Stabilization of the Dimeric State of SARS-CoV-2 Main Protease by GC376 and Nirmatrelvir
Abstract
1. Introduction
2. Results
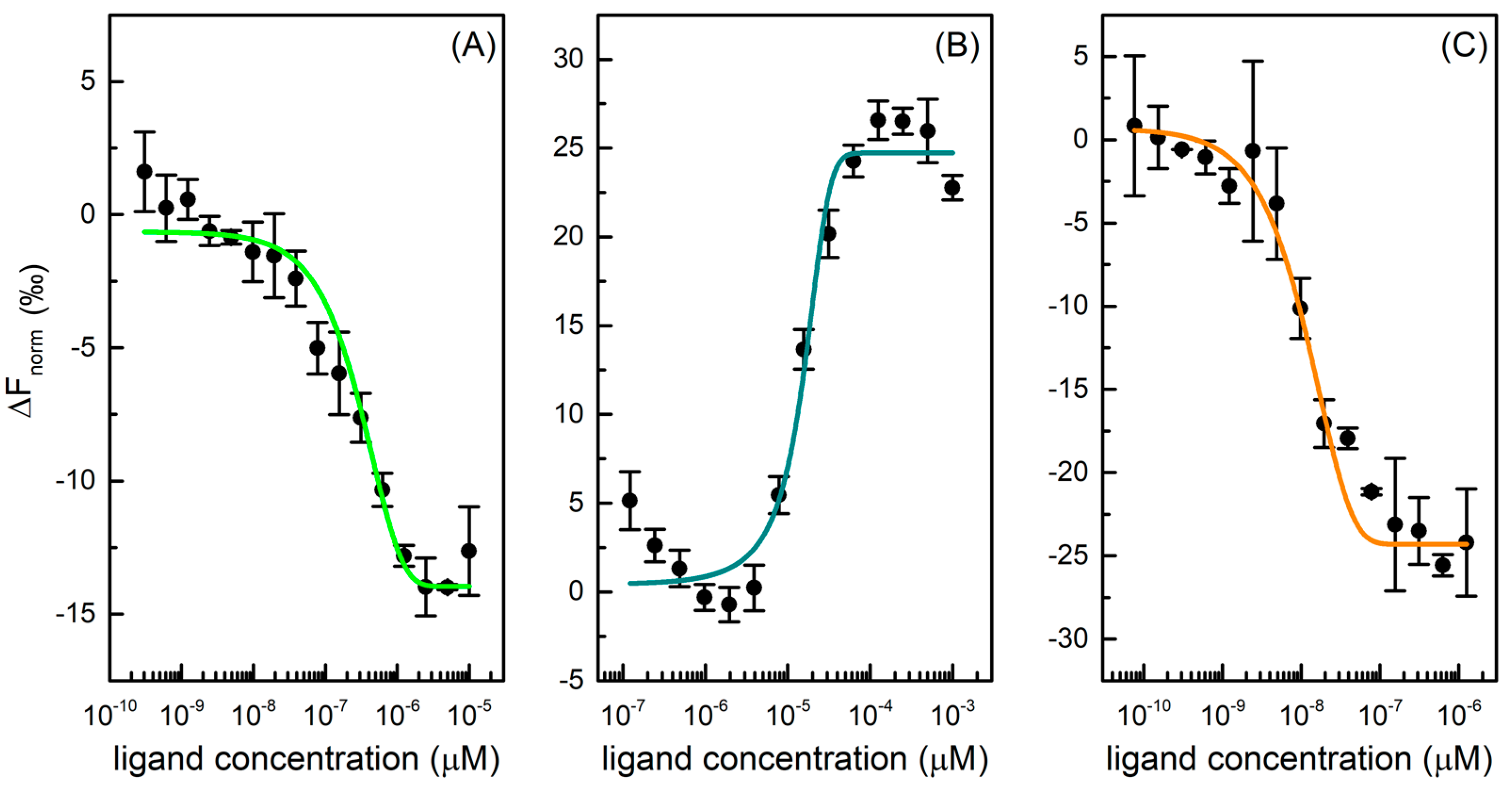
2.1. MicroScale Thermoforesis
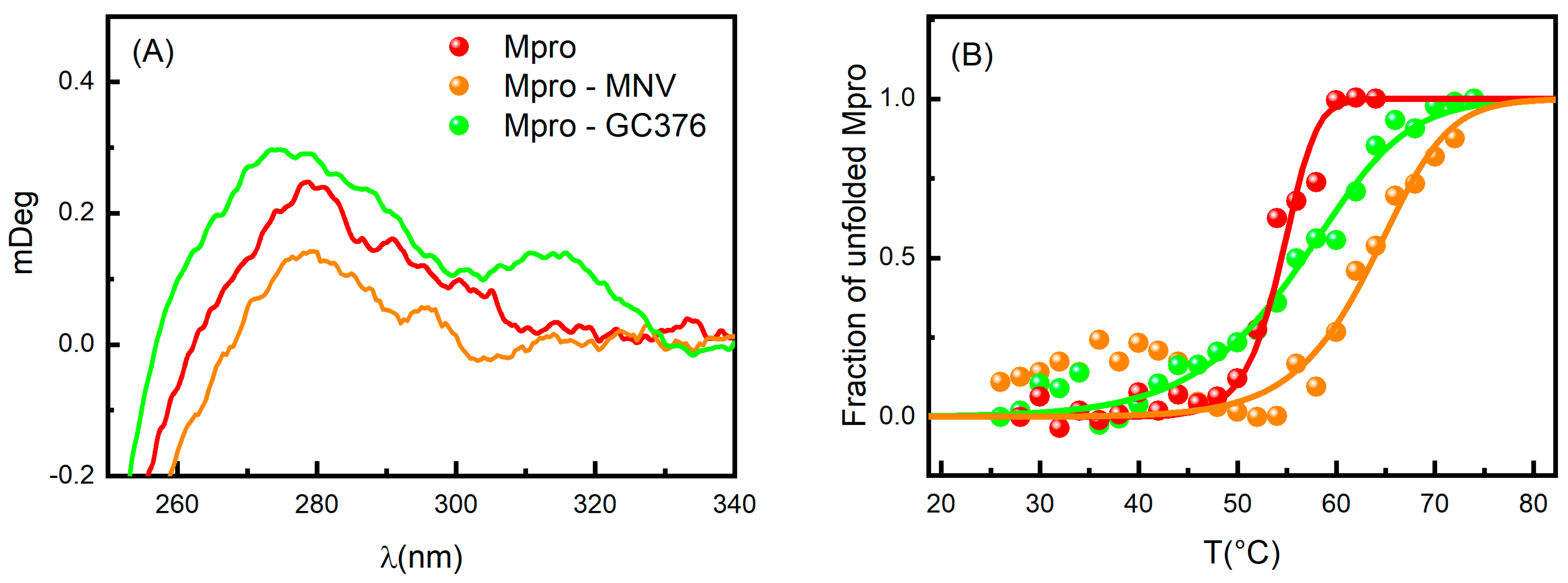
2.2. Circular Dichroism and UV-Vis Absorption
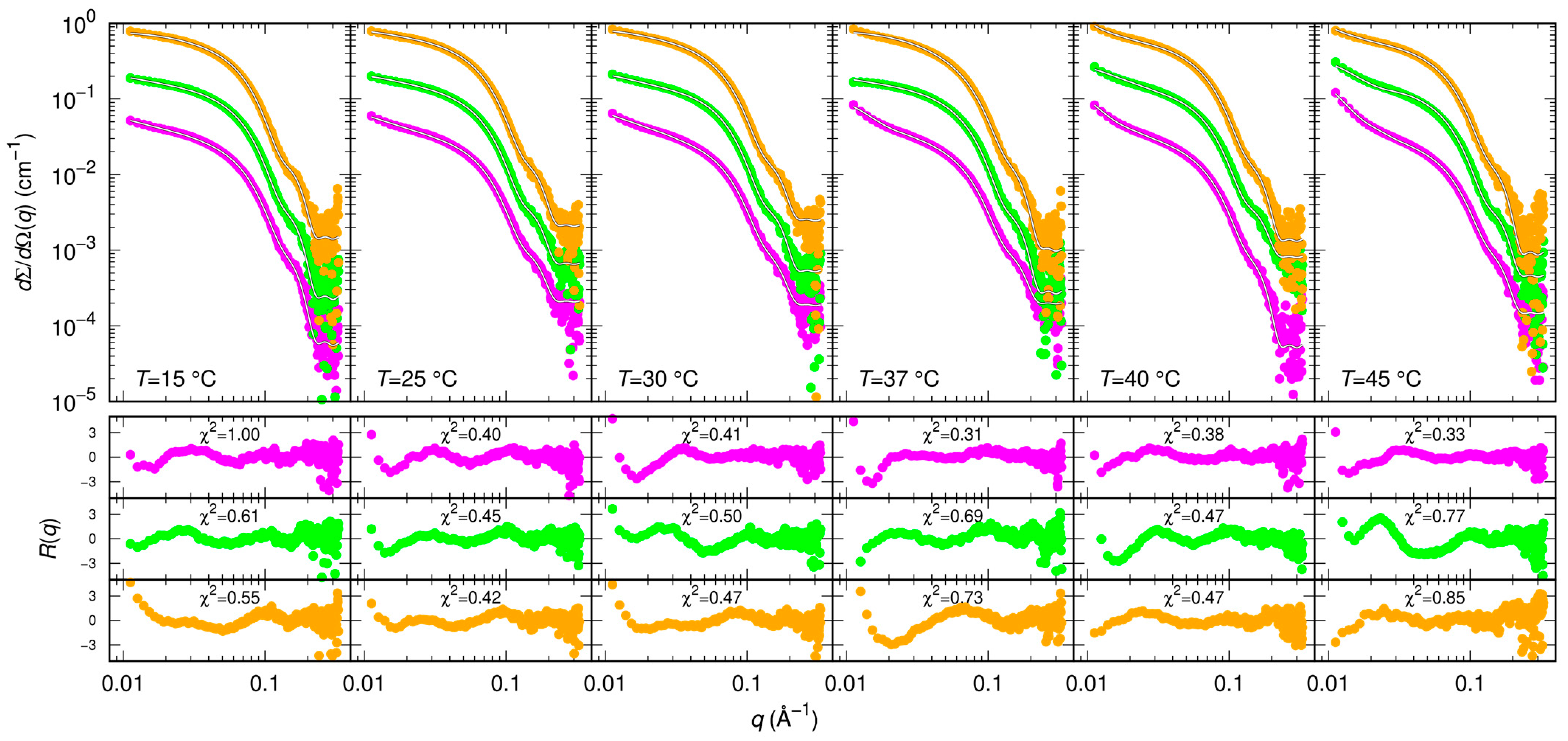
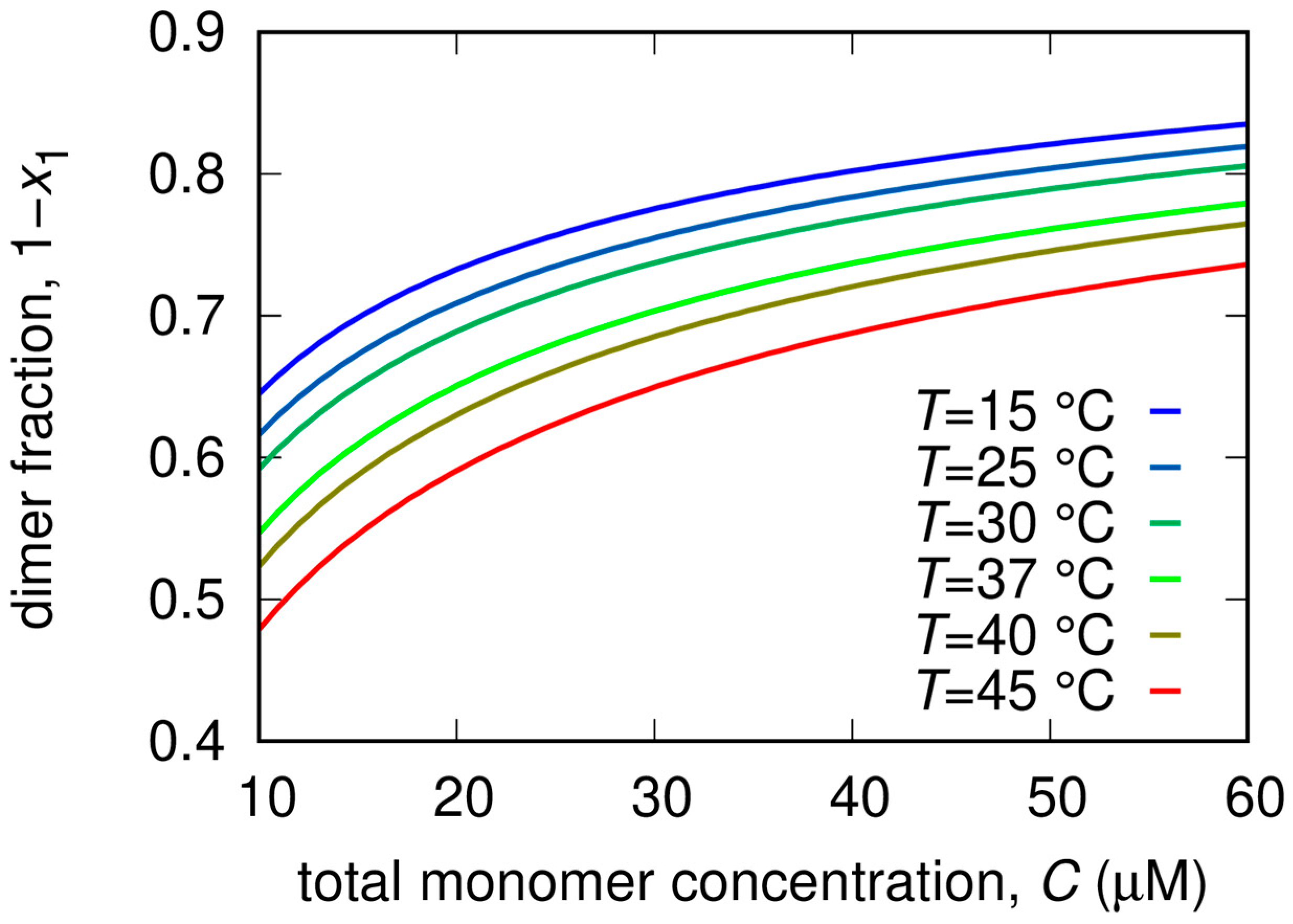
2.3. Small Angle X-ray Scattering
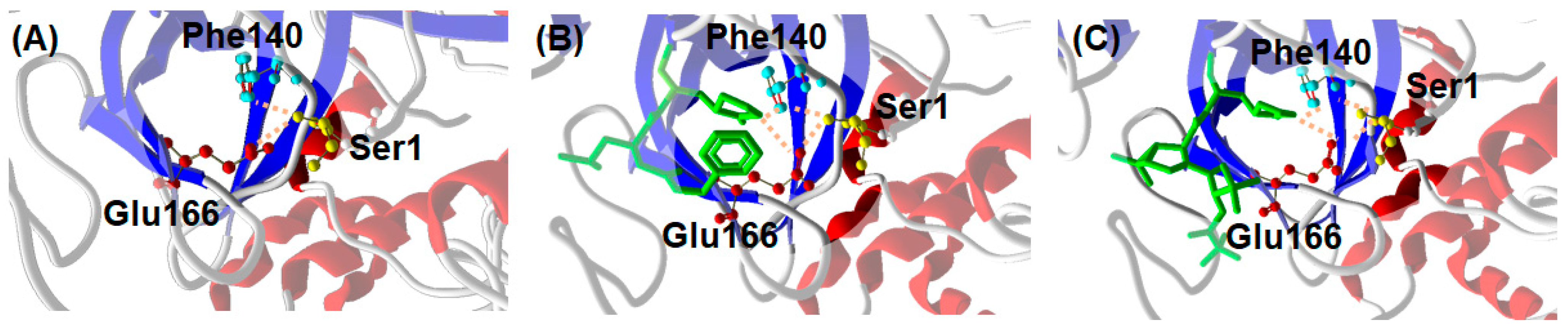
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.2. Microscale Thermophoresis (MST) Standard Assay
4.3. Circular Dichroism and UV-Vis Absorption
4.4. Small Angle X-ray Scattering
4.5. Model for SAXS Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Acter, T.; Uddin, N.; Das, J.; Akhter, A.; Choudhury, T.R.; Kim, S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: A global health emergency. Sci. Total Environ. 2020, 730, 138996. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, C.; Alsafi, Z.; O’neill, N.; Khan, M.; Kerwan, A.; Al-Jabir, A.; Agha, R. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int. J. Surg. 2020, 76, 71–76. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Jiang, T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Morse, J.S.; Lalonde, T.; Xu, S.; Liu, W.R. Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem 2020, 21, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lovell, S.; Tiew, K.C.; Mandadapu, S.R.; Alliston, K.R.; Battaile, K.P.; Chang, K.O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2021, 86, 11754–11762. [Google Scholar] [CrossRef]
- Ahn, D.G.; Shin, H.J.; Kim, M.H.; Lee, S.; Kim, H.S.; Myoung, J.; Kim, S.J. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19). J. Microbiol. Biotechnol. 2020, 30, 313–324. [Google Scholar] [CrossRef]
- Hu, T.; Zhang, Y.; Li, L.; Wang, K.; Chen, S.; Chen, J.; Ding, J.; Jiang, H.; Shen, X. Two adjacent mutations on the dimer interface of SARS coronavirus 3C-like protease cause different conformational changes in crystal structure. Virology 2009, 388, 324–334. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, J.; Hu, T.; Chen, K.; Jiang, H.; Shen, X. Residues on the dimer interface of SARS coronavirus 3C-like protease: Dimer stability characterization and enzyme catalytic activity analysis. J. Biochem. 2008, 143, 525–536. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef]
- La Monica, G.; Bono, A.; Lauria, A.; Martorana, A. Targeting SARS-CoV-2 main protease for treatment of COVID-19: Covalent inhibitors structure–activity relationship insights and evolution perspectives. J. Med. Chem. 2022, 65, 12500–12534. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.; Schiffer, C.; Lee, S.K.; Swanstrom, R. Viral protease inhibitors. In Antiviral Strategies; Springer: Berlin/Heidelberg, Germany, 2009; pp. 1–24. [Google Scholar]
- Gidari, A.; Sabbatini, S.; Schiaroli, E.; Bastianelli, S.; Pierucci, S.; Busti, C.; Francisci, D. The combination of molnupiravir with nirmatrelvir or GC376 has a synergic role in the inhibition of SARS-CoV-2 replication in vitro. Microorganisms 2022, 10, 1475. [Google Scholar] [CrossRef] [PubMed]
- Oerlemans, R.; Ruiz-Moreno, A.J.; Cong, Y.; Kumar, N.D.; Velasco-Velazquez, M.A.; Neochoritis, C.G.; Dömling, A. Repurposing the HCV NS3–4A protease drug boceprevir as COVID-19 therapeutics. RSC Med. Chem. 2021, 12, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, L.; Tan, J.; Chen, J.; Du, L.; Sun, T.; Shen, X. Severe acute respiratory syndrome coronavirus 3C-like proteinase N terminus is indispensable for proteolytic activity but not for enzyme dimerization: Biochemical and thermodynamic investigation in conjunction with molecular dynamics simulations. J. Biol. Chem. 2005, 280, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.Y.; Chang, H.C.; Hsu, W.C.; Lin, T.Z.; Lin, C.H.; Chang, G.G. Quaternary structure of the severe acute respiratory syndrome (SARS) coronavirus main protease. Biochemistry 2004, 43, 14958–14970. [Google Scholar] [CrossRef] [PubMed]
- Silvestrini, L.; Belhaj, N.; Comez, L.; Gerelli, Y.; Lauria, A.; Libera, V.; Spinozzi, F. The dimer-monomer equilibrium of SARS-CoV-2 main protease is affected by small molecule inhibitors. Sci. Rep. 2021, 11, 9283. [Google Scholar] [CrossRef]
- Goyal, B.; Goyal, D. Targeting the dimerization of the main protease of coronaviruses: A potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020, 22, 297–305. [Google Scholar] [CrossRef]
- Arutyunova, E.; Khan, M.B.; Fischer, C.; Lu, J.; Lamer, T.; Vuong, W.; Lemieux, M.J. N-terminal finger stabilizes the S1 pocket for the reversible feline drug GC376 in the SARS-CoV-2 Mpro dimer. J. Mol. Bio. 2021, 433, 167003. [Google Scholar] [CrossRef]
- Halford, B. The path to Paxlovid. ACS Cent. Sci. 2022, 8, 405–407. [Google Scholar] [CrossRef]
- COVID-19: EMA Recommends Conditional Marketing Authorization for Paxlovid. Available online: https://www.ema.europa.eu/en/news/covid-19-ema-recommends-conditional-marketing-authorisation-paxlovid (accessed on 10 February 2023).
- Owen, D.R.; Allerton, C.M.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Zhu, Y. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- Vuong, W.; Khan, M.B.; Fischer, C.; Arutyunova, E.; Lamer, T.; Shields, J.; Lemieux, M.J. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020, 11, 4282. [Google Scholar] [CrossRef] [PubMed]
- Duhr, S.; Braun, D. Why molecules move along a temperature gradient. Proc. Nat. Acad. Sci. USA 2006, 103, 19678–19682. [Google Scholar] [CrossRef] [PubMed]
- Jerabek-Willemsen, M.; André, T.; Wanner, R.; Roth, H.M.; Duhr, S.; Baaske, P.; Breitsprecher, D. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014, 1077, 101–113. [Google Scholar] [CrossRef]
- Nashed, N.T.; Aniana, A.; Ghirlando, R.; Chiliveri, S.C.; Louis, J.M. Modulation of the monomer-dimer equilibrium and catalytic activity of SARS-CoV-2 main protease by a transition-state analog inhibitor. Commun. Biol. 2022, 5, 160. [Google Scholar] [CrossRef]
- Wang, Y.C.; Yang, W.H.; Yang, C.S.; Hou, M.H.; Tsai, C.L.; Chou, Y.Z.; Chen, Y. Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug. Am. J. Cancer Res. 2020, 10, 2535. [Google Scholar]
- El-Baba, T.J.; Lutomski, C.A.; Kantsadi, A.L.; Malla, T.R.; John, T.; Mikhailov, V.; Robinson, C.V. Allosteric inhibition of the SARS-CoV-2 main protease: Insights from mass spectrometry based assays. Angew. Chem. Int. Ed. 2020, 59, 23544–23548. [Google Scholar] [CrossRef]
- Macchiagodena, M.; Pagliai, M.; Procacci, P. Characterization of the non-covalent interaction between the PF-07321332 inhibitor and the SARS-CoV-2 main protease. J. Mol. Graph. Model. 2022, 110, 108042. [Google Scholar] [CrossRef]
- Mahdi, M.; Mótyán, J.A.; Szojka, Z.I.; Golda, M.; Miczi, M.; Tőzsér, J. Analysis of the efficacy of HIV protease inhibitors against SARS-CoV-2′ s main protease. Virol. J. 2020, 17, 1–8. [Google Scholar] [CrossRef]
- Siligardi, G.; Hussain, R. Biomolecules interactions and competitions by non-immobilised ligand interaction assay by circular dichroism. Enantiomer 1998, 3, 77–87. [Google Scholar]
- Kelly, S.M.; Price, N.C. The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 2000, 1, 349–384. [Google Scholar] [CrossRef]
- Graziano, V.; McGrath, W.J.; Yang, L.; Mangel, W.F. SARS CoV main proteinase: The monomer− dimer equilibrium dissociation constant. Biochemistry 2006, 45, 14632–14641. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, T.S.; Okimoto, N.; Koyama, Y.M.; Hirano, Y.; Morimoto, G.; Ohno, Y.; Taiji, M. Drug binding dynamics of the dimeric SARS-CoV-2 main protease, determined by molecular dynamics simulation. Sci. Rep. 2020, 10, 16986. [Google Scholar] [CrossRef] [PubMed]
- Kidera, A.; Moritsugu, K.; Ekimoto, T.; Ikeguchi, M. Allosteric regulation of 3CL protease of SARS-CoV-2 and SARS-CoV observed in the crystal structure ensemble. J. Mol. Biol. 2021, 433, 167324. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Fang, C.; Zhang, Q.; Zhang, R.; Zhao, X.; Duan, Y.; Yang, H. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell 2022, 13, 689–693. [Google Scholar] [CrossRef]
- Terse, V.L.; Gosavi, S. The molecular mechanism of domain swapping of the C-terminal domain of the SARS-Coronavirus Main Protease. Biophys. J. 2021, 120, 504–516. [Google Scholar] [CrossRef]
- Spinozzi, F.; Ferrero, C.; Ortore, M.G.; Antolinos, A.D.M.; Mariani, P. GENFIT: Software for the analysis of small-angle X-ray and neutron scattering data of macromolecules in-solution. J. App. Cryst. 2014, 47, 1132–1139. [Google Scholar] [CrossRef]


| Compound | Kd (μM) |
|---|---|
| Lopinavir | >200 |
| RTV | >250 |
| Carmofur | 17 ± 4 |
| GC376 | 0.17 ± 0.04 |
| Nelfinavir | >1000 |
| NMV | 0.004 ± 0.001 |
| Sample | (μM) | (kJ mol−1) | (J K−1mol−1) | (kJ K−1mol−1) |
|---|---|---|---|---|
| Mpro | 5 ± 1 | 21 ± 4 | −30 ± 10 | 1.3 ± 0.4 |
| Mpro-RTV | 3 ± 1 | 11 ± 5 | −70 ± 50 | 4 ± 1 |
| Mpro-GC376 | 0.4 ± 1 | −15 ± 8 | −170 ± 60 | 2.1 ± 0.8 |
| Mpro-NMV | 0.7 ± 1 | 64 ± 9 | 100 ± 30 | −0.9 ± 0.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paciaroni, A.; Libera, V.; Ripanti, F.; Orecchini, A.; Petrillo, C.; Francisci, D.; Schiaroli, E.; Sabbatini, S.; Gidari, A.; Bianconi, E.; et al. Stabilization of the Dimeric State of SARS-CoV-2 Main Protease by GC376 and Nirmatrelvir. Int. J. Mol. Sci. 2023, 24, 6062. https://doi.org/10.3390/ijms24076062
Paciaroni A, Libera V, Ripanti F, Orecchini A, Petrillo C, Francisci D, Schiaroli E, Sabbatini S, Gidari A, Bianconi E, et al. Stabilization of the Dimeric State of SARS-CoV-2 Main Protease by GC376 and Nirmatrelvir. International Journal of Molecular Sciences. 2023; 24(7):6062. https://doi.org/10.3390/ijms24076062
Chicago/Turabian StylePaciaroni, Alessandro, Valeria Libera, Francesca Ripanti, Andrea Orecchini, Caterina Petrillo, Daniela Francisci, Elisabetta Schiaroli, Samuele Sabbatini, Anna Gidari, Elisa Bianconi, and et al. 2023. "Stabilization of the Dimeric State of SARS-CoV-2 Main Protease by GC376 and Nirmatrelvir" International Journal of Molecular Sciences 24, no. 7: 6062. https://doi.org/10.3390/ijms24076062
APA StylePaciaroni, A., Libera, V., Ripanti, F., Orecchini, A., Petrillo, C., Francisci, D., Schiaroli, E., Sabbatini, S., Gidari, A., Bianconi, E., Macchiarulo, A., Hussain, R., Silvestrini, L., Moretti, P., Belhaj, N., Vercelli, M., Roque, Y., Mariani, P., Comez, L., & Spinozzi, F. (2023). Stabilization of the Dimeric State of SARS-CoV-2 Main Protease by GC376 and Nirmatrelvir. International Journal of Molecular Sciences, 24(7), 6062. https://doi.org/10.3390/ijms24076062










