, 51, 299–304. [Google Scholar] [CrossRef]
- Phakeenuya, V.; Ratanakhanokchai, K.; Kosugi, A.; Tachaapaikoon, C. A novel multifunctional GH9 enzyme from Paenibacillus curdlanolyticus B-6 exhibiting endo/exo functions of cellulase, mannanase and xylanase activities. Appl. Microbiol. Biotechnol. 2020, 104, 2079–2096. [Google Scholar] [CrossRef]
- Heng, S.; Sutheeworapong, S.; Prommeenate, P.; Cheevadhanarak, S.; Kosugi, A.; Pason, P.; Waeonukul, R.; Ratanakhanokchai, K.; Tachaapaikoon, C. Complete genome sequence of Halocella sp. strain SP3-1, an extremely halophilic, glycoside hydrolase- and bacteriocin-producing bacterium isolated from a salt evaporation pond. Microbiol. Resour. Announc. 2019, 8, e01696-18. [Google Scholar] [CrossRef] [PubMed]
- Oshima, H.; Kimura, I.; Izumori, K. Psicose contents in various food products and its origin. Food Sci. Technol. Res. 2006, 12, 137–143. [Google Scholar] [CrossRef]
- Hossain, A.; Yamaguchi, F.; Matsuo, T.; Tsukamoto, I.; Toyoda, Y.; Ogawa, M.; Nagata, Y.; Tokuda, M. Rare sugar D-allulose: Potential role and therapeutic monitoring in maintaining obesity and type 2 diabetes mellitus. Pharmacol. Ther. 2015, 155, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Beerens, K.; Desmet, T.; Soetaert, W. Enzymes for the biocatalytic production of rare sugars. J. Ind. Microbiol. Biotechnol. 2012, 39, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.H.; Kwon, M.; Kim, S.W. Advanced whole-cell conversion for D-allulose production using an engineered Corynebacterium glutamicum. Biotechnol. Bioprocess Eng. 2022, 27, 276–285. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Phitsuwan, P.; Tachaapaikoon, C.; Kosugi, A.; Mori, Y.; Kyu, K.L.; Ratanakhanokchai, K. A cellulolytic and xylanolytic enzyme complex from an alkalothermoanaerobacterium, Tepidimicrobium xylanilyticum BT14. J. Microbiol. Biotechnol. 2010, 20, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Høie, M.H.; Kiehl, E.N.; Petersen, B.; Nielsen, M.; Winther, O.; Nielsen, H.; Hallgren, J.; Marcatili, P. NetSurfP-3.0: Accurate and fast prediction of protein structural features by protein language models and deep learning. Nucleic Acids Res. 2022, 50, W510–W515. [Google Scholar] [CrossRef]


| Source | Total Number of Aspartic and Glutamic Acids | Aspartic and Glutamic Acids on the Protein Surface (NetsurfP-3.0) | Aspartic and Glutamic Acids on the Protein Surface (PredictProtein) | Percentage of Aspartic and Glutamic Acids on the Protein Surface (%) | pI | Similarity of IfDPEase Compared to Other 3-Epimerases (%) |
|---|---|---|---|---|---|---|
| Iocasia fonsfrigidae strain SP3-1 | 40 from 269 aa | 34 | 34 | 85.00 | 4.95 | 100 |
| Bacillus sp. KCTC 13219 | 46 from 288 aa | 37 | 37 | 80.43 | 4.98 | 24.32 |
| Clostridium cellulolyticum H10 | 43 from 293 aa | 35 | 35 | 81.39 | 5.41 | 23.55 |
| Agrobacterium sp. ATCC 31749 | 37 from 289 aa | 27 | 27 | 72.97 | 5.94 | 22.01 |
| Ruminococcus sp. | 43 from 291 aa | 32 | 32 | 74.41 | 5.24 | 21.84 |
| Agrobacterium tumefaciens | 38 from 289 aa | 24 | 24 | 63.15 | 5.88 | 21.24 |
| Rhodobacter sphaeroides | 40 from 295 aa | 33 | 33 | 82.50 | 5.20 | 20.93 |
| Treponema primitia ZAS-1 | 41 from 295 aa | 31 | 31 | 75.60 | 5.93 | 20.69 |
| Clostridium. scindens 35704 | 47 from 289 aa | 35 | 35 | 74.46 | 5.14 | 20.46 |
| Pseudomonas cichorii | 46 from 290 aa | 38 | 38 | 82.60 | 5.21 | 20.38 |
| Desmospora sp. 8437 | 49 from 289 aa | 38 | 38 | 77.55 | 5.24 | 20.08 |
| Caballeronia fortuita | 44 from 291 aa | 33 | 33 | 75.00 | 5.19 | 20.00 |
| Clostridium bolteae | 49 from 301 aa | 39 | 39 | 79.59 | 5.04 | 19.92 |
| Dorea sp. CAG317 | 46 from 289 aa | 34 | 34 | 73.91 | 4.99 | 18.92 |
| Active Site | I. fonsfrigidae | Bacillus sp. | C. cellulolyticum | C. boltae | A. tumefaciens | P. cichorii | C. fortuita | R. sphaeroides |
|---|---|---|---|---|---|---|---|---|
| Catalytic site | Glu153 | Glu150 | Glu150 | Glu155 | Glu150 | Glu152 | Glu152 | Glu156 |
| Glu247 | Glu244 | Glu244 | Glu256 | Glu244 | Glu246 | Glu246 | Glu250 | |
| Metal-coordinating site | Glu153 | Glu150 | Glu150 | Glu162 | Glu150 | Glu152 | Glu152 | Glu156 |
| Asp186 | Asp183 | Asp183 | Asp195 | Asp183 | Asp185 | Asp185 | Asp189 | |
| His212 | His209 | His209 | His221 | His209 | His211 | His211 | Gln215 | |
| Glu247 | Glu244 | Glu244 | Glu256 | Glu244 | Glu246 | Glu246 | Glu250 | |
| Binding site on D-fructose for O-1 | Glu153 | Glu150 | Glu150 | Glu162 | Glu150 | Glu152 | Glu152 | Glu156 |
| Gly218 | Arg215 | Arg215 | Arg227 | Arg215 | Arg217 | Arg217 | Arg221 | |
| O-2 | Asp186 | Asp183 | Asp183 | Asp195 | Asp183 | Asp185 | Asp185 | Asp189 |
| His189 | His186 | His186 | His198 | His186 | His188 | His188 | His192 | |
| Glu247 | Glu244 | Glu244 | Glu256 | Glu244 | Glu246 | Glu246 | Glu250 | |
| O-3 OE1 | Glu153 | Glu150 | Glu150 | Glu162 | Glu150 | Glu152 | Glu152 | Glu156 |
| OE2 | Glu153 | Glu150 | Glu150 | Glu162 | Glu150 | Glu152 | Glu152 | Glu156 |
| O-4 | His212 | His209 | His209 | His221 | His209 | His211 | His211 | Gln215 |
| O-6 | Cys6 | Tyr6 | Tyr6 | Tyr16 | Tyr6 | Phe7 | Phe7 | Ile8 |
| Reference | This study | [2] | [11] | [13] | [14] | [15] | [3] | [16] |
| Enzyme | Source | Specific Activity (U mg−1) | Epimerization Reaction Ratio (D-Fructose/D-Psicose) | Reference | ||
|---|---|---|---|---|---|---|
| D-Fructose | D-Psicose | D-Tagatose | ||||
| DPEase | I. fonsfrigidae SP3-1 | 67.62 ± 2.10 | 96.35 ± 1.70 | ND | 0.70 | This study |
| C. bolteae | 150.70 | 226.90 | 52.70 | 0.66 | [13] | |
| Dorea sp. CAG317 | 803.00 | 1310.00 | 64.18 | 0.61 | [17] | |
| Ruminococcus sp. | 8.95 | 16.00 | 0.15 | 0.56 | [18] | |
| A. tumefaciens | 8.89 | 17.50 | 5.93 | 0.51 | [14] | |
| C. cellulolyticum H10 | 287.00 | 595.40 | 29.20 | 0.48 | [19] | |
| B. produca | 1.76 | 5.27 | 3.76 | 0.33 | [4] | |
| DTEase | P. cichorii | 4.00 | 12.00 | 20.00 | 0.33 | [20] |
| C. fortuita | 270.00 | 450.00 | 801.00 | 0.60 | [3] | |
| DFEase | R. sphaeroides | 380.70 | 209.90 | 230.80 | 1.81 | [16] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wulansari, S.; Heng, S.; Ketbot, P.; Baramee, S.; Waeonukul, R.; Pason, P.; Ratanakhanokchai, K.; Uke, A.; Kosugi, A.; Tachaapaikoon, C. A Novel D-Psicose 3-Epimerase from Halophilic, Anaerobic Iocasia fonsfrigidae and Its Application in Coconut Water. Int. J. Mol. Sci. 2023, 24, 6394. https://doi.org/10.3390/ijms24076394
Wulansari S, Heng S, Ketbot P, Baramee S, Waeonukul R, Pason P, Ratanakhanokchai K, Uke A, Kosugi A, Tachaapaikoon C. A Novel D-Psicose 3-Epimerase from Halophilic, Anaerobic Iocasia fonsfrigidae and Its Application in Coconut Water. International Journal of Molecular Sciences. 2023; 24(7):6394. https://doi.org/10.3390/ijms24076394
Chicago/Turabian StyleWulansari, Shinta, Sobroney Heng, Prattana Ketbot, Sirilak Baramee, Rattiya Waeonukul, Patthra Pason, Khanok Ratanakhanokchai, Ayaka Uke, Akihiko Kosugi, and Chakrit Tachaapaikoon. 2023. "A Novel D-Psicose 3-Epimerase from Halophilic, Anaerobic Iocasia fonsfrigidae and Its Application in Coconut Water" International Journal of Molecular Sciences 24, no. 7: 6394. https://doi.org/10.3390/ijms24076394
APA StyleWulansari, S., Heng, S., Ketbot, P., Baramee, S., Waeonukul, R., Pason, P., Ratanakhanokchai, K., Uke, A., Kosugi, A., & Tachaapaikoon, C. (2023). A Novel D-Psicose 3-Epimerase from Halophilic, Anaerobic Iocasia fonsfrigidae and Its Application in Coconut Water. International Journal of Molecular Sciences, 24(7), 6394. https://doi.org/10.3390/ijms24076394