Abstract
Phytophthora root and stem rot caused by Phytophthora sojae Kaufmann and Gerdemann is a soil-borne disease severely affecting soybean production worldwide. Losses caused by P. sojae can be controlled by both major genes and quantitative trait locus. Here, we tested 112 short-season soybean cultivars from Northeast China for resistance to P. sojae. A total of 58 germplasms were resistant to 7–11 P. sojae strains. Among these, Mengdou 28 and Kejiao 10-262 may harbor either Rps3a or multiple Rps genes conferring resistance to P. sojae. The remaining 110 germplasms produced 91 reaction types and may contain new resistance genes or gene combinations. Partial resistance evaluation using the inoculum layer method revealed that 34 soybean germplasms had high partial resistance, with a mean disease index lower than 30. Combining the results of resistance and partial resistance analyses, we identified 35 excellent germplasm resources as potential elite materials for resistance and tolerance in future breeding programs. In addition, we compared the radicle inoculation method with the inoculum layer method to screen for partial resistance to P. sojae. Our results demonstrate that the radicle inoculation method could potentially replace the inoculum layer method to identify partial resistance against P. sojae, and further verification with larger samples is required in the future.
1. Introduction
Phytophthora root and stem rot (PRSR) caused by Phytophthora sojae Kaufmann and Gerdemann is a destructive disease of soybean (Glycine max (L.) Merr.) worldwide [1]. P. sojae can infect soybean at various plant growth stages, especially in low-lying fields with continuous cropping, heavy soil, and poor drainage [2]. Infection results in seed rot, seedlings damping off, root and stem rot, and even soybean plant death [3]. This disease severely limits soybean production and causes economic losses of up to USD 1–2 billion annually [4].
It has been clear that the pathogenetic variation of P. sojae has been complex since it was first described [5], and to date, at least 55 races have been reported [6,7,8,9]. Although several measures, such as seed fungicides, application of calcium-containing compounds, and soil drainage condition improvement, etc. have been reported to mitigate the damage of P. sojae [2,10,11], the deployment of resistant cultivars remains the most environmentally friendly and effective strategy to limit losses caused by the disease.
Single dominant resistance genes to P. sojae (Rps genes), also known as race-specific resistance genes, have been used extensively in soybean to manage P. sojae [12]. Since the first identification of Rps1 [13], more than 35 Rps genes/alleles have been mapped to nine chromosomes, including the newly discovered Rps13 [14]. Based on the typical gene-to-gene theory, the effectiveness of each Rps gene depends on the presence of the corresponding avirulence gene (Avr) in P. sojae [4]. However, due to the emergence of new virulent races in response to selection pressure exerted by the continuous use of specific resistant cultivars, the exploitation of these Rps genes has often been short-lived, as their effectiveness is limited to 8–15 years [15].
Partial resistance, also known as field tolerance, is a highly heritable quantitative trait controlled by multiple genes such as Rps1a, Rps1k, and Rps3a. It does not exert selection pressure on pathogens and produces more durable resistance to P. sojae [12]. Currently, the focus of breeding research is on conferring partial resistance in soybean. For example, Schneider et al. [16] evaluated the resistance of 1395 plant introductions (PIs) to two highly virulent P. sojae isolates and screened several QTLs for partial resistance to P. sojae. Meanwhile, some soybean germplasm with high partial resistance were also selected as candidate parental resources [17,18]. This strategy can result in longer-lasting defense response and, to a certain extent, attenuate the incidence of disease in soybean plants following infection by P. sojae and thereby lessen any impact on soybean yield [19,20]. Combining partial resistance and single-gene resistance is essential to improve broad-spectrum resistance in soybean and necessitates screening for new resistance sources.
As soybean is native to China, the country possesses abundant germplasm resources. Northeastern provinces, including Heilongjiang, Inner Mongolia, Liaoning, and Jilin, are the main regions of soybean cultivation, accounting for up to 50% of the total cultivated area and yield of soybean in China, where most of the short-season soybean is grown. Since PRR was first reported in Heilongjiang Province in 1989 [21], it has become a major disease in most soybean-producing regions in China [22,23,24,25,26,27]. The virulence structure of P. sojae is complex, with different dominant virulence types of P. sojae in different regions [23]. Therefore, it is imperative to evaluate soybean germplasm resources for resistance to a range of virulence types of P. sojae and identify their resistance genotypes to obtain effective disease-resistant cultivars and new sources of disease resistance.
The objectives of the present study were to assess resistance and partial resistance to P. sojae races in early-maturing soybean cultivars grown in the short-season regions of China. In addition, we aimed to identify candidate excellent soybean germplasm resources that contain new disease resistance genes or multi-resistance gene combinations and identify candidate parental lines for PRR resistance breeding.
2. Results
2.1. Resistance of the Soybean Germplasm Resources
We first used hypocotyl wound inoculation to identify resistance to 12 P. sojae strains on 112 soybean cultivars from Northeastern China. Five days after inoculation, the cultivar responses to each race were classified as resistant, intermediate, and susceptible reactions based on mortality levels of ≤30, 30–70, and ≥70%. Figure 1A shows the number of soybean cultivars showing resistance, intermediate reactions, and susceptibility against different P. sojae strains. The results indicate that resistance to P. sojae races was relatively common in the tested soybean cultivars (lines) from Northeastern China. Figure 1B shows the number of soybean germplasms resistant to different numbers of strains. Collectively, 110 germplasms were resistant to 1–11 P. sojae strains, accounting for 98.21% of the tested material. The percentage of cultivars with resistant and intermediate reactions (combined) to races 1, 3, 4, 5, 9, 13, 44, 54, PsJs2, PsMC1, Ps41-1, and USAR2 is shown in Figure 1C. The highest percentage of accessions with resistant and intermediate reactions was obtained in response to race 54, followed by races 1, 9, 13, 5, PsMC1, 4, 3, PsJs2, 44, USAR2, and Ps41-1. Only 25% of the total accessions were resistant to race Ps41-1.
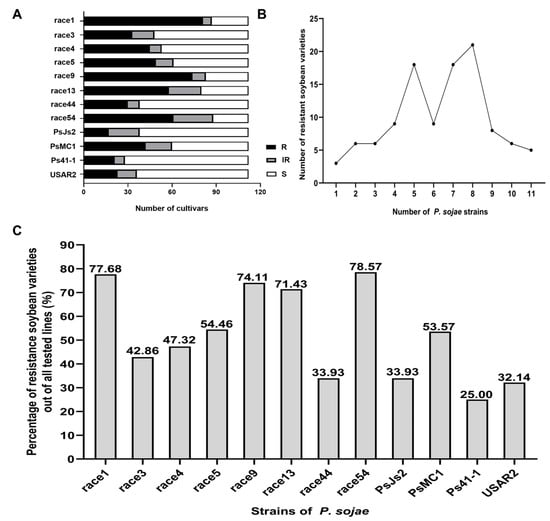
Figure 1.
Resistance analysis of 112 soybean cultivars to 12 P. sojae strains. (A), The number of soybean cultivars showing resistance, intermediate reactions, and susceptibility against different P. sojae strains. R = resistance, IR = intermediate resistance, S = susceptible. (B), The number of germplasm resistance to different numbers of P. sojae strains in R + IR levels. (C), The percentage of germplasm resistance to different P. sojae strains.
Among the 112 soybean cultivars, a total of 58 germplasms were resistant to 7–11 P. sojae strains. Dengke 4, Mengdou 28, Kejiao 10-262, Heinong 57, and Suinong 35 were resistant to 11 P. sojae strains; Jiyu 35, Henong 75, Dengke 3, Beidou 48, Kejiao 07-584, and Suinong 32 were resistant to 10 P. sojae strains; Dengke 1, Suinong 33, Suinong 36, Henong 67, Suinong 29, Fengshou 23, Neidou 4, and Mengdou 15 were resistant to 9 P. sojae strains; 21 germplasms including Mengdou 11, Mengdou 37, and Mengdou 38 were resistant to 8 P. sojae strains; 18 germplasms including Dengke 6, Mengdou 26, and Mengdou 34 were resistant to 7 P. sojae strains. Except for Suinong 28 and Henong 60, which were not resistant to 12 P. sojae strains, the remaining 52 germplasms, including Heihe 22, Heinong 67, and Dengke 5, were resistant to 1-6 P. sojae strains.
Gene postulation of the above resistance results yielded 92 resistance response types (Table S1). Mengdou 28 and Kejiao 10-262 exhibited RRRRRRRRSRRR for race1, race3, race4, race5, race9, race13, race44, race54, PsJs2, PsMC1, Ps41-1, and USAR2, and may possess either the Rps3a or multiple resistance Rps gene Rps1a + Rps3a, Rps1a + Rps1b, Rps1b + Rps1c, Rps1b + Rps1d, Rps1b + Rps3a, Rps1b + Rps6, Rps1c + Rps1d, Rps1c + Rps3a, Rps1d + Rps1k, Rps1d + Rps3a, Rps1d + Rps6 to P. sojae. Furthermore, 110 germplasms had unspecified genotypes, yielding 91 reaction types that differed both from lines containing a single known disease resistance gene and from those with a combination of two known disease resistance genes, thus potentially containing new disease resistance genes or gene combinations.
2.2. Partial Resistance of the Soybean Germplasm Resources
Once a germplasm was identified as susceptible to P. sojae strains from the hypocotyl inoculation test, it was evaluated for partial resistance to the strains using the inoculum layer method. The scoring range of disease index is 0–100, where the mean disease index ≤ 30 is a highly tolerant germplasm. The results of disease resistance in germplasms inoculated with different strains are shown in Table S2. Of all tested cultivars (lines), 11 show a high tolerance to race 1, 42 to race 3, 25 to race 4, 18 to race 5, 11 to race 9, 15 to race 13, 33 to race 44, 12 to race 54, 23 to PsJS2, 24 to PsMC1, 22 to Ps41-1, and 38 to USAR2. The percentage of highly resistant germplasm in the test material ranged from 26.19% to 65.63% (Figure 2). Moreover, the average value of plant disease index in 34 soybean germplasms, including Heinong 51, Hefeng 46, Hefeng 52, Suinong 27, Suinong 29, Suinong 35, Suinong 38, Suizhongzuo 40, Kejiao 10–262, Beidou 42, Heihe 4, Heihe 29, Heihe 35, Heihe 43, Heihe 48, Heihe 52, Mengdou 9, Mengdou 11, Mengdou 12, Mengdou 13, Mengdou 14, Mengdou 28, Mengdou 32, Mengdou 33, Mengdou 35, Mengdou 37, Mengdou 38, Dengke 1, Dengke 4, Dengke 6, Dengke 9, Dengke 10, Kenfeng 16, and Jiyu 35 for disease tolerance identification was lower than 30. Among these 34 highly tolerant soybean germplasms, a total of 25 germplasms were resistant to 7–11 P. sojae strains in previous disease resistance identification and could be used as elite resistance and tolerance materials for breeding in the future. The other nine soybean germplasms were resistant to two to six P. sojae strains and can also be introduced into the genetic background of highly resistant cultivars for breeding and application. These results indicate that the Northeast region is rich in disease-tolerant resources and that these highly tolerant germplasms can provide excellent parents and carriers of superior genes for breeding soybean lines resistant to P. sojae in China.
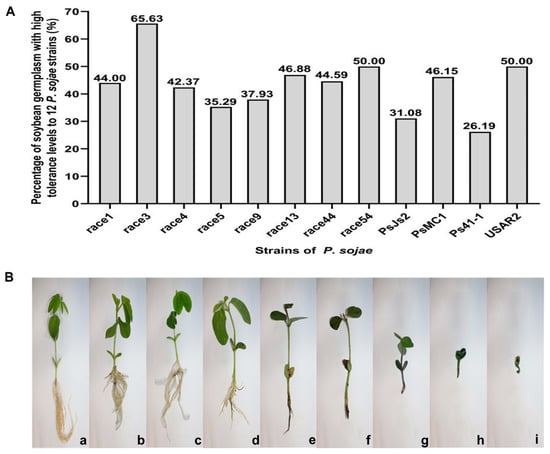
Figure 2.
Partial resistance of the soybean germplasm resources. (A) Percentage of soybean germplasm with high tolerance levels to 12 P. sojae strains. (B) The reaction of roots after inoculation with P. sojae strain using inoculum layer method. a—The roots are not rotted, and the plant grows normally (denoted as 1); b—The roots of the plant are slightly rotted (denoted as 2); c—The roots of the plant are 1/3 rotted (denoted as 3); d—The roots of the plant are 2/3 rotted (denoted as 4); e—All roots are rotted, 10% of plants die (denoted as 5); f—50% of the plants died, and the plants were stunted (denoted as 6); g—75% of plants die, severe development of plants (denoted as 7); h—90% of plants die (denoted as 8); i—All plants die (denoted as 9).
2.3. Acquisition of Germplasm Resources with Resistance and Partial Resistance
The results of comprehensive resistance and partial resistance identification showed that Mengdou 28, Kejiao 10–262, Suinong 35, and Dengke 4 were resistant to 11 P. sojae strains, and the plant response was high disease resistance when partial resistance was identified; Heinong 57 was resistant to 11 P. sojae strains and was a better multi-resistant germplasm, although it was not resistant when inoculated with race 9; Jiyu 35 was resistant to 10 P. sojae strains, and the plant response was high disease resistance when partial resistance was identified; 29 soybean germplasms, including Heinong 51, Hefeng 46, and Suinong 27, were resistant to 2–9 strains of P. sojae strains, and the results of partial resistance identification showed high disease resistance (Table 1). The above 35 soybean germplasms were identified as suitably resistant and tolerant materials and could be used as excellent parents for breeding against PRR in China.

Table 1.
List of 35 candidate soybean cultivars that showed suitable resistance and partial resistance.
2.4. Comparison of the Radicle Inoculation and Inoculum Layer Methods
The radicle inoculation method was first used to evaluate the pathogenicity of Fusarium graminearum on soybean. It has several advantages, including being a simple time- and space-saving operation that produces stable results. In the present study, this technique was applied to identify partial resistance against P. sojae. We randomly selected race1, race13, and race54 for further identification of partial resistance by the radicle inoculation method. Ten days after inoculation, the roots of the plants were observed for different disease resistance reactions, some of which are shown in Figure 3. Similarly, the radicle inoculation method was graded on a scale of 0–7 and converted into a disease index for evaluating soybean cultivars for partial resistance. Soybean cultivars with consistent results for tolerance in both methods were counted, and their percentage of the total was calculated. The results showed that the consistency of the inoculum layer method and the radicle inoculation method for races 1, 13, and 54 were 96.00%, 84.38%, and 83.33%. Table S3 lists the disease index comparison of partial resistance evaluation by the inoculum layer method and radicle inoculation method. The above results suggest that the radicle inoculation method could potentially replace the inoculum layer method to identify partial resistance against P. sojae, and further verification with larger samples is required in the future.

Figure 3.
The reaction of roots after inoculation with P. sojae strain using radicle inoculation method. a—No visible disease symptoms on taproot and lateral roots (denoted as 0); b—Trace of rot on taproot and lateral roots (denoted as 1); c—Less than the bottom third of taproot mass rotted (denoted as 2); d—Bottom third of taproot mass rotted (denoted as 3); e—Bottom two-thirds of taproot mass rotted (denoted as 4); f—More than the bottom two-thirds of taproot mass rotted (denoted as 5); g—Taproot completely rotted with only a few lateral roots (denoted as 6); h—Taproot completely rotted without lateral roots + plant death (denoted as 7).
3. Discussion
Since P. sojae was first discovered and isolated in Northeastern China in 1989 [21], many scholars have devoted themselves to the screening of germplasm resources for resistance to P. sojae and have confirmed the existence of abundant germplasm resources for resistance and concurrent resistance in China [27,28,29,30,31,32]. In the present study, 12 P. sojae strains were used to identify 112 short-season soybean cultivars from Northeast China for their resistance to PRR. Among them, 58 germplasms were resistant to 7–11 strains, accounting for 51.79% of the tested species. This result indicates abundant germplasm resources for disease resistance and multiple resistance in the Northeastern soybean production area and supports earlier reports from China [29,30,31,33]. Furthermore, it is noteworthy that only 23 germplasms showed resistance to the exotic strain USAR2. A possible explanation is that soybean resources from the same region may be similar in terms of genetic background and level of resistance [34]. Moreover, long-term coexistence and co-evolution of pathogens and hosts in the same place of origin could potentially result in fewer resistant resources in the absence of selection pressure. Therefore, using excellent resistant planting resources with different genetic backgrounds when cultivating disease-resistant cultivars can enrich the resistance diversity in Northeast China.
Among the tested materials in this study, 110 germplasms were resistant to 1–11 P. sojae strains, accounting for 98.21% of the total number of identifications. This information further shows that soybean cultivars commonly grown in Northeastern China are resistant to almost all P. sojae races. Previous studies have shown that Rps1k, Rps1c, and Rps1a have been widely used in breeding against P. sojae due to their stable broad-spectrum resistance, with Rps1k having the most stable and the highest broad-spectrum resistance [12,35,36,37]. Gene mining was performed by inoculating the hypocotyl to identify the results of resistance to susceptibility, producing a total of 92 types of anti-inductive responses. Mengdou 28 and Kejiao 10–262 may contain the Rps1k, Rps1c, and Rps1a genes. To the best of our knowledge, this is the first time that soybean germplasm containing the Rps1k gene has been derived from germplasm resources in Northeast China. Furthermore, 110 germplasms had unspecified genotypes, producing 91 reactivity types that were different from those containing a single known disease resistance gene or a combination of two known disease resistance genes. These cultivars may contain a new disease resistance gene or a combination of resistance genes. Using these cultivars in breeding programs will enable gene pyramiding in subsequent generations to develop multiple gene resistance for broader effectiveness against the pathogen.
Partial resistance is a highly heritable quantitative trait that is a valuable complement to major gene resistance. It limits the pathogen’s spot growth rate in the host tissue and reduces the severity of disease caused by P. sojae, thus limiting yield loss [19,20]. Further identification of partial resistance in the present study showed that the percentage of germplasm with high disease resistance accounted for 26.19–65.63% of the tested materials, confirming the presence of disease-resistant germplasm resources in Northeast China. However, partial resistance may not provide adequate control against a high number of pathogens [12]. The massive deployment of qualitative traits controlled by a single gene has resulted in higher selection pressure on the pathogen, thus shortening its available time. Therefore, incorporating Rps resistance into soybean genetic backgrounds with high levels of partial resistance may prolong the effective longevity of the Rps gene. Mining new disease resistance genes or combinations of disease resistance genes can provide excellent parents and vectors of superior genes for breeding cultivars resistant to PRR.
Since root resistance to P. sojae is a very important index in soybean breeding, the evaluation of root resistance is practical and valuable for the soybean industry. However, the research on soybean resistance to P. sojae is mainly focused on inoculating aboveground plant parts, usually hypocotyls, primarily for the inconvenience of investigating the roots in soil [38]. To date, there were only hydroponic inoculation procedure, aeroponics system, and inoculum layer test [38,39,40,41] reported to screen soybean root reaction to P. sojae, but all the above-mentioned methods were laborious and time-consuming when a large number of genotypes need to be evaluated against different P. sojae isolates. Therefore, an effective, fast, and reliable method to measure root resistance under controlled conditions would be very beneficial. In this study, the radicle inoculation method invented by Xue et al. [42] was also applied for the first time to screen soybean germplasm for partial resistance to P. sojae. We randomly selected race1, race13, and race54 for further identification of partial resistance by the radicle inoculation method and obtained more than 80% concordance. Benefits of the radicle inoculation method include better control over inoculation conditions, as well as over temperature and water levels for the disease, reduced infestation of foreign pests and diseases, and the possibility to record disease resistance in plants more accurately and conveniently. In addition, it can considerably reduce the amount of bacteria used during the procedure, effectively shorten the cycle of disease resistance, and save space. Therefore, this technique may become an alternative method for identifying disease resistance in future breeding programs; however, further validation with larger samples is still required.
4. Materials and Methods
4.1. Short-Season Soybean Cultivars
A total of 112 commercial soybean cultivars (lines) (Maturity Groups 000, 00, 0, and I; Table 2) were used to evaluate resistance to the 12 P. sojae races under greenhouse conditions. The breeding units that kindly provided these seeds are also listed in Table 2.

Table 2.
Source, maturity group, and heat unit of tested soybean genotypes.
4.2. P. sojae Races
Isolates of 12 P. sojae races were used as inoculum for this work. P. sojae races 1, 3, 4, 5, 9, 13, 44, and 54 were isolated from soybean fields in Heilongjiang Province [28]. Races PsJs2, PsMC1, Ps41-1, and USAR2 were obtained from Dr. Zhendong Zhu (Institute of Crop Science, Chinese Academy of Agricultural Sciences). Throughout this investigation, isolates were kept on V8 juice agar plates at 25 °C for 7 days before being moved to fresh plates every 2 months. Additionally, all isolates were retested for virulence pre-inoculation. Table 3 contains a list of the virulence pathotypes against different cultivars.

Table 3.
Virulence reaction of P. sojae strains on differential hosts.
4.3. Resistance Identification
Ten seeds of each cultivar (line) were grown in plastic pots (diameter = 10 cm) containing a soil: perlite:peat moss mixture (in a 1:1:1 volume ratio) in a greenhouse with a temperature range of 22–25 °C. Metal halide lamps of 300 W were used as additional lighting to maintain a 16 h photoperiod.
Seedlings at the first-node stage (V1) [35] were inoculated with P. sojae isolates using a hypocotyl wound technique described by Kaufmann and Gerdmann [5]. A blade was used to make a shallow cut along the hypocotyls of the seedlings, 1 cm below the cotyledon node. Next, the aerial mycelium was inserted into the longitudinal wound, which was taken from the edge of the pre-cultured P. sojae isolates.
Following inoculation, the plants were kept in a moist chamber for 3 days before being placed back in the growth chambers to monitor disease progression. Three replication pots with every ten plants were used in the experiment to gauge the response of soybean cultivars (lines) to different races. In order to test the adequacy of the environment for the development of infection and illness as well as the potential harm brought on by wounding plants, Williams (rpsrps) plants from three pots were wounded and inoculated with blank V8 juice agar without P. sojae isolates in each experiment. Five days after inoculation, the disease level of various soybean cultivars was examined. If a differential exhibited ≤30% seedling mortality, the reaction was considered resistance. If a differential exhibited ≥70% seedling mortality, the reaction was considered susceptible. Seedling mortality from 30% to 70% was considered an intermediate reaction [43].
4.4. Partial Resistance Reaction
Once a germplasm was identified as susceptible to P. sojae strains from the hypocotyl inoculation test, it was further evaluated for partial resistance to the strains using the inoculums layer method. Soybean plants were grown in plastic houses under natural conditions to ensure normal growth. Seeds of different soybean cultivars were inoculated with P. sojae isolates that involved placing the corresponding agar cultures underneath the seeds, according to the method described by Walker and Schmitthenner [44]. After three weeks, the degree of partial resistance of the different cultivars was assessed. The rating system for the layer test used a scale of 1 to 9, in which 1 = no root rot, 2 = a trace of root rot, 3 = the bottom third of root mass rotted, 4 = the bottom two-thirds of root mass rotted, 5 = all roots rooted + 10% of seedlings dead, 6 = 50% of seedlings dead + moderate stunting on tops, 7 = 75% of seedlings dead + severe stunting of tops, 8 = 90% of seedling dead, 9 = all seedlings dead. Referring to the identification criteria of Dorrance and Schmitthenner [45] and using the disease level formula of Liu [46] to transform into a disease index, it was concluded that a disease index ≤ 30 has high tolerance, a disease index from 30 to 60 has moderate tolerance, and a disease index ≥ 60 has a low tolerance.
4.5. Radicle Inoculation Techniques
The radicle inoculation techniques described by Xue et al. [42] for evaluating the pathogenicity of different Fusarium isolates on soybean seedlings were applied to evaluate the partial resistance of soybean to P. sojae in the present study. Each experiment’s seeds were sterilized by dipping them into 0.5% NaClO for 45 s and then giving them two rinses in sterile distilled water. After spreading the seeds evenly on two layers of sterile paper towels and moistening them with enough sterile water, the other two layers of sterile paper towels were laid on top to allow the seeds to germinate. When plants reached the early V1 growth stage [35] and root hairs became apparent, they were transferred to dark conditions at 25 °C and kept for 18 h. Then, visually sound seedlings were chosen and subjected to the same surface sterilization procedures as previously mentioned. Using a sterile metal needle, a 0.6 cm diameter agar plug was removed from the edge of the pre-cultured P. sojae isolates and inoculated approximately 1.5 cm behind the main root tip of the soybean seedling. Every inoculated plant was set on a pre-cut covering sheet, which was made of two layers of sterile tissue paper placed on a sheet of aluminum foil. The aluminum foil sheet was used to keep each unit separate and retain moisture. The trays were then placed in a growth chamber, and the water level in the tray was checked daily, and water was added as needed.
After 10 days of inoculation, the severity of root rot of different cultivars was assessed. The rating system employed a scale of 0–7, in which 0 = no visible disease symptoms on taproot and lateral roots, 1 = trace of rot on taproot and lateral roots, 2 = less than the bottom third of taproot mass rotted, 3 = bottom third of taproot mass rotted, 4 = bottom two-thirds of taproot mass rotted, 5 = more than the bottom two-thirds of taproot mass rotted, 6 = taproot completely rotted with only a few lateral roots, 7 = taproot completely rotted without lateral roots + plant death. Referring to the rating criteria of the inoculums layer method with slight modifications, the disease rating formula of Liu [46] was transformed into a disease index, and it was concluded that a disease index ≤20 has high tolerance, a disease index from 20 to 50 has moderate tolerance, and a disease index ≥50 has a low tolerance.
4.6. Statistical Analysis
Three replications of a randomized complete block design were used to arrange every pot. Residuals for each parameter in each experimental parameter were examined for normality and homogeneity of variances. The SAS UNIVARIATE technique was used to check the Shapiro–Wilk test’s assumption of normality, and the PLOT function was used to evaluate the random and homogeneous distribution of residuals (SAS Institute Inc., Cary, NC, USA, 2008).
5. Conclusions
In this study, we tested 112 short-season soybean cultivars from Northeast China for resistance to P. sojae. Combining the results of hypocotyl inoculation for resistance and the inoculum layer method for partial resistance, we screened a total of 35 superior soybean germplasm. Among these, Mengdou 28 and Kejiao 10–262 may harbor either Rps3a or multiple Rps genes conferring resistance to P. sojae. Furthermore, we used radicle inoculation for the first time to identify partial resistance to P. sojae and confirmed that the results were in high agreement with the inoculum layer method, which may serve as a time- and labor-saving method to identify partial resistance against P. sojae in the future. Our findings indicate that Northeast China is rich in excellent soybean germplasm resources resistant to P. sojae and provides a theoretical basis for screening candidate parental lines for PRR resistance breeding.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24076027/s1.
Author Contributions
P.X. and S.Z. designed the experiments. S.H., X.W., X.S., M.Z., S.C., Y.Z., J.W. and X.C. performed the experiments. C.Z., X.F., Y.S., B.S., S.L. and Y.L. analyzed the data. S.H., P.X. and S.Z. wrote the manuscript. S.H., XW. and X.S. contributed equally to this research. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2021YFD1201103), NSFC Projects (31971972), Natural Science Foundation of Heilongjiang Province (ZD2019C001), Key Research and Development Program of Heilongjiang Province (GX18B032), and Outstanding Talents and Innovative Team of Agricultural Scientific Research.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are represented in the article’s Supplementary tables.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wrather, J.A.; Anderson, T.R.; Arsyad, D.M.; Gai, J.; Ploper, L.D.; Porta-Puglia, A.; Ram, H.H.; Yorinori, J.T. Soybean disease loss estimates for the top 10 soybean producing countries in 1994. Plant Dis. 1997, 81, 107–110. [Google Scholar] [CrossRef]
- Dorrance, A.E.; Mills, D.; Robertson, A.E.; Draper, M.A.; Giesler, L.; Tenuta, A. Phytophthora Root and Stem Rot of Soybean. Plant Health Instr. 2007, 1–10. [Google Scholar] [CrossRef]
- Schmitthenner, A.F. Problems and progress in control of Phytophthora root rot of soybean. Plant Dis. 1985, 69, 362–368. [Google Scholar] [CrossRef]
- Tyler, B.M. Phytophthora sojae: Root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 2007, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, M.J.; Gerdemann, J.W. Root and stem rot of soybean caused by Phytophthora sojae. Phytopathology 1958, 48, 201–208. [Google Scholar] [CrossRef]
- Morgan, F.L.; Hartwig, E.E. Physiologic specialization in Phytophthora megasperma var. sojae. Phytopathology 1965, 55, 1277–1279. [Google Scholar]
- Leitz, R.A.; Hartman, G.L.; Pedersen, W.L.; Nickell, C.D. Races of Phytophthora sojae on soybean in Illinois. Plant Dis. 2000, 84, 487. [Google Scholar] [CrossRef]
- Burnham, K.D.; Dorrance, A.E.; Vantoai, T.T.; Martin, S.S. Quantitative trait loci for partial resistance to Phytophthora sojae in soybean. Crop Sci. 2003, 43, 673–675. [Google Scholar] [CrossRef]
- Jackson, T.A.; Kirkpatrick, T.L.; Rupe, J.C. Races of Phytophthora sojae in Arkansas soybean fields and their effects on commonly grown soybean cultivars. Plant Dis. 2004, 88, 345–351. [Google Scholar] [CrossRef]
- Sugimoto, T.; Watanabe, K.; Yoshida, S.; Aino, M.; Furiki, M.; Shiono, M.; Matoh, T.; Biggs, A.R. Field application of calcium to reduce Phytophthora stem rot of soybean, and calcium distribution in plants. Plant Dis. 2010, 94, 812–819. [Google Scholar] [CrossRef]
- Dorrance, A.E. Management of Phytophthora sojae of soybean: A review and future perspectives. Can. J. Plant Pathol. 2018, 40, 210–219. [Google Scholar] [CrossRef]
- Dorrance, A.E.; McClure, S.A.; Martin, S.K. Effect of partial resistance on Phytophthora stem rot incidence and yield of soybean in Ohio. Plant Dis. 2003, 87, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Bernard, R.L.; Smith, P.E.; Kaufmann, M.J.; Schmitthenner, A.F. Inheritance of resistance to Phytophthora root and stem rot in soybean. Agron. J. 1957, 49, 391. [Google Scholar] [CrossRef]
- Sahoo, D.K.; Das, A.; Huang, X.; Cianzio, S.; Bhattacharyya, M.K. Tightly linked Rps12 and Rps13 genes provide broad-spectrum Phytophthora resistance in soybean. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Schmitthenner, A.F.; Hobe, M.; Bhat, R.G. Phytophthora sojae races in Ohio over a 10-year interval. Plant Dis. 1994, 78, 269–276. [Google Scholar] [CrossRef]
- Schneider, R.; Rolling, W.; Song, Q.; Cregan, P.; Dorrance, A.E.; McHale, L.K. Genome-wide association mapping of partial resistance to Phytophthora sojae in soybean plant introductions from the Republic of Korea. BMC Genom. 2016, 17, 607. [Google Scholar] [CrossRef]
- Jia, H.; Kurle, J.E. Resistance and partial resistance to Phytophthora sojae in early maturity group soybean plant introductions. Euphytica 2008, 159, 27–34. [Google Scholar] [CrossRef]
- Wu, X.L.; Zhao, J.M.; Sun, S.; Yang, F.; Wang, Y.C.; Gai, J.Y.; Xing, H. A survey of soybean germplasm for resistance to Phytophthora sojae. Euphytica 2010, 176, 261–268. [Google Scholar] [CrossRef]
- Tooley, P.W.; Grau, C.R. Field characterization of rate-reducing resistance to Phytophthora megasperma f. sp. glycinea in soybean. Phytopathology 1984, 74, 1201–1208. [Google Scholar] [CrossRef]
- Tooley, P.W.; Grau, C.R. The relationship between rate-reducing resistance to Phytophthora megasperma f. sp. glycinea and yield of soybean. Phytopathology 1984, 74, 1209–1216. [Google Scholar] [CrossRef]
- Shen, C.Y.; Su, Y.C. Discovery and preliminary studies of Phytophthora megasperma on soybean in China. Acta Phytopathol. Sin. 1991, 21, 198. [Google Scholar]
- Zhu, Z.D.; Wang, X.O.; Wang, H.B.; Wu, X.F.; Zhang, L. Identification and race of Phytophthora sojae isolates collected in Mengcheng, Anhui province. Acta Phytopathol. Sin. 2001, 31, 236–240. [Google Scholar] [CrossRef]
- Zhu, Z.D.; Wang, H.B.; Wang, X.O.; Chang, R.Z.; Wu, X.F. Distribution and virulence diversity of Phytophthora sojae in China. Chin. Agric. Sci. 2003, 36, 793–799. [Google Scholar] [CrossRef]
- Chen, S.K.; Yan, R.P.; Wang, Q.R.; Wu, Y.H. Study on technology of preventative curing and incidence of Phytophthora Megasperma in Holonboir league. J. Inner Mongo Agric. Univ. 2002, 17, 223–227. [Google Scholar] [CrossRef]
- Tang, Q.H.; Cui, L.K.; Li, D.L.; Dai, T.T.; Yin, S.M.; Xing, G.; Zheng, X.B.; Wang, Y.C. Resistance evaluation of soybean germplasm from Huanghuai valley to Phytophthora root rot. Chin. Agric. Sci. 2010, 43, 2246–2252. [Google Scholar] [CrossRef]
- Cheng, Y.B.; Ma, Q.B.; Mu, Y.H.; Tan, Z.Y.; Wu, H.; Nian, H. Analysis of resistance genes of soybean accessions from South China to Phytophthora root rot. Sci. Agric. Sin. 2015, 48, 2296–2305. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, Y.; Yu, H.; Chen, Y.; Gu, X.; Wen, J. Pathotypes of Phytophthora sojae and their distribution in Jilin, China. J. Plant. Pathol. 2021, 103, 241–248. [Google Scholar] [CrossRef]
- Zhang, S.Z.; Xu, P.F.; Wu, J.J.; Xue, A.G.; Zhang, J.X.; Li, W.B.; Chen, C.; Chen, W.Y.; Lv, H.Y. Races of Phytophthora sojae and their virulences on soybean cultivars in Heilongjiang, China. Plant Dis. 2010, 94, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.M.; Li, B.Y.; Ding, J.J. Selection of soybean germplasms with resistance to Phytophthora root rot and its use in breeding for resistance. Soybean Sci. 2001, 20, 197–199. [Google Scholar] [CrossRef]
- Lv, H.Y.; Kong, F.J.; Xu, X.H.; Yuan, X.L.; Yang, Q.K. Response of soybean germplasm from the Northeast China to Phytophthora root rot caused by Phytophthora sojae. Chin. J. Oil Crop Sci. 2001, 23, 16–18. [Google Scholar] [CrossRef]
- Xu, X.H.; Lv, H.Y.; Qu, J.J.; Yang, Q.K. Identification and virulence analysis of physiological races of Phytophthora sojae. Acta Phytopathol. Sin. 2003, 30, 125–128. [Google Scholar] [CrossRef]
- Yang, J.; Ye, W.; Wang, X.; Ren, L.; Yao, Y.; Wang, X.; Wang, Y. An improved method for the identification of soybean resistance to Phytophthora sojae applied to germplasm resources from the Huanghuaihai and Dongbei regions of China. Plant Dis. 2020, 104, 408–413. [Google Scholar] [CrossRef]
- Zhu, Z.D.; Wang, H.B.; Wang, X.M.; Chang, R.Z.; Wu, X.F. Response of soybean cultivars or lines developed in Heilongjiang province to five strains of Phytophthora sojae. J. Plant Genet. Resour. 2004, 5, 22–25. [Google Scholar] [CrossRef]
- Kyle, D.E.; Nickell, C.D.; Nelson, R.L.; Pederson, W.L. Response of soybean accessions from provinces in Southern China to Phytophthora sojae. Plant Dis. 1998, 82, 555–559. [Google Scholar] [CrossRef]
- Fehr, W.R.; Caviness, C.E.; Burmood, D.T.; Pennington, J.S. Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Sci. 1971, 11, 929–931. [Google Scholar] [CrossRef]
- Ferro, C.R.; Hill, C.B.; Miles, M.R.; Hartman, G.L. Evaluation of soybean cultivars with the Rps1k gene for partial resistance or field tolerance to Phytophthora sojae. Crop Sci. 2006, 46, 2427–2436. [Google Scholar] [CrossRef]
- Yang, J.; Zheng, S.J.; Wang, X.M.; Ye, W.W.; Zheng, X.B.; Wang, Y.C. Identification of resistance genes to Phytophthora sojae in domestic Soybean cultivars from China using particle bombardment. Plant Dis. 2020, 104, 1888–1893. [Google Scholar] [CrossRef]
- Wagner, R.E.; Wilkinson, H.T. An aeroponics system for investigating disease development on soybean taproots infected with Phytophthora sojae. Plant Dis. 1992, 76, 610–614. [Google Scholar] [CrossRef]
- Kilen, T.C.; Hartwig, E.E.; Keeling, B.L. Inheritance of a second major gene for resistance to Phytophthora root rot in soybeans. Crop Sci. 1974, 14, 260–262. [Google Scholar] [CrossRef]
- Thomison, P.R.; Thomas, C.A.; Kenworthy, W.J. Tolerant and root-resistant soybean cultivars: Reactions to Phytophthora rot in inoculum-layer tests. Crop Sci. 1991, 31, 73–75. [Google Scholar] [CrossRef]
- Lohnes, D.G.; Wagner, R.E.; Bernard, R.L. Soybean genes, Rj2, Rmd, and Rps2 in linkage group 19. J. Hered. 1993, 84, 109–111. [Google Scholar] [CrossRef]
- Xue, A.G.; Cober, H.D.; Voldeng, H.D.; Babcock, C.; Clear, R.M. Evaluation of the pathogenicity of Fusarium graminearum and Fusarium pseudograminearum on soybean seedlings under controlled conditions. Plant Pathol. 2007, 29, 35–40. [Google Scholar] [CrossRef]
- Yang, X.; Ruff, R.L.; Meng, X.Q.; Workneh, F. Race of Phytophthora sojae in lowa soybean fields. Plant Dis. 1996, 80, 1418–1420. [Google Scholar] [CrossRef]
- Walker, A.K.; Schmitthenner, A.F. Comparation of field and greenhouse evaluation for tolerance to Phytophthora rot in soybean. Crop Sci. 1984, 24, 487–489. [Google Scholar] [CrossRef]
- Dorrance, A.E.; Schmitthenner, A.F. New sources of resistance to Phytophthora sojae in the soybean plant introductions. Plant Dis. 2000, 84, 1303–1308. [Google Scholar] [CrossRef]
- Liu, L. Discussion of errors in the calculation formula of disease index and the correction method. J. Zhejiang Agric. Sci. 1987, 2, 88–92. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).