Recent Advances in Generation of In Vitro Cardiac Organoids
Abstract
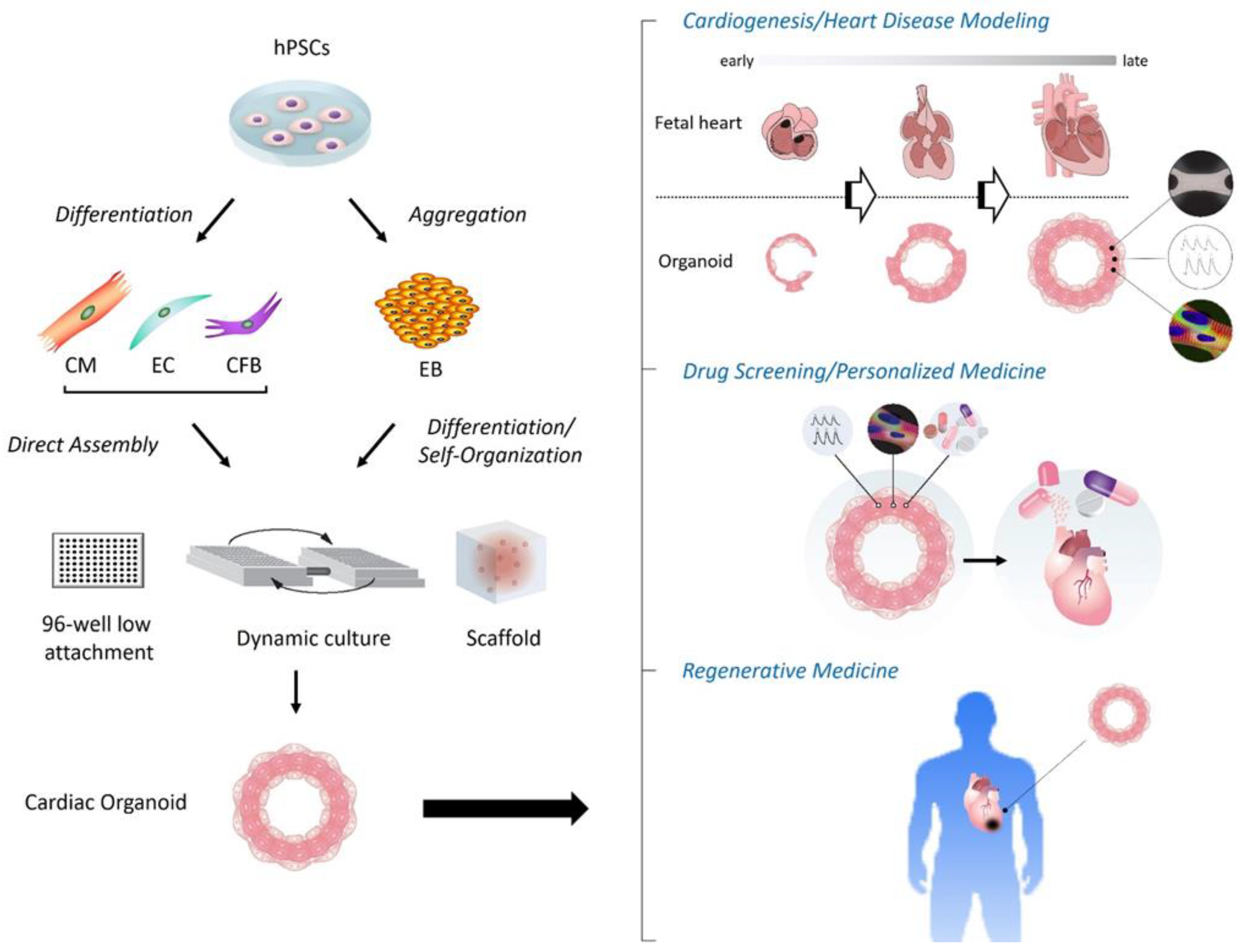
1. Introduction
2. Various Types of Cardiac Organoids
2.1. Cardiac Organoids in an Early Era
2.2. Latest Cardiac Organoids since 2021
| Authors & Years | CO Models/Platform | Sp. | Cells | Scaffold | Chemicals | Features | Chambers/Cavities | Applications | Refs |
|---|---|---|---|---|---|---|---|---|---|
| Rossi et al., 2021 | Gatruloid-derived CO | M | mESC aggregates | CHIR, FGF2, Ascorbic Acid, VEGF-A | - Gastruloids containing the three germ layer derivatives - Induction of cardiac crescent-like FHF/SHF structure - Formation of primitive gut-like structures with a codeveloped CM heart tube with a vascular/endocardium-like network | (−) | Cardiogenesis modeling | [44] | |
| Silva et al., 2021 | Multilineage CO | H | hiPSC-derived mes-endoderm progenitors’ aggregates | CHIR, IWP2, Ascorbic Acid, | - Reconstitution (force aggregation) of hiPSC-derived mesendoderm progenitors - Co-emergence of cardiac core and gut-like tube cells with epicardial lining, promoting CM compaction and maturation | (−) | Cardiogenesis modeling | [45] | |
| Drakhlis et al., 2021 | Heart-Forming Organoid | H | hPSC aggregates | Matrigel | CHIR, IWP2 | - Formation of three-layered self-assembly: (inner) endothelial/endocardial/foregut cells; (middle) myocardial/epicardial cells; (outer) mesenchyme/liver cells - Recapitulation of early heart and foregut development | (+) | Cardiogenesis & disease modeling (non-compact HCM by NKX2-5 KO) | [46] |
| Hofbauer et al., 2021 | Cardioid | H | hPSC aggregates | Vitronectin Laminin | CHIR, BMP4, FGF2, Activin A, LY294002, Insulin, IWP2, VEGF-A, RA | - Identification of mesodermal Wnt-BMP signaling axis (with HAND1)-modulated cavity formation principles, assembled by epicardium and myocardium and lined by a layer of ECs | (+) | Cardiogenesis & disease modeling (cryoinjury & CHD by NKX2-5 or HAND1 KO) | [48] |
| Lewis-Israeli et al., 2021 | Scaffold-free self-assembling CO | H | hPSC aggregates | CHIR (On/Off/On), BMP4, Activin A, C59 | - Three-step Wnt signaling modulation for induction of cardiac mesoderm and epicardial cells - Recapitulation of internal chambers formed by multi-lineage cardiac cell types with well-organized sarcomeres in CMs and developed vasculature | (+) | Cardiogenesis & disease modeling (pregestational diabetes-induced cardiomyopathy) | [49] | |
| Ormsted et al., 2022 | EMLOC-induced CO | H | hPSC aggregates | CHIR, FGF2, HGF, IGF-1, VEGF-A, Ascorbic Acid | - Interconnected neuro-cardiac lineages in a single gastruloid model - Induction of heart tube formation, chamber-like structures, formation of a putative OFT, and innervated heart-like structure populated by neurons | (+) | Cardiogenesis modeling | [50] | |
| Branco et al., 2022 | Epicardium-myocardium organoid (EMO) | H | hPSC aggregate-derived CM aggregates and PE/STM/PFH organoids | CHIR, BMP4, RA, Ascorbic Acid | - Wnt/BMP4/RA-mediated hPSC-PE/STM/PFH organoids - EMO generated by reaggregating hPSC-derived CM aggregates and PE/STM/PFH-dissociated cells - EMO comprising an epicardium layer fully surrounding a myocardium layer | (−) | Cardiogenesis modeling | [52] | |
| Lee et al., 2022 | Chamber-forming CO | H | hiPSC aggregates | Matrigel | CHIR, C59 | - Manufacturing of chamber-forming hiPSC-derived COs based on Matrigel (10%) in anti-adherent dishes with dynamic culture | (+) | Cardiogenesis modeling & in vivo transplantation | [53] |
| Feng et al., 2022 | Chamber (atrium/ventricle)-specific CO | H | hiPSC aggregates | CHIR, C59, RA (+ or −) | - Generation of atrial-lineage and ventricular-lineage COs with or without RA treatment, enabling the study of heart disease with a specific chamber defect | (+) | Cardiogenesis & disease modeling (Ebstein’s anomaly by NKX2-5 mutant) | [54] | |
| Meier et al., 2023 | Epicardioid | H | hPSC aggregates | Collagen I | CHIR, BMP4, FGF2, Activin A, LY294002, Insulin, IWP2, RA | - Generation of self-organizing COs displaying morphological and functional patterning of the epicardium and myocardium typical of the left ventricular wall - Elucidation of fundamental roles and cellular heterogeneity of epicardial cells during ventricular development | (−) | Cardiogenesis & disease modeling (CHD using Noonan syndrome Pt. hiPSCs & ET1-induced HCM) | [55] |
3. Applications of Cardiac Organoids
4. Current Limitations and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.A.; Gonçalves, J.T.; Bloyd, C.W.; Li, H.; Fernandes, S.; Quang, D.; Johnston, S.; Parylak, S.L.; Jin, X.; Gage, F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018, 36, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Gehart, H.; Artegiani, B.; LÖpez-Iglesias, C.; Dekkers, F.; Basak, O.; van Es, J.; Chuva de Sousa Lopes, S.M.; Begthel, H.; Korving, J.; et al. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 2018, 175, 1591–1606. [Google Scholar] [CrossRef]
- Serra, D.; Mayr, U.; Boni, A.; Lukonin, I.; Rempfler, M.; Challet Meylan, L.; Stadler, M.B.; Strnad, P.; Papasaikas, P.; Vischi, D.; et al. Self-organization and symmetry breaking in intestinal organoid development. Nature 2019, 569, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Matano, M.; Toshimitsu, K.; Takano, A.; Mikami, Y.; Nishikori, S.; Sugimoto, S.; Sato, T. Human Intestinal Organoids Maintain Self-Renewal Capacity and Cellular Diversity in Niche-Inspired Culture Condition. Cell Stem Cell 2018, 23, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Takasato, M.; Er, P.X.; Chiu, H.S.; Maier, B.; Baillie, G.J.; Ferguson, C.; Parton, R.G.; Wolvetang, E.J.; Roost, M.S.; Chuva de Sousa Lopes, S.M.; et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 2015, 526, 564–568. [Google Scholar] [CrossRef]
- Thomas, D.; Choi, S.; Alamana, C.; Parker, K.K.; Wu, J.C. Cellular and Engineered Organoids for Cardiovascular Models. Circ. Res. 2022, 130, 1780–1802. [Google Scholar] [CrossRef]
- Rao, K.S.; Kameswaran, V.; Bruneau, B.G. Modeling congenital heart disease: Lessons from mice, hPSC-based models, and organoids. Genes Dev. 2022, 36, 652–663. [Google Scholar] [CrossRef]
- Xuan, W.; Tipparaju, S.M.; Ashraf, M. Transformational Applications of Human Cardiac Organoids in Cardiovascular Diseases. Front. Cell Dev. Biol. 2022, 10, 936084. [Google Scholar] [CrossRef]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Bellin, M.; Casini, S.; Davis, R.P.; D’Aniello, C.; Haas, J.; Ward-van Oostwaard, D.; Tertoolen, L.G.; Jung, C.B.; Elliott, D.A.; Welling, A.; et al. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. 2013, 32, 3161–3175. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.G.W.; Owen, T.; Bhagwan, J.R.; Mosqueira, D.; Scott, E.; Mannhardt, I.; Patel, A.; Barriales-Villa, R.; Monserrat, L.; Hansen, A.; et al. Isogenic Pairs of hiPSC-CMs with Hypertrophic Cardiomyopathy/LVNC-Associated ACTC1 E99K Mutation Unveil Differential Functional Deficits. Stem Cell Rep. 2018, 11, 1226–1243. [Google Scholar] [CrossRef] [PubMed]
- Prondzynski, M.; Lemoine, M.D.; Zech, A.T.; Horváth, A.; Di Mauro, V.; Koivumäki, J.T.; Kresin, N.; Busch, J.; Krause, T.; Krämer, E.; et al. Disease modeling of a mutation in α-actinin 2 guides clinical therapy in hypertrophic cardiomyopathy. EMBO Mol. Med. 2019, 11, e11115. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Aulner, N.; Bickle, M.; Davies, A.M.; Nery, E.D.; Ebner, D.; Montoya, M.C.; Östling, P.; Pietiäinen, V.; Price, L.S.; et al. Screening out irrelevant cell-based models of disease. Nat. Rev. Drug Discov. 2016, 15, 751–769. [Google Scholar] [CrossRef]
- Florian, W.; Ingra, M.; Thomas, E. Engineering Cardiac Muscle Tissue: A Maturating Field of Research. Circ. Res. 2017, 120, 1487–1500. [Google Scholar]
- Wang, G.; McCain, M.L.; Yang, L.; He, A.; Pasqualini, F.S.; Agarwal, A.; Yuan, H.; Jiang, D.; Zhang, D.; Zangi, L.; et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014, 20, 616–623. [Google Scholar] [CrossRef]
- Mosqueira, D.; Mannhardt, I.; Bhagwan, J.R.; Lis-Slimak, K.; Katili, P.; Scott, E.; Hassan, M.; Prondzynski, M.; Harmer, S.C.; Tinker, A.; et al. CRISPR/Cas9 editing in human pluripotent stem cell-cardiomyocytes highlights arrhythmias, hypocontractility, and energy depletion as potential therapeutic targets for hypertrophic cardiomyopathy. Eur. Heart J. 2018, 39, 3879–3892. [Google Scholar] [CrossRef]
- Hinson, J.T.; Chopra, A.; Nafissi, N.; Polacheck, W.J.; Benson, C.C.; Swist, S.; Gorham, J.; Yang, L.; Schafer, S.; Sheng, C.C.; et al. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 2015, 349, 982–986. [Google Scholar] [CrossRef]
- Tenreiro, M.F.; Louro, A.F.; Alves, P.M.; Serra, M. Next generation of heart regenerative therapies: Progress and promise of cardiac tissue engineering. NPJ Regen. Med. 2021, 6, 30. [Google Scholar] [CrossRef]
- Mohr, E.; Thum, T.; Bär, C. Accelerating cardiovascular research: Recent advances in translational 2D and 3D heart models. Eur. J. Heart Fail. 2022, 24, 1778–1791. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.; Löbel, W.; Finklea, F.; Halloin, C.; Ritzenhoff, K.; Manstein, F.; Mohammadi, S.; Hashemi, M.; Zweigerdt, R.; Lipke, E.; et al. Prediction of Human Induced Pluripotent Stem Cell Cardiac Differentiation Outcome by Multifactorial Process Modeling. Front. Bioeng. Biotechnol. 2020, 8, 851. [Google Scholar] [CrossRef]
- Wang, K.L.; Xue, Q.; Xu, X.H.; Hu, F.; Shao, H. Recent progress in induced pluripotent stem cell-derived 3D cultures for cardiac regeneration. Cell Tissue Res. 2021, 384, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.; Imran Alsous, J.; Guggenheim, J.W.; Mak, M.; Munera, J.; Wells, J.M.; Kamm, R.D.; Asada, H.H.; Shvartsman, S.Y.; Griffith, L.G. A process engineering approach to increase organoid yield. Development 2017, 144, 1128–1136. [Google Scholar] [CrossRef]
- Zhu, L.; Liu, K.; Feng, Q.; Liao, Y. Cardiac Organoids: A 3D Technology for Modeling Heart Development and Disease. Stem Cell Rev. Rep. 2022, 18, 2593–2605. [Google Scholar] [CrossRef] [PubMed]
- Häneke, T.; Sahara, M. Progress in Bioengineering Strategies for Heart Regenerative Medicine. Int. J. Mol. Sci. 2022, 23, 3482. [Google Scholar] [CrossRef]
- Vunjak-Novakovic, G.; Ronaldson-Bouchard, K.; Radisic, M. Organs-on-a-chip models for biological research. Cell 2021, 184, 4597–4611. [Google Scholar] [CrossRef]
- Saorin, G.; Caligiuri, I.; Rizzolio, F. Microfluidic organoids-on-a-chip: The future of human models. Semin. Cell Dev. Biol. 2023, 144, 41–54. [Google Scholar] [CrossRef]
- Qasim, M.; Haq, F.; Kang, M.H.; Kim, J.H. 3D printing approaches for cardiac tissue engineering and role of immune modulation in tissue regeneration. Int. J. Nanomed. 2019, 14, 1311–1333. [Google Scholar] [CrossRef]
- Kozaniti, F.K.; Metsiou, D.N.; Manara, A.E.; Athanassiou, G.; Deligianni, D.D. Recent Advancements in 3D Printing and Bioprinting Methods for Cardiovascular Tissue Engineering. Bioengineering 2021, 8, 133. [Google Scholar] [CrossRef]
- MacQueen, L.A.; Sheehy, S.P.; Chantre, C.O.; Zimmerman, J.F.; Pasqualini, F.S.; Liu, X.; Goss, J.A.; Campbell, P.H.; Gonzalez, G.M.; Park, S.J.; et al. A tissue-engineered scale model of the heart ventricle. Nat. Biomed. Eng. 2018, 2, 930–941. [Google Scholar] [CrossRef]
- Joddar, B.; Natividad-Diaz, S.L.; Padilla, A.E.; Esparza, A.A.; Ramirez, S.P.; Chambers, D.R.; Ibaroudene, H. Engineering approaches for cardiac organoid formation and their characterization. Transl. Res. 2022, 250, 46–67. [Google Scholar] [CrossRef] [PubMed]
- Moroni, L.; Burdick, J.A.; Highley, C.; Lee, S.J.; Morimoto, Y.; Takeuchi, S.; Yoo, J.J. Biofabrication strategies for 3D in vitro models and regenerative medicine. Nat. Rev. Mater. 2018, 3, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Zuppinger, C. 3D Cardiac Cell Culture: A Critical Review of Current Technologies and Applications. Front. Cardiovasc. Med. 2019, 6, 87. [Google Scholar] [CrossRef]
- Mills, R.J.; Titmarsh, D.M.; Koenig, X.; Parker, B.L.; Ryall, J.G.; Quaife-Ryan, G.A.; Voges, H.K.; Hodson, M.P.; Ferguson, C.; Drowley, L.; et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. USA 2017, 114, E8372–E8381. [Google Scholar] [CrossRef] [PubMed]
- Voges, H.K.; Mills, R.J.; Elliott, D.A.; Parton, R.G.; Porrello, E.R.; Hudson, J.E. Development of a human cardiac organoid injury model reveals innate regenerative potential. Development 2017, 144, 1118–1127. [Google Scholar] [CrossRef]
- Mills, R.J.; Parker, B.L.; Quaife-Ryan, G.A.; Voges, H.K.; Needham, E.J.; Bornot, A.; Ding, M.; Andersson, H.; Polla, M.; Elliott, D.A.; et al. Drug Screening in Human PSC-Cardiac Organoids Identifies Pro-proliferative Compounds Acting via the Mevalonate Pathway. Cell Stem Cell 2019, 24, 895–907. [Google Scholar] [CrossRef]
- Hoang, P.; Wang, J.; Conklin, B.R.; Healy, K.E.; Ma, Z. Generation of spatial-patterned early-developing cardiac organoids using human pluripotent stem cells. Nat. Protoc. 2018, 13, 723–737. [Google Scholar] [CrossRef]
- Hoang, P.; Kowalczewski, A.; Sun, S.; Winston, T.S.; Archilla, A.M.; Lemus, S.M.; Ercan-Sencicek, A.G.; Gupta, A.R.; Liu, W.; Kontaridis, M.I.; et al. Engineering spatial-organized cardiac organoids for developmental toxicity testing. Stem Cell Rep. 2021, 16, 1228–1244. [Google Scholar] [CrossRef]
- Lee, J.; Sutani, A.; Kaneko, R.; Takeuchi, J.; Sasano, T.; Kohda, T.; Ihara, K.; Takahashi, K.; Yamazoe, M.; Morio, T.; et al. In vitro generation of functional murine heart organoids via FGF4 and extracellular matrix. Nat. Commun. 2020, 11, 4283. [Google Scholar] [CrossRef]
- Richards, D.J.; Coyle, R.C.; Tan, Y.; Jia, J.; Wong, K.; Toomer, K.; Menick, D.R.; Mei, Y. Inspiration from heart development: Biomimetic development of functional human cardiac organoids. Biomaterials 2017, 142, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Richards, D.J.; Li, Y.; Kerr, C.M.; Yao, J.; Beeson, G.C.; Coyle, R.C.; Chen, X.; Jia, J.; Damon, B.; Wilson, R.; et al. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat. Biomed. Eng. 2020, 4, 446–462. [Google Scholar] [CrossRef] [PubMed]
- Filippo Buono, M.; von Boehmer, L.; Strang, J.; Hoerstrup, S.P.; Emmert, M.Y.; Nugraha, B. Human Cardiac Organoids for Modeling Genetic Cardiomyopathy. Cells 2020, 9, 1733. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Broguiere, N.; Miyamoto, M.; Boni, A.; Guiet, R.; Girgin, M.; Kelly, R.G.; Kwon, C.; Lutolf, M.P. Capturing Cardiogenesis in Gastruloids. Cell Stem Cell 2021, 28, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.C.; Matthys, O.B.; Joy, D.A.; Kauss, M.A.; Natarajan, V.; Lai, M.H.; Turaga, D.; Blair, A.P.; Alexanian, M.; Bruneau, B.G.; et al. Co-emergence of cardiac and gut tissues promotes cardiomyocyte maturation within human iPSC-derived organoids. Cell Stem Cell 2021, 28, 2137–2152. [Google Scholar] [CrossRef]
- Drakhlis, L.; Biswanath, S.; Farr, C.M.; Lupanow, V.; Teske, J.; Ritzenhoff, K.; Franke, A.; Manstein, F.; Bolesani, E.; Kempf, H.; et al. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 2021, 39, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Pashmforoush, M.; Lu, J.T.; Chen, H.; Amand, T.S.; Kondo, R.; Pradervand, S.; Evans, S.M.; Clark, B.; Feramisco, J.R.; Giles, W.; et al. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 2004, 117, 373–386. [Google Scholar] [CrossRef]
- Hofbauer, P.; Jahnel, S.M.; Papai, N.; Giesshammer, M.; Deyett, A.; Schmidt, C.; Penc, M.; Tavernini, K.; Grdseloff, N.; Meledeth, C.; et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell 2021, 184, 3299–3317. [Google Scholar] [CrossRef]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Gabalski, M.A.; Volmert, B.D.; Ming, Y.; Ball, K.A.; Yang, W.; Zou, J.; Ni, G.; Pajares, N.; et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat. Commun. 2021, 12, 5142. [Google Scholar] [CrossRef]
- Olmsted, Z.T.; Paluh, J.L. A combined human gastruloid model of cardiogenesis and neurogenesis. iScience 2022, 25, 104486. [Google Scholar] [CrossRef]
- Olmsted, Z.T.; Paluh, J.L. Co-development of central and peripheral neurons with trunk mesendoderm in human elongating multi-lineage organized gastruloids. Nat. Commun. 2021, 12, 3020. [Google Scholar] [CrossRef] [PubMed]
- Branco, M.A.; Dias, T.P.; Cabral, J.M.S.; Pinto-do-Ó, P.; Diogo, M.M. Human multilineage pro-epicardium/foregut organoids support the development of an epicardium/myocardium organoid. Nat. Commun. 2022, 13, 6981. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.G.; Kim, Y.J.; Son, M.Y.; Oh, M.S.; Kim, J.; Ryu, B.; Kang, K.R.; Baek, J.; Chung, G.; Woo, D.H.; et al. Generation of human iPSCs derived heart organoids structurally and functionally similar to heart. Biomaterials 2022, 290, 121860. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Schriever, H.; Jiang, S.; Bais, A.; Wu, H.; Kostka, D.; Li, G. Computational profiling of hiPSC-derived heart organoids reveals chamber defects associated with NKX2-5 deficiency. Commun. Biol. 2022, 5, 399. [Google Scholar] [CrossRef]
- Meier, A.B.; Zawada, D.; de Angelis, M.T.; Martens, L.D.; Santamaria, G.; Zengerle, S.; Nowak-Imialek, M.; Kornherr, J.; Zhang, F.; Tian, Q.; et al. Epicardioid single-cell genomics uncover principles of human epicardium biology in heart development and disease. Nat. Biotechnol. 2023, in press. [Google Scholar]
- Varzideh, F.; Pahlavan, S.; Ansari, H.; Halvaei, M.; Kostin, S.; Feiz, M.S.; Latifi, H.; Aghdami, N.; Braun, T.; Baharvand, H. Human cardiomyocytes undergo enhanced maturation in embryonic stem cell-derived organoid transplants. Biomaterials 2019, 192, 537–550. [Google Scholar] [CrossRef]
- Long, C.; Li, H.; Tiburcy, M.; Rodriguez-Caycedo, C.; Kyrychenko, V.; Zhou, H.; Zhang, Y.; Min, Y.L.; Shelton, J.M.; Mammen, P.P.A.; et al. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci. Adv. 2018, 4, eaap9004. [Google Scholar] [CrossRef]
- Mills, R.J.; Humphrey, S.J.; Fortuna, P.R.J.; Lor, M.; Foster, S.R.; Quaife-Ryan, G.A.; Johnston, R.L.; Dumenil, T.; Bishop, C.; Rudraraju, R.; et al. BET inhibition blocks inflammation-induced cardiac dysfunction and SARS-CoV-2 infection. Cell 2021, 184, 2167–2182. [Google Scholar] [CrossRef]
- Kim, H.; Kamm, R.D.; Vunjak-Novakovic, G.; Wu, J.C. Progress in multicellular human cardiac organoids for clinical applications. Cell Stem Cell 2022, 29, 503–514. [Google Scholar] [CrossRef]
- Jiang, S.; Feng, W.; Chang, C.; Li, G. Modeling Human Heart Development and Congenital Defects Using Organoids: How Close Are We? J. Cardiovasc. Dev. Dis. 2022, 9, 125. [Google Scholar] [CrossRef]
- Sesena-Rubfiaro, A.; Prajapati, N.J.; Paolino, L.; Lou, L.; Cotayo, D.; Pandey, P.; Shaver, M.; Hutcheson, J.D.; Agarwal, A.; He, J. Membrane Remodeling of Human-Engineered Cardiac Tissue by Chronic Electric Stimulation. ACS Biomater. Sci. Eng. 2023, 9, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Sewanan, L.R.; Shao, S.; Halder, S.S.; Stankey, P.; Li, X.; Campbell, S.G. Physiological calcium combined with electrical pacing accelerates maturation of human engineered heart tissue. Stem Cell Rep. 2022, 17, 2037–2049. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wan, Z.; Kamm, R.D. Vascularized organoids on a chip: Strategies for engineering organoids with functional vasculature. Lab Chip 2021, 21, 473–488. [Google Scholar] [CrossRef] [PubMed]
| Authors & Years | CO Models/Platform | Sp. | Cells | Scaffold | Chemicals | Features | Chambers/Cavities | Applications | Refs |
|---|---|---|---|---|---|---|---|---|---|
| Mills et al., 2017 Mills et al., 2019 | Heart Dynamometer-engineered CO | H | hPSC-derived cardiac cells | Collagen I Matrigel | CHIR, BMP4, Activin A, FGF2, IWP4 | - A 96-well device for high-throughput functional screening of hiPSC-derived CO to facilitate testing for maturation conditions - Identification of two pro-proliferative molecules without side effects on cardiac function, acting via the Mevalonate pathway | (−) | Maturation & drug screening (pro-proliferation) | [35,37] |
| Voges et al., 2017 | Circular CO engineered in the mold | H | hPSC-derived cardiac cells | Collagen I | CHIR, BMP4, Activin A, FGF2, IWP4 | - Human CO exhibiting an endogenous and full regenerative response 2 weeks after acute injury | (−) | Disease modeling (cryoinjury) | [36] |
| Hoang et al., 2018 Hoang et al., 2021 | Spacially-patterned CO | H | hPSCs | PDMS stencils with aligned holes | CHIR, IWP4 | - Biomaterial-based cell patterning combined with stem cell organoid engineering - Optimization of CO geometries for efficient CO production, reflecting high consistency and large morphology - Quantification of the embryotoxic potential of 9 pharmaceutical compounds | (−) | Cardiogenesis modeling & drug screening (embryo toxicity) | [38,39] |
| Lee et al., 2020 | Murine CO | M | mESC-EB | Laminin Entactin | FGF4, BIO, BMP4 | - Innovative approach to generate CO with chamber formation (i.e., both atria- and ventricle-like parts) from mESC EBs via FGF4 and ECM | (+) | Cardiogenesis modeling | [40] |
| Richards et al., 2017 Richards et al., 2020 | Multicellular CO | H | hiPSC-CM, hCFB, HUVEC, hADSC | Agarose | Not applicable * | - Generation of human CO that resembled the lumenized vascular network in the developing heart - Modeling of human heart structure after MI by oxygen diffusion gradient and NA stimulation | (−) | Cardiogenesis & disease modeling (ischemia) | [41,42] |
| Buono et al., 2020 | Multicellular CO | H | hiPSC-CM, HCMEC, hCFB | CHIR, C59 | - CO generation with a tri-culture approach - Clear differences in structures and beating behavior in between HCM Pt. (MYH7-mutant) hiPSC-derived and control COs | (−) | Disease modeling (HCM) | [43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sahara, M. Recent Advances in Generation of In Vitro Cardiac Organoids. Int. J. Mol. Sci. 2023, 24, 6244. https://doi.org/10.3390/ijms24076244
Sahara M. Recent Advances in Generation of In Vitro Cardiac Organoids. International Journal of Molecular Sciences. 2023; 24(7):6244. https://doi.org/10.3390/ijms24076244
Chicago/Turabian StyleSahara, Makoto. 2023. "Recent Advances in Generation of In Vitro Cardiac Organoids" International Journal of Molecular Sciences 24, no. 7: 6244. https://doi.org/10.3390/ijms24076244
APA StyleSahara, M. (2023). Recent Advances in Generation of In Vitro Cardiac Organoids. International Journal of Molecular Sciences, 24(7), 6244. https://doi.org/10.3390/ijms24076244







