Membrane Dynamics Regulated by Cytoskeleton in Plant Immunity
Abstract
1. Introduction
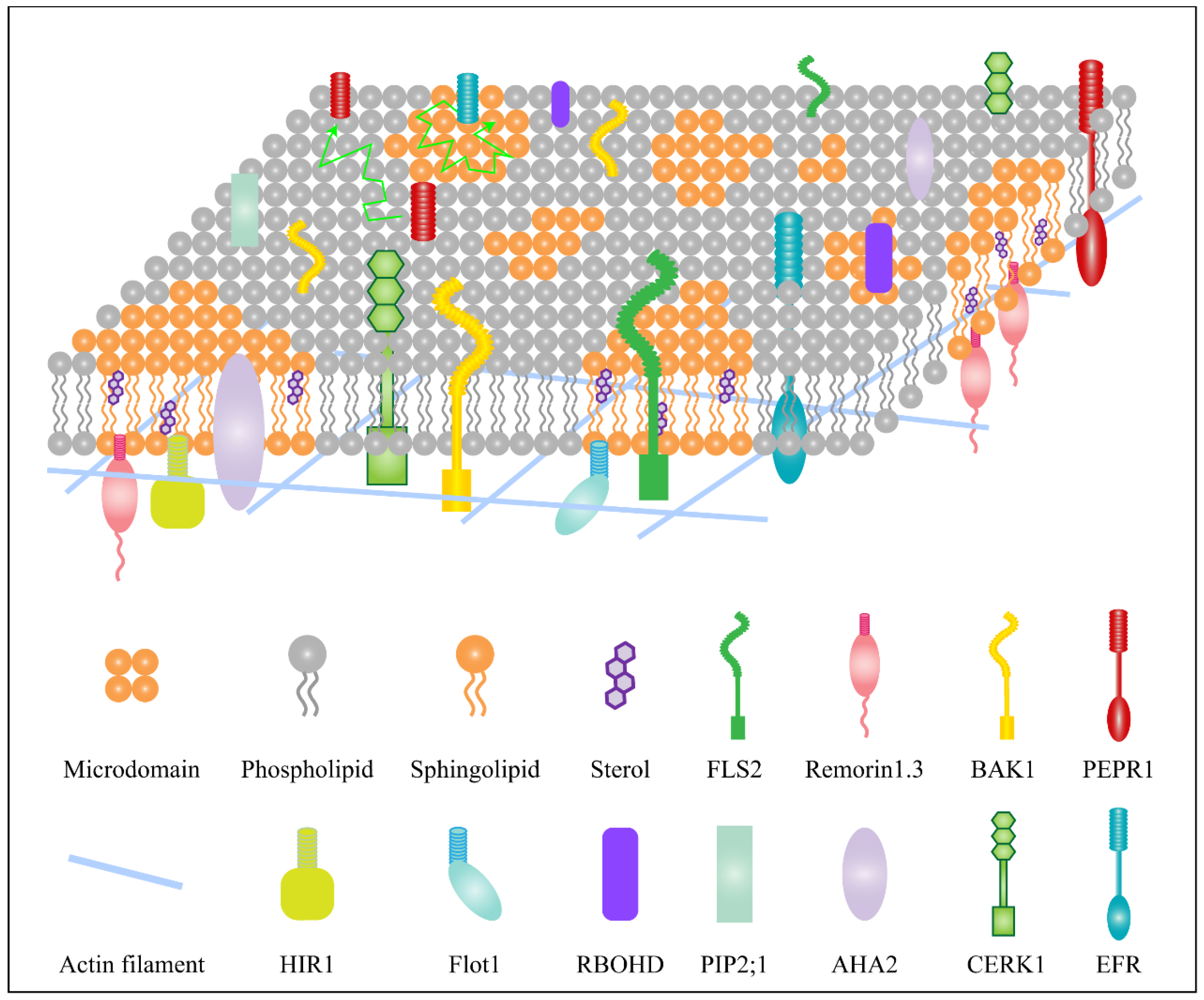
2. Membrane Lipids and Proteins Involved in Plant Immunity
2.1. Functional Diversity of the Membrane Lipids That Are Involved in Immunity Response
2.2. Functional Diversity of the Membrane Proteins That Are Involved in Immunity Response
3. Dynamics of Membrane Lipids and Proteins Are Regulated by Cytoskeleton
4. The Cytoskeleton Regulates Endo-Membrane Trafficking of PRRs Involved in Plant Immunity
4.1. The Role of the Cytoskeleton in Exocytosis of Plant Immune Processes
4.2. The Role of the Cytoskeleton in Endocytosis of Plant Immune Processes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, J.M.; Zhang, Y. Plant Immunity: Danger Perception and Signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; He, S.Y. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef]
- Henry, G.; Thonart, P.; Ongena, M. PAMPs, MAMPs, DAMPs and others: An update on the diversity of plant immunity elicitors. Biotechnol. Agron. Soc. 2012, 16, 257–268. [Google Scholar]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Song, Y.; Liu, J.; Xu, G.; Dou, D. Ubiquitination of Receptorsomes, Frontline of Plant Immunity. Int. J. Mol. Sci. 2022, 23, 2937. [Google Scholar] [CrossRef] [PubMed]
- Bjornson, M.; Zipfel, C. Plant immunity: Crosstalk between plant immune receptors. Curr. Biol. 2021, 31, R796–R798. [Google Scholar] [CrossRef]
- Peng, Y.; van Wersch, R.; Zhang, Y. Convergent and Divergent Signaling in PAMP-Triggered Immunity and Effector-Triggered Immunity. Mol. Plant-Microbe Interact. 2018, 31, 403–409. [Google Scholar] [CrossRef]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef]
- Roux, M.; Schwessinger, B.; Albrecht, C.; Chinchilla, D.; Jones, A.; Holton, N.; Malinovsky, F.G.; Tor, M.; de Vries, S.; Zipfel, C. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 2011, 23, 2440–2455. [Google Scholar] [CrossRef]
- Chang, M.; Chen, H.; Liu, F.; Fu, Z.Q. PTI and ETI: Convergent pathways with diverse elicitors. Trends Plant Sci. 2022, 27, 113–115. [Google Scholar] [CrossRef]
- Li, P.; Day, B. Battlefield Cytoskeleton: Turning the Tide on Plant Immunity. Mol. Plant-Microbe Interact. 2019, 32, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, D.; Zheng, J.; Feng, Y.; Zhang, Y.; Liu, W. Actin cytoskeleton-dependent pathways for ADMA-induced NF-kappaB activation and TGF-beta high expression in human renal glomerular endothelial cells. Acta Biochim. Biophys. Sin. 2012, 44, 918–923. [Google Scholar] [CrossRef] [PubMed]
- Henty-Ridilla, J.L.; Shimono, M.; Li, J.; Chang, J.H.; Day, B.; Staiger, C.J. The plant actin cytoskeleton responds to signals from microbe-associated molecular patterns. PLoS Pathog. 2013, 9, e1003290. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Wang, W.; Zhang, W.; Staiger, C.J. Lipid Signaling Requires ROS Production to Elicit Actin Cytoskeleton Remodeling during Plant Innate Immunity. Int. J. Mol. Sci. 2022, 23, 2447. [Google Scholar] [CrossRef]
- Yang, L.; Qin, L.; Liu, G.; Peremyslov, V.V.; Dolja, V.V.; Wei, Y. Myosins XI modulate host cellular responses and penetration resistance to fungal pathogens. Proc. Natl. Acad. Sci. USA 2014, 111, 13996–14001. [Google Scholar] [CrossRef]
- Cai, G.; Faleri, C.; Del, C.C.; Emons, A.M.; Cresti, M. Distribution of callose synthase, cellulose synthase, and sucrose synthase in tobacco pollen tube is controlled in dissimilar ways by actin filaments and microtubules. Plant Physiol. 2011, 155, 1169–1190. [Google Scholar] [CrossRef]
- Pollard, T.D.; Goldman, R.D. Overview of the Cytoskeleton from an Evolutionary Perspective. Cold Spring Harb. Perspect. Biol. 2018, 10, a30288. [Google Scholar] [CrossRef]
- Akil, C.; Kitaoku, Y.; Tran, L.T.; Liebl, D.; Choe, H.; Muengsaen, D.; Suginta, W.; Schulte, A.; Robinson, R.C. Mythical origins of the actin cytoskeleton. Curr. Opin. Cell Biol. 2021, 68, 55–63. [Google Scholar] [CrossRef]
- Wang, X.; Mao, T. Understanding the functions and mechanisms of plant cytoskeleton in response to environmental signals. Curr. Opin. Plant Biol. 2019, 52, 86–96. [Google Scholar] [CrossRef]
- Li, S.; Sun, T.; Ren, H. The functions of the cytoskeleton and associated proteins during mitosis and cytokinesis in plant cells. Front. Plant Sci. 2015, 6, 282. [Google Scholar] [CrossRef]
- Ren, Z.; Zhang, Y.; Zhang, Y.; He, Y.; Du, P.; Wang, Z.; Sun, F.; Ren, H. Cryo-EM Structure of Actin Filaments from Zea mays Pollen. Plant Cell 2019, 31, 2855–2867. [Google Scholar] [CrossRef] [PubMed]
- Cacas, J.L.; Bure, C.; Grosjean, K.; Gerbeau-Pissot, P.; Lherminier, J.; Rombouts, Y.; Maes, E.; Bossard, C.; Gronnier, J.; Furt, F.; et al. Revisiting Plant Plasma Membrane Lipids in Tobacco: A Focus on Sphingolipids. Plant Physiol. 2016, 170, 367–384. [Google Scholar] [CrossRef] [PubMed]
- Ukawa, T.; Banno, F.; Ishikawa, T.; Kasahara, K.; Nishina, Y.; Inoue, R.; Tsujii, K.; Yamaguchi, M.; Takahashi, T.; Fukao, Y.; et al. Sphingolipids with 2-hydroxy fatty acids aid in plasma membrane nanodomain organization and oxidative burst. Plant Physiol. 2022, 189, 839–857. [Google Scholar] [CrossRef] [PubMed]
- Nagano, M.; Ishikawa, T.; Fujiwara, M.; Fukao, Y.; Kawano, Y.; Kawai-Yamada, M.; Shimamoto, K. Plasma Membrane Microdomains Are Essential for Rac1-RbohB/H-Mediated Immunity in Rice. Plant Cell 2016, 28, 1966–1983. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Fu, X.; Chu, Y.; Wu, P.; Liu, Y.; Ma, L.; Tian, H.; Zhu, B. Biosynthesis and the Roles of Plant Sterols in Development and Stress Responses. Int. J. Mol. Sci. 2022, 23, 2332. [Google Scholar] [CrossRef]
- Ferrer, A.; Altabella, T.; Arro, M.; Boronat, A. Emerging roles for conjugated sterols in plants. Prog. Lipid Res. 2017, 67, 27–37. [Google Scholar] [CrossRef]
- Szymanski, W.G.; Zauber, H.; Erban, A.; Gorka, M.; Wu, X.N.; Schulze, W.X. Cytoskeletal Components Define Protein Location to Membrane Microdomains. Mol. Cell. Proteom. 2015, 14, 2493–2509. [Google Scholar] [CrossRef]
- Cui, Y.; Li, X.; Yu, M.; Li, R.; Fan, L.; Zhu, Y.; Lin, J. Sterols regulate endocytic pathways during flg22-induced defense responses in Arabidopsis. Development 2018, 145, v165688. [Google Scholar] [CrossRef]
- Perraki, A.; Gronnier, J.; Gouguet, P.; Boudsocq, M.; Deroubaix, A.F.; Simon, V.; German-Retana, S.; Legrand, A.; Habenstein, B.; Zipfel, C.; et al. REM1.3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog. 2018, 14, e1007378. [Google Scholar] [CrossRef]
- Monaghan, J.; Zipfel, C. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 2012, 15, 349–357. [Google Scholar] [CrossRef]
- Sun, Y.; Li, L.; Macho, A.P.; Han, Z.; Hu, Z.; Zipfel, C.; Zhou, J.M.; Chai, J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 2013, 342, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Macho, A.P.; Zipfel, C. Plant PRRs and the Activation of Innate Immune Signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Boutrot, F.; Zipfel, C. Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef]
- Bartels, S.; Boller, T. Quo vadis, Pep? Plant elicitor peptides at the crossroads of immunity, stress, and development. J. Exp. Bot. 2015, 66, 5183–5193. [Google Scholar] [CrossRef] [PubMed]
- Xiang, T.; Zong, N.; Zou, Y.; Wu, Y.; Zhang, J.; Xing, W.; Li, Y.; Tang, X.; Zhu, L.; Chai, J.; et al. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 2008, 18, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395. [Google Scholar] [CrossRef]
- Jubic, L.M.; Saile, S.; Furzer, O.J.; El, K.F.; Dangl, J.L. Help wanted: Helper NLRs and plant immune responses. Curr. Opin. Plant Biol. 2019, 50, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Roux, B.; Feng, F.; Guy, E.; Li, L.; Li, N.; Zhang, X.; Lautier, M.; Jardinaud, M.F.; Chabannes, M.; et al. The Decoy Substrate of a Pathogen Effector and a Pseudokinase Specify Pathogen-Induced Modified-Self Recognition and Immunity in Plants. Cell Host Microbe 2015, 18, 285–295. [Google Scholar] [CrossRef]
- Bi, G.; Su, M.; Li, N.; Liang, Y.; Dang, S.; Xu, J.; Hu, M.; Wang, J.; Zou, M.; Deng, Y.; et al. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 2021, 184, 3528–3541. [Google Scholar] [CrossRef]
- Lapin, D.; Kovacova, V.; Sun, X. A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 2019, 31, 2430–2455. [Google Scholar] [CrossRef]
- Shoucai, M.; Dmitry, L.; Li, L.; Yue, S.; Wen, S.; Xiaoxiao, Z.; Elke, L.; Dongli, Y.; Jia, W.; Jan, J.; et al. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 2020, 370, 6521. [Google Scholar]
- Fichman, Y.; Miller, G.; Mittler, R. Whole-Plant Live Imaging of Reactive Oxygen Species. Mol. Plant 2019, 12, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Pfeilmeier, S.; Petti, G.C.; Bortfeld-Miller, M.; Daniel, B.; Field, C.M.; Sunagawa, S.; Vorholt, J.A. The plant NADPH oxidase RBOHD is required for microbiota homeostasis in leaves. Nat. Microbiol. 2021, 6, 852–864. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Wang, X.; Li, P.; Wang, H.; Ji, H.; Xie, J.; Qiu, Q.; Shen, D.; Dong, H. Plant Aquaporin AtPIP1;4 Links Apoplastic H2O2 Induction to Disease Immunity Pathways. Plant Physiol. 2016, 171, 1635–1650. [Google Scholar] [CrossRef]
- Rodrigues, O.; Reshetnyak, G.; Grondin, A.; Saijo, Y.; Leonhardt, N.; Maurel, C.; Verdoucq, L. Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proc. Natl. Acad. Sci. USA 2017, 114, 9200–9205. [Google Scholar] [CrossRef]
- Li, P.; Zhang, L.; Mo, X.; Ji, H.; Bian, H.; Hu, Y.; Majid, T.; Long, J.; Pang, H.; Tao, Y.; et al. Rice aquaporin PIP1;3 and harpin Hpa1 of bacterial blight pathogen cooperate in a type III effector translocation. J. Exp. Bot. 2019, 70, 3057–3073. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, Y.; Li, P.; Wang, X.; Dong, H. Silencing of an aquaporin gene diminishes bacterial blight disease in rice. Australas. Plant Pathol. 2019, 48, 143–158. [Google Scholar] [CrossRef]
- Li, S.; Zhao, J.; Zhai, Y.; Yuan, Q.; Zhang, H.; Wu, X.; Lu, Y.; Peng, J.; Sun, Z.; Lin, L.; et al. The hypersensitive induced reaction 3 (HIR3) gene contributes to plant basal resistance via an EDS1 and salicylic acid-dependent pathway. Plant J. 2019, 98, 783–797. [Google Scholar] [CrossRef]
- Zhou, L.; Cheung, M.Y.; Li, M.W.; Fu, Y.; Sun, Z.; Sun, S.M.; Lam, H.M. Rice hypersensitive induced reaction protein 1 (OsHIR1) associates with plasma membrane and triggers hypersensitive cell death. BMC Plant Biol. 2010, 10, 290. [Google Scholar] [CrossRef]
- Gronnier, J.; Crowet, J.M.; Habenstein, B.; Nasir, M.N.; Bayle, V.; Hosy, E.; Platre, M.P.; Gouguet, P.; Raffaele, S.; Martinez, D.; et al. Structural basis for plant plasma membrane protein dynamics and organization into functional nanodomains. eLife 2017, 6, e26404. [Google Scholar] [CrossRef]
- Jamann, T.M.; Luo, X.; Morales, L.; Kolkman, J.M.; Chung, C.L.; Nelson, R.J. A remorin gene is implicated in quantitative disease resistance in maize. Theor. Appl. Genet. 2016, 129, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.G.; Da, C.L.; McFall, A.J.; Belkhadir, Y.; DebRoy, S.; Dangl, J.L.; Mackey, D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 2005, 121, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Sun, Y.; Zhu, X.; Yang, L.; Chen, X.; Miao, Y. Membrane nanodomains modulate formin condensation for actin remodeling in Arabidopsis innate immune responses. Plant Cell 2022, 34, 374–394. [Google Scholar] [CrossRef]
- Yu, M.J.; Pisitkun, T.; Wang, G.; Aranda, J.F.; Gonzales, P.A.; Tchapyjnikov, D.; Shen, R.F.; Alonso, M.A.; Knepper, M.A. Large-scale quantitative LC-MS/MS analysis of detergent-resistant membrane proteins from rat renal collecting duct. Am. J. Physiol. Cell Physiol. 2008, 295, C661–C678. [Google Scholar] [CrossRef] [PubMed]
- Yanagida, M.; Nakayama, H.; Yoshizaki, F.; Fujimura, T.; Takamori, K.; Ogawa, H.; Iwabuchi, K. Proteomic analysis of plasma membrane lipid rafts of HL-60 cells. Proteomics 2007, 7, 2398–2409. [Google Scholar] [CrossRef]
- Kusumi, A.; Koyama-Honda, I.; Suzuki, K. Molecular dynamics and interactions for creation of stimulation-induced stabilized rafts from small unstable steady-state rafts. Traffic 2004, 5, 213–230. [Google Scholar] [CrossRef]
- Kusumi, A.; Fujiwara, T.K.; Chadda, R.; Xie, M.; Tsunoyama, T.A.; Kalay, Z.; Kasai, R.S.; Suzuki, K.G. Dynamic organizing principles of the plasma membrane that regulate signal transduction: Commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu. Rev. Cell Dev. Biol. 2012, 28, 215–250. [Google Scholar] [CrossRef]
- Pralle, A. Modulation and dynamics of cell membrane heterogeneities. Chem. Phys. Lipids 2020, 233, 105006. [Google Scholar] [CrossRef]
- Treanor, B.; Depoil, D.; Gonzalez-Granja, A.; Barral, P.; Weber, M.; Dushek, O.; Bruckbauer, A.; Batista, F.D. The membrane skeleton controls diffusion dynamics and signaling through the B cell receptor. Immunity 2010, 32, 187–199. [Google Scholar] [CrossRef]
- Lv, X.; Jing, Y.; Xiao, J.; Zhang, Y.; Zhu, Y.; Julian, R.; Lin, J. Membrane microdomains and the cytoskeleton constrain AtHIR1 dynamics and facilitate the formation of an AtHIR1-associated immune complex. Plant J. 2017, 90, 3–16. [Google Scholar] [CrossRef]
- Gui, J.; Zheng, S.; Shen, J.; Li, L. Grain setting defect1 (GSD1) function in rice depends on S-acylation and interacts with actin 1 (OsACT1) at its C-terminal. Front. Plant Sci. 2015, 6, 804. [Google Scholar] [CrossRef] [PubMed]
- Bücherl, C.A.; Jarsch, I.K.; Schudoma, C.; Segonzac, C.; Mbengue, M.; Robatzek, S.; MacLean, D.; Ott, T.; Zipfel, C. Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 2017, 6, e25114. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, D.D.; Brandizzi, F. Plant endomembranes and cytoskeleton: Moving targets in immunity. Curr. Opin. Plant Biol. 2020, 58, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Ali, G.S.; Prasad, K.V.; Day, I.; Reddy, A.S. Ligand-dependent reduction in the membrane mobility of FLAGELLIN SENSITIVE2, an Arabidopsis receptor-like kinase. Plant Cell Physiol. 2007, 48, 1601–1611. [Google Scholar] [CrossRef]
- McKenna, J.F.; Rolfe, D.J.; Webb, S.E.D.; Tolmie, A.F.; Botchway, S.W.; Martin-Fernandez, M.L.; Hawes, C.; Runions, J. The cell wall regulates dynamics and size of plasma-membrane nanodomains in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 12857–12862. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhao, Y.; Lu, Y.; Su, X.; Chen, Y.; Shen, Y.; Lin, J.; Li, X. In vivo single-particle tracking of the aquaporin AtPIP2;1 in stomata reveals cell type-specific dynamics. Plant Physiol. 2021, 185, 1666–1681. [Google Scholar] [CrossRef]
- Groves, J.T.; Parthasarathy, R.; Forstner, M.B. Fluorescence imaging of membrane dynamics. Annu. Rev. Biomed. Eng. 2008, 10, 311–338. [Google Scholar] [CrossRef]
- Hao, H.; Fan, L.; Chen, T.; Li, R.; Li, X.; He, Q.; Botella, M.A.; Lin, J. Clathrin and Membrane Microdomains Cooperatively Regulate RbohD Dynamics and Activity in Arabidopsis. Plant Cell 2014, 26, 1729–1745. [Google Scholar] [CrossRef]
- Su, B.; Zhang, X.; Li, L.; Abbas, S.; Yu, M.; Cui, Y.; Baluska, F.; Hwang, I.; Shan, X.; Lin, J. Dynamic spatial reorganization of BSK1 complexes in the plasma membrane underpins signal-specific activation for growth and immunity. Mol. Plant 2021, 14, 588–603. [Google Scholar] [CrossRef]
- Day, B.; Henty, J.L.; Porter, K.J.; Staiger, C.J. The pathogen-actin connection: A platform for defense signaling in plants. Annu. Rev. Phytopathol. 2011, 49, 483–506. [Google Scholar] [CrossRef]
- Gu, Y.; Zavaliev, R.; Dong, X. Membrane Trafficking in Plant Immunity. Mol. Plant 2017, 10, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Wollman, R.; Meyer, T. Coordinated oscillations in cortical actin and Ca2+ correlate with cycles of vesicle secretion. Nat. Cell Biol. 2012, 14, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wei, G.; Cao, Y.; Deng, Q.; Han, X.; Huang, X.; Huo, Y.; He, Y.; Chen, L.; Luo, J. Myosin IIa is critical for cAMP-mediated endothelial secretion of von Willebrand factor. Blood 2018, 131, 686–698. [Google Scholar] [CrossRef]
- Han, X.; Li, P.; Yang, Z.; Huang, X.; Wei, G.; Sun, Y.; Kang, X.; Hu, X.; Deng, Q.; Chen, L.; et al. Zyxin regulates endothelial von Willebrand factor secretion by reorganizing actin filaments around exocytic granules. Nat. Commun. 2017, 8, 14639. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wen, Y.; Berkey, R.; Xiao, S. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 2009, 21, 2898–2913. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Shang, Y.; Fan, B.; Yu, J.Q.; Chen, Z. Arabidopsis LIP5, a positive regulator of multivesicular body biogenesis, is a critical target of pathogen-responsive MAPK cascade in plant basal defense. PLoS Pathog. 2014, 10, e1004243. [Google Scholar] [CrossRef]
- Bohlenius, H.; Morch, S.M.; Godfrey, D.; Nielsen, M.E.; Thordal-Christensen, H. The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell 2010, 22, 3831–3844. [Google Scholar] [CrossRef]
- Underwood, W.; Somerville, S.C. Perception of conserved pathogen elicitors at the plasma membrane leads to relocalization of the Arabidopsis PEN3 transporter. Proc. Natl. Acad. Sci. USA 2013, 110, 12492–12497. [Google Scholar] [CrossRef]
- Li, L.; Zhu, X.M.; Zhang, Y.R.; Cai, Y.Y.; Wang, J.Y.; Liu, M.Y.; Wang, J.Y.; Bao, J.D.; Lin, F.C. Research on the Molecular Interaction Mechanism between Plants and Pathogenic Fungi. Int. J. Mol. Sci. 2022, 23, 4658. [Google Scholar] [CrossRef]
- Zhang, X.; Man, Y.; Zhuang, X.; Shen, J.; Zhang, Y.; Cui, Y.; Yu, M.; Xing, J.; Wang, G.; Lian, N.; et al. Plant multiscale networks: Charting plant connectivity by multi-level analysis and imaging techniques. Sci. China Life Sci. 2021, 64, 1392–1422. [Google Scholar] [CrossRef]
- Sampathkumar, A.; Gutierrez, R.; McFarlane, H.E.; Bringmann, M.; Lindeboom, J.; Emons, A.M.; Samuels, L.; Ketelaar, T.; Ehrhardt, D.W.; Persson, S. Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiol. 2013, 162, 675–688. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, H.E.; Doring, A.; Persson, S. The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 2014, 65, 69–94. [Google Scholar] [CrossRef] [PubMed]
- Paredez, A.R.; Somerville, C.R.; Ehrhardt, D.W. Visualization of cellulose synthase demonstrates functional association with microtubules. Science 2006, 312, 1491–1495. [Google Scholar] [CrossRef] [PubMed]
- Wightman, R.; Turner, S. Trafficking of the plant cellulose synthase complex. Plant Physiol. 2010, 153, 427–432. [Google Scholar] [CrossRef]
- Crowell, E.F.; Bischoff, V.; Desprez, T.; Rolland, A.; Stierhof, Y.D.; Schumacher, K.; Gonneau, M.; Hofte, H.; Vernhettes, S. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 2009, 21, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Ioki, M.; Braybrook, S.; Li, S.; Ye, Z.H.; Julie, L.Y.; Hotta, T.; Chang, A.; Tian, J.; Wang, G.; et al. Kinesin-4 Functions in Vesicular Transport on Cortical Microtubules and Regulates Cell Wall Mechanics during Cell Elongation in Plants. Mol. Plant 2015, 8, 1011–1023. [Google Scholar] [CrossRef]
- Li, J.; Henty-Ridilla, J.L.; Staiger, B.H.; Day, B.; Staiger, C.J. Capping protein integrates multiple MAMP signalling pathways to modulate actin dynamics during plant innate immunity. Nat. Commun. 2015, 6, 7206. [Google Scholar] [CrossRef] [PubMed]
- Henty-Ridilla, J.L.; Li, J.; Day, B.; Staiger, C.J. ACTIN DEPOLYMERIZING FACTOR4 regulates actin dynamics during innate immune signaling in Arabidopsis. Plant Cell 2014, 26, 340–352. [Google Scholar] [CrossRef]
- Guo, M.; Kim, P.; Li, G.; Elowsky, C.G.; Alfano, J.R. A Bacterial Effector Co-opts Calmodulin to Target the Plant Microtubule Network. Cell Host Microbe 2016, 19, 67–78. [Google Scholar] [CrossRef]
- Lee, A.H.; Hurley, B.; Felsensteiner, C.; Yea, C.; Ckurshumova, W.; Bartetzko, V.; Wang, P.W.; Quach, V.; Lewis, J.D.; Liu, Y.C.; et al. A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog. 2012, 8, e1002523. [Google Scholar] [CrossRef]
- Bibeau, J.P.; Kingsley, J.L.; Furt, F.; Tuzel, E.; Vidali, L. F-Actin Mediated Focusing of Vesicles at the Cell Tip Is Essential for Polarized Growth. Plant Physiol. 2018, 176, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Renna, L.; Stefano, G.; Brandizzi, F. SYP73 Anchors the ER to the Actin Cytoskeleton for Maintenance of ER Integrity and Streaming in Arabidopsis. Curr. Biol. 2016, 26, 3245–3254. [Google Scholar] [CrossRef] [PubMed]
- Thurmond, D.C.; Gonelle-Gispert, C.; Furukawa, M.; Halban, P.A.; Pessin, J.E. Glucose-stimulated insulin secretion is coupled to the interaction of actin with the t-SNARE (target membrane soluble N-ethylmaleimide-sensitive factor attachment protein receptor protein) complex. Mol. Endocrinol. 2003, 17, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, M.; Uemura, T.; Ebine, K.; Nishimori, Y.; Ueda, T.; Nakano, A.; Sato, M.H.; Fukao, Y. Interactomics of Qa-SNARE in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 781–789. [Google Scholar] [CrossRef]
- Meunier, F.A.; Gutierrez, L.M. Captivating New Roles of F-Actin Cortex in Exocytosis and Bulk Endocytosis in Neurosecretory Cells. Trends Neurosci. 2016, 39, 605–613. [Google Scholar] [CrossRef]
- Yang, F.; Kimberlin, A.N.; Elowsky, C.G.; Liu, Y.; Gonzalez-Solis, A.; Cahoon, E.B.; Alfano, J.R. A Plant Immune Receptor Degraded by Selective Autophagy. Mol. Plant 2019, 12, 113–123. [Google Scholar] [CrossRef]
- Yeh, Y.H.; Panzeri, D.; Kadota, Y.; Huang, Y.C.; Huang, P.Y.; Tao, C.N.; Roux, M.; Chien, H.C.; Chin, T.C.; Chu, P.W.; et al. The Arabidopsis Malectin-Like/LRR-RLK IOS1 Is Critical for BAK1-Dependent and BAK1-Independent Pattern-Triggered Immunity. Plant Cell 2016, 28, 1701–1721. [Google Scholar] [CrossRef]
- Beck, M.; Zhou, J.; Faulkner, C.; MacLean, D.; Robatzek, S. Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 2012, 24, 4205–4219. [Google Scholar] [CrossRef]
- Ortiz-Morea, F.A.; Savatin, D.V.; Dejonghe, W.; Kumar, R.; Luo, Y.; Adamowski, M.; Van den Begin, J.; Dressano, K.; Pereira, D.O.G.; Zhao, X.; et al. Danger-associated peptide signaling in Arabidopsis requires clathrin. Proc. Natl. Acad. Sci. USA 2016, 113, 11028–11033. [Google Scholar] [CrossRef]
- Mbengue, M.; Bourdais, G.; Gervasi, F.; Beck, M.; Zhou, J.; Spallek, T.; Bartels, S.; Boller, T.; Ueda, T.; Kuhn, H.; et al. Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc. Natl. Acad. Sci. USA 2016, 113, 11034–11039. [Google Scholar] [CrossRef]
- Lu, D.; Lin, W.; Gao, X.; Wu, S.; Cheng, C.; Avila, J.; Heese, A.; Devarenne, T.P.; He, P.; Shan, L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 2011, 332, 1439–1442. [Google Scholar] [CrossRef] [PubMed]
- Göhre, V.; Spallek, T.; Häweker, H.; Mersmann, S.; Mentzel, T.; Boller, T.; de Torres, M.; Mansfield, J.W.; Robatzek, S. Plant Pattern-Recognition Receptor FLS2 Is Directed for Degradation by the Bacterial Ubiquitin Ligase AvrPtoB. Curr. Biol. 2008, 18, 1824–1832. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xing, J.; Lin, J. At the intersection of exocytosis and endocytosis in plants. New Phytol. 2019, 224, 1479–1489. [Google Scholar] [CrossRef]
- Smith, J.M.; Leslie, M.E.; Robinson, S.J.; Korasick, D.A.; Zhang, T.; Backues, S.K.; Cornish, P.V.; Koo, A.J.; Bednarek, S.Y.; Heese, A. Loss of Arabidopsis thaliana Dynamin-Related Protein 2B reveals separation of innate immune signaling pathways. PLoS Pathog. 2014, 10, e1004578. [Google Scholar] [CrossRef]
- Smith, J.M.; Salamango, D.J.; Leslie, M.E.; Collins, C.A.; Heese, A. Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol. 2014, 164, 440–454. [Google Scholar] [CrossRef]
- Smith, S.M.; Smith, C.J. Capturing the mechanics of clathrin-mediated endocytosis. Curr. Opin. Struct. Biol. 2022, 75, 102427. [Google Scholar] [CrossRef] [PubMed]
- Merrifield, C.J.; Feldman, M.E.; Wan, L.; Almers, W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat. Cell Biol. 2002, 4, 691–698. [Google Scholar] [CrossRef]
- Akamatsu, M.; Vasan, R.; Serwas, D.; Ferrin, M.A.; Rangamani, P.; Drubin, D.G. Principles of self-organization and load adaptation by the actin cytoskeleton during clathrin-mediated endocytosis. eLife 2020, 9, e49840. [Google Scholar] [CrossRef]
- Hemalatha, A.; Mayor, S. Recent advances in clathrin-independent endocytosis. F1000Research 2019, 8, 16549. [Google Scholar] [CrossRef]
- Serwas, D.; Akamatsu, M.; Moayed, A.; Vegesna, K.; Vasan, R.; Hill, J.M.; Schoneberg, J.; Davies, K.M.; Rangamani, P.; Drubin, D.G. Mechanistic insights into actin force generation during vesicle formation from cryo-electron tomography. Dev. Cell 2022, 57, 1132–1145. [Google Scholar] [CrossRef]
- Yoshida, A.; Sakai, N.; Uekusa, Y.; Imaoka, Y.; Itagaki, Y.; Suzuki, Y.; Yoshimura, S.H. Morphological changes of plasma membrane and protein assembly during clathrin-mediated endocytosis. PLoS Biol. 2018, 16, e2004786. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Zhang, Y.; Lian, N.; Li, X. Membrane Dynamics Regulated by Cytoskeleton in Plant Immunity. Int. J. Mol. Sci. 2023, 24, 6059. https://doi.org/10.3390/ijms24076059
Lu Y, Zhang Y, Lian N, Li X. Membrane Dynamics Regulated by Cytoskeleton in Plant Immunity. International Journal of Molecular Sciences. 2023; 24(7):6059. https://doi.org/10.3390/ijms24076059
Chicago/Turabian StyleLu, Yuqing, Yuan Zhang, Na Lian, and Xiaojuan Li. 2023. "Membrane Dynamics Regulated by Cytoskeleton in Plant Immunity" International Journal of Molecular Sciences 24, no. 7: 6059. https://doi.org/10.3390/ijms24076059
APA StyleLu, Y., Zhang, Y., Lian, N., & Li, X. (2023). Membrane Dynamics Regulated by Cytoskeleton in Plant Immunity. International Journal of Molecular Sciences, 24(7), 6059. https://doi.org/10.3390/ijms24076059



