1,4-α-Glucosidase from Fusarium solani for Controllable Biosynthesis of Silver Nanoparticles and Their Multifunctional Applications
Abstract
1. Introduction
2. Results
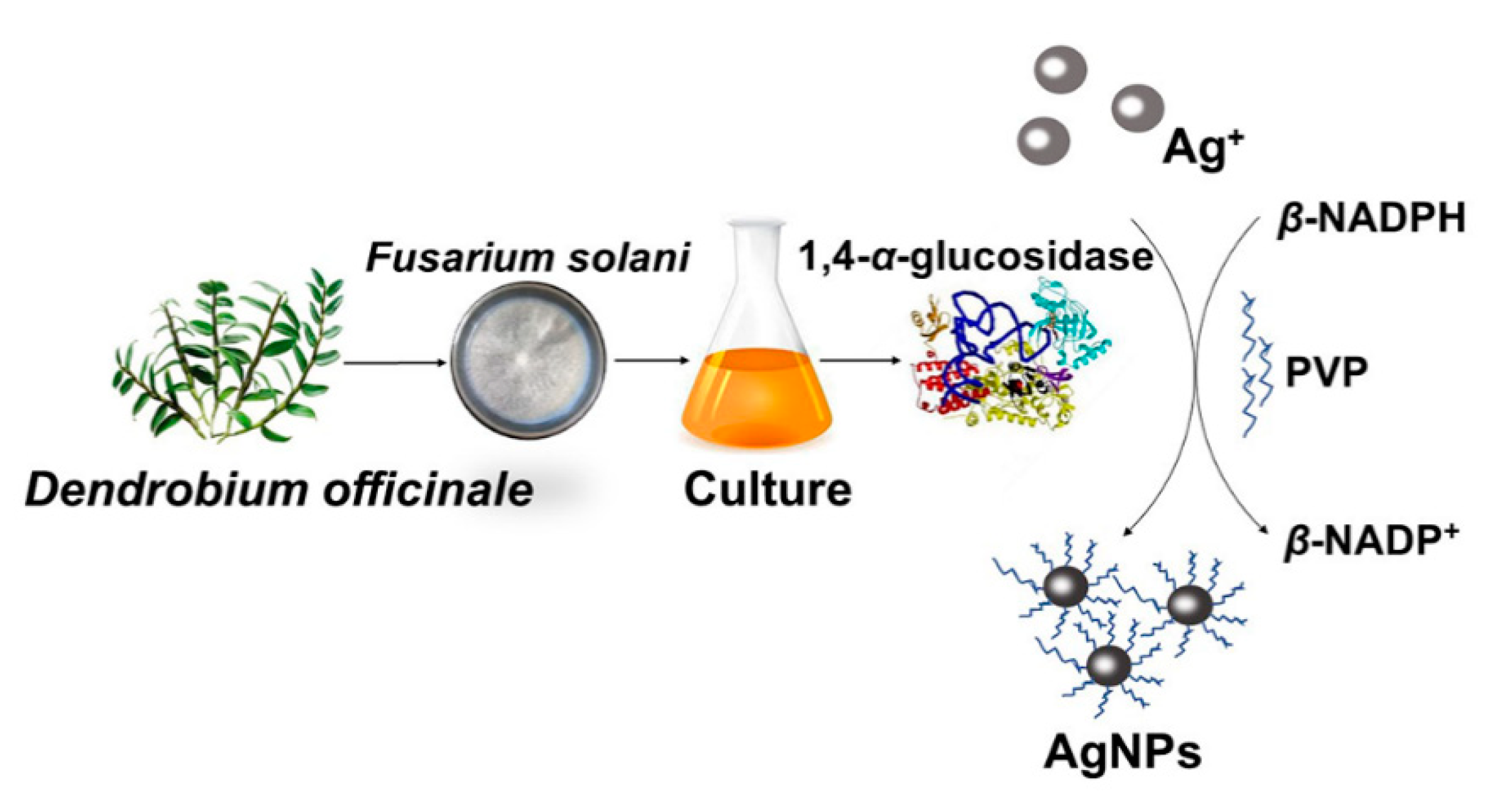
2.1. Preparation of AgNPs
2.2. Assay of the Compositions Responsible for AgNPs Synthesis
2.3. Controllable Synthesis of AgNPs
2.4. Characteristics of AgNPs
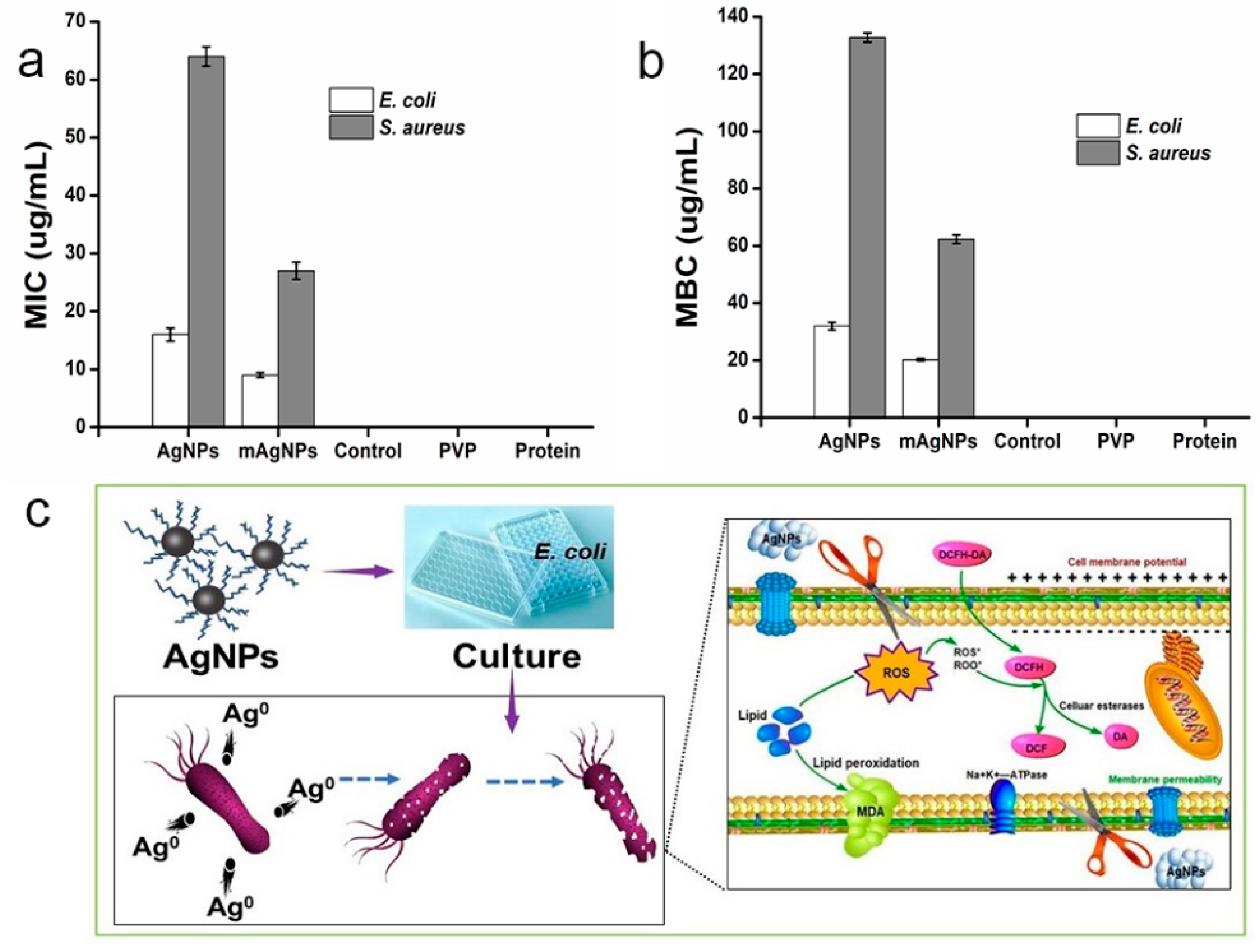
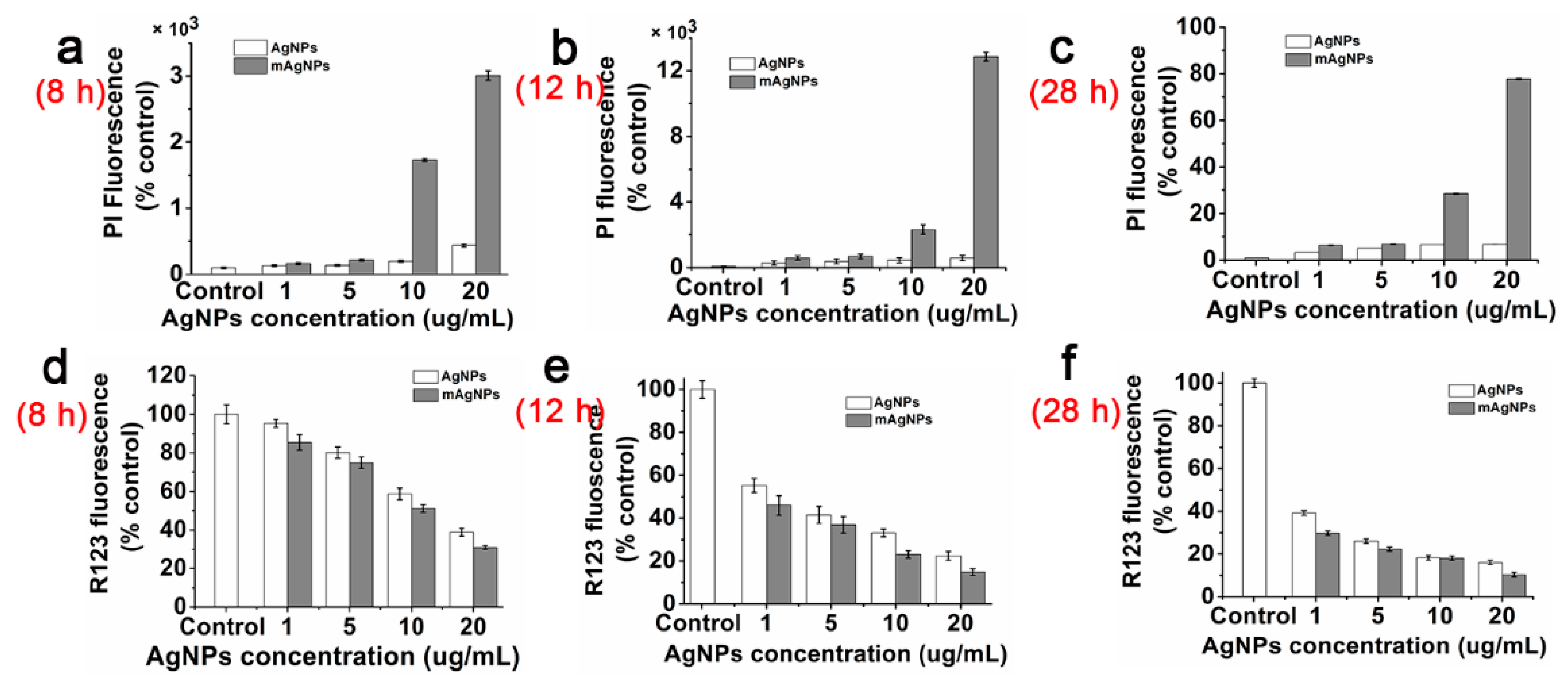
2.5. Antibacterial Mechanism of AgNPs
2.6. Catalytic Reduction of 4−NA
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Preparation of AgNPs
4.3. Isolation and Identification of Protein
4.4. Characteristics of AgNPs
4.5. Size and Shape Control of AgNPs
4.6. Antibacterial Activity and Mechanism Assay
4.7. Cell Morphology Observation
4.8. Catalytic Reduction Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.; Huang, Y.; Liu, K.; Guo, L.; Yuan, Q.; Dong, B. Multichannel-improved charge-carrier dynamics in well-designed hetero-nanostructural plasmonic photocatalysts toward highly efficient solar-to-fuels conversion. Adv. Mater. 2015, 27, 5906–5914. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.-Q.; Ru, S.; Zhang, Q.; Jang, M.; Kurade, M.B.; Kim, S.-H.; Jeon, B.-H. Insights into the effect of cerium oxide nanoparticle on microalgal degradation of sulfonamides. Bioresour. Technol. 2020, 309, 123452. [Google Scholar] [CrossRef] [PubMed]
- Mota, D.A.; Rajan, D.; Heinzl, G.C.; Osório, N.M.; Gominho, J.; Krause, L.C.; Soares, C.M.F.; Nampoothiri, K.M.; Sukumaran, R.K.; Ferreira-Dias, S. Production of low-calorie structured lipids from spent coffee grounds or olive pomace crude oils catalyzed by immobilized lipase in magnetic nanoparticles. Bioresour. Technol. 2020, 307, 123223. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Lee, D.-J. Impact of adding metal nanoparticles on anaerobic digestion performance—A review. Bioresour. Technol. 2019, 292, 121926. [Google Scholar] [CrossRef]
- Arduini, F.; Cinti, S.; Scognamiglio, V.; Moscone, D. Nanomaterials in electrochemical biosensors for pesticide detection: Advances and challenges in food analysis. Microchim. Acta 2016, 183, 2063–2083. [Google Scholar] [CrossRef]
- Rashidzadeh, H.; Danafar, H.; Rahimi, H.; Mozafari, F.; Salehiabar, M.; Rahmati, M.A.; Rahamooz-Haghighi, S.; Mousazadeh, N.; Mohammadi, A.; Ertas, Y.N.; et al. Nanotechnology against the novel coronavirus (severe acute respiratory syndrome Thanks.coronavirus 2): Diagnosis, treatment, therapy and future perspectives. Nanomedicine 2021, 16, 497–516. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Xiao, J.; Chen, M.; Cao, C.; Yan, C.; Ma, Y.; Huang, M.; Wang, M. Fate of silver nanoparticles in constructed wetlands and its influence on performance and microbiome in the ecosystems after a 450-day exposure. Bioresour. Technol. 2019, 281, 107–117. [Google Scholar] [CrossRef]
- Barman, S.R.; Nain, A.; Jain, S.; Punjabi, N.; Mukherji, S.; Satija, J. Dendrimer as a multifunctional capping agent for metal nanoparticles for use in bioimaging, drug delivery and sensor applications. J. Mater. Chem. B 2018, 6, 2368–2384. [Google Scholar] [CrossRef]
- Panacek, A.; Kvítek, L.; Prucek, R.; Kolar, M.; Vecerova, R.; Pizúrova, N.; Sharma, V.K.; Nevecna, T.; Zboril, R. Silver colloid nanoparticles: Synthesis, characterization, and their antibacterial activity. J. Phys. Chem. B 2006, 110, 16248–16253. [Google Scholar] [CrossRef]
- Chun Kiat, N.; Krishnakumar, S.; Xin, L.; Munusamy, M.; Lianghui, J.; Liang, Y.; Chuyang, T.; Hao, S.; Staffan, K.; Bin, C. Influence of outer membrane c-type cytochromes on particle size and activity of extracellular nanoparticles produced by Shewanella oneidensis. Biotechnol. Bioeng. 2013, 110, 1831–1837. [Google Scholar]
- Oliver, S.; Wagh, H.; Liang, Y.; Yang, S.; Boyer, C. Enhancing the antimicrobial and antibiofilm effectiveness of silver nanoparticles prepared by green synthesis. J. Mater. Chem. B 2018, 6, 4124–4138. [Google Scholar] [CrossRef] [PubMed]
- Escárcega-González, C.E.; Garza-Cervantes, J.A.; Vázquez-Rodríguez, A.; Montelongo-Peralta, L.Z.; Treviño-González, M.T.; Díaz Barriga Castro, E.; Saucedo-Salazar, E.M.; Chávez Morales, R.M.; Regalado Soto, D.I.; Treviño González, F.M.; et al. In vivo antimicrobial activity of silver nanoparticles produced via a green chemistry synthesis using Acacia rigidula as a reducing and capping agent. Int. J. Nanomed. 2018, 13, 2349–2363. [Google Scholar] [CrossRef] [PubMed]
- Desireddy, A.; Conn, B.E.; Guo, J.; Yoon, B.; Barnett, R.N.; Monahan, B.M.; Kirschbaum, K.; Griffith, W.P.; Whetten, R.L.; Landman, U. Ultrastable silver nanoparticles. Nature 2017, 501, 399–402. [Google Scholar] [CrossRef]
- Zhong, Y.; Ma, S.; Chen, K.; Wang, P.-F.; Qiu, Y.-H.; Liang, S.; Zhou, L.; Chen, Y.; Wang, Q.-Q. Controlled growth of plasmonic heterostructures and their applications. Sci. China Mater. 2020, 63, 1398–1417. [Google Scholar] [CrossRef]
- Feng, Q.L.; Wu, J.; Chen, G.Q.; Cui, F.Z.; Kim, T.N.; Kim, J.O. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mater. Res. 2000, 52, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Morones, J.R.; Elechiguerra, J.L.; Camacho, A.; Holt, K.; Kouri, J.B.; Ramírez, J.T.; Yacaman, M.J. The bactericidal effect of silver nanoparticles. Nanotechnology 2005, 16, 2346–2353. [Google Scholar] [CrossRef]
- Suryoprabowo, S.; Liu, L.; Kuang, H.; Cui, G.; Xu, C. Gold immunochromatographic assay for simultaneous detection of sibutramine and sildenafil in slimming tea and coffee. Sci. China Mater. 2020, 63, 654–659. [Google Scholar] [CrossRef]
- Qiao, Z.; Yao, Y.; Song, S.; Yin, M.; Luo, J. Silver nanoparticles with pH induced surface charge switchable properties for antibacterial and antibiofilm applications. J. Mater. Chem. B 2019, 7, 830–840. [Google Scholar] [CrossRef]
- Dong, C.; Lu, J.; Qiu, B.; Shen, B.; Xing, M.; Zhang, J. Developing stretchable and graphene-oxide-based hydrogel for the removal of organic pollutants and metal ions. Appl. Catal. B Environ. 2018, 222, 146–156. [Google Scholar] [CrossRef]
- Karaolia, P.; Michael-Kordatou, I.; Hapeshi, E.; Drosou, C.; Bertakis, Y.; Christofilos, D.; Armatas, G.S.; Sygellou, L.; Schwartz, T.; Xekoukoulotakis, N.P.; et al. Removal of antibiotics, antibiotic-resistant bacteria and their associated genes by graphene-based TiO2 composite photocatalysts under solar radiation in urban wastewaters. Appl. Catal. B Environ. 2018, 224, 810–824. [Google Scholar] [CrossRef]
- Wang, P.-T.; Song, Y.-H.; Fan, H.-C.; Yu, L. Bioreduction of azo dyes was enhanced by in-situ biogenic palladium nanoparticles. Bioresour. Technol. 2018, 266, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Ai, L.; Jiang, J. Catalytic reduction of 4-nitrophenol by silver nanoparticles stabilized on environmentally benign macroscopic biopolymer hydrogel. Bioresour. Technol. 2013, 132, 374–377. [Google Scholar] [CrossRef]
- Ingle, A.; Gade, A.; Pierrat, S.; Sonnichsen, C.; Rai, M. Mycosynthesis of silver nanoparticles using the fungus Fusarium acuminatum and its activity against some human pathogenic bacteria. Curr. Nanosci. 2008, 4, 141–144. [Google Scholar] [CrossRef]
- Zhang, J.L.; Han, B.X.; Chen, J.; Li, Z.H.; Liu, Z.; Wu, W. Synthesis of Ag/BSA composite nanospheres from water-in-oil microemulsion using compressed CO2 as antisolvent. Biotechnol. Bioeng. 2005, 89, 274–279. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Q.; Lu, Y.; Sun, D.; Lin, X.; Deng, X.; He, N.; Zheng, S. Biosorption and bioreduction of diamine silver complex by Corynebacterium. J. Chem. Technol. Biotechnol. 2005, 80, 285–290. [Google Scholar] [CrossRef]
- Verma, V.C.; Kharwar, R.N.; Gange, A.C. Biosynthesis of antimicrobial silver nanoparticles by the endophytic fungus Aspergillus clavatus. Nanomedicine 2010, 5, 33–40. [Google Scholar] [CrossRef]
- Rai, M.; Yadav, A.; Gade, A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 2009, 27, 76–83. [Google Scholar] [CrossRef]
- Pradhan, N.; Pal, A.; Pal, T. Silver nanoparticle catalyzed reduction of aromatic nitro compounds. Colloids Surf. Physicochem. Eng. 2002, 196, 247–257. [Google Scholar] [CrossRef]
- Liu, H.; Ma, H.; Joo, J.; Yin, Y. Contribution of multiple reflections to light utilization efficiency of submicron hollow TiO2 photocatalyst. Sci. China Mater. 2016, 59, 1017–1026. [Google Scholar] [CrossRef]
- Doi, S.; Clark, J.H.; Macquarrie, D.J.; Milkowski, K. New materials based on renewable resources: Chemically modified expanded corn starches as catalysts for liquid phase organic reactions. Chem. Commun. 2002, 34, 2632–2633. [Google Scholar] [CrossRef]
- Sun, Y.; Xia, Y. Shape-controlled synthesis of gold and silver nanoparticles. Science 2003, 34, 2176. [Google Scholar]
- Kvítek, L.; Panáček, A.; Soukupová, J.; Kolář, M.; Večeřová, R.; Prucek, R.; Mirka Holecová, A.; Zbořil, R. Effect of surfactants and polymers on stability and antibacterial activity of silver nanoparticles (NPs). J. Phys. Chem. C 2008, 112, 5825–5834. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, B.; Hu, L. PVP protective mechanism of ultrafine silver powder synthesized by chemical reduction processes. J. Solid State Chem. 1996, 121, 105–110. [Google Scholar] [CrossRef]
- Gole, A.; Dash, C.; Ramakrishnan, V.; Sainkar, S.R.; Mandale, A.B.; Rao, M.; Sastry, M. Pepsin−gold colloid conjugates: preparation, characterization, and enzymatic activity. Langmuir 2006, 17, 1674–1679. [Google Scholar] [CrossRef]
- Sanghi, R.; Verma, P. Biomimetic synthesis and characterisation of protein capped silver nanoparticles. Bioresour. Technol. 2009, 100, 501–504. [Google Scholar] [CrossRef]
- Koczkur, K.M.; Mourdikoudis, S.; Polavarapu, L.; Skrabalak, S.E. Polyvinylpyrrolidone (PVP) in nanoparticle synthesis. Dalton T. 2015, 44, 17883–17905. [Google Scholar] [CrossRef]
- Richter, A.P.; Brown, J.S.; Bhuvnesh, B.; Amy, W.; Sumit, G.; Keith, H.; Hubal, C.E.A.; Paunov, V.N.; Stoyanov, S.D.; Velev, O.D. An environmentally benign antimicrobial nanoparticle based on a silver-infused lignin core. Nat. Nanotechnol. 2015, 10, 817–823. [Google Scholar] [CrossRef]
- Edison, T.N.J.I.; Baral, E.R.; Lee, Y.R.; Kim, S.H. Biogenic synthesis of silver nanoparticles using Cnidium officinale extract and their catalytic reduction of 4-Nitroaniline. J. Cluster Sci. 2016, 27, 285–298. [Google Scholar] [CrossRef]
- Ingle, A.; Rai, M.; Gade, A.; Bawaskar, M. Fusarium solani: A novel biological agent for the extracellular synthesis of silver nanoparticles. J. Nanopart. Res. 2009, 11, 2079–2085. [Google Scholar] [CrossRef]
- Zeng, Y.-J.; Yang, H.-R.; Zong, M.-H.; Yang, J.-G.; Lou, W.-Y. Novel antibacterial polysaccharides produced by endophyte Fusarium solani DO7. Bioresour. Technol. 2019, 288, 121596. [Google Scholar] [CrossRef]
- Malhotra, A.; Dolma, K.; Kaur, N.; Rathore, Y.S.; Ashish, S.; Mayilraj, A.R. Choudhury, Biosynthesis of gold and silver nanoparticles using a novel marine strain of Stenotrophomonas. Bioresour. Technol. 2013, 142, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhang, Y.; Du, B.; Wei, D.; Wei, Q.; Zhao, Y. Enhancement effect of silver nanoparticles on fermentative biohydrogen production using mixed bacteria. Bioresour. Technol. 2013, 142, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kuk, E.; Yu, K.N.; Kim, J.-H.; Park, S.J.; Lee, H.J.; Kim, S.H.; Park, Y.K.; Park, Y.H.; Hwang, C.-Y. Antimicrobial effects of silver nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 95. [Google Scholar] [CrossRef] [PubMed]
- Dong, A.; Lan, S.; Huang, J.; Wang, T.; Zhao, T.; Xiao, L.; Wang, W.; Zheng, X.; Liu, F.; Gao, G. Modifying Fe3O4-functionalized nanoparticles with N-halamine and their magnetic/antibacterial properties. ACS Appl. Mater. Interfaces 2011, 3, 4228. [Google Scholar] [CrossRef]
- Yang, H.; Zong, X.; Xu, Y.; Zeng, Y.; Zhao, H. Improvement of multiple-stress tolerance and ethanol production in yeast during very-high-gravity fermentation by supplementation of wheat-gluten hydrolysates and their ultrafiltration fractions. J. Agric. Food. Chem. 2018, 66, 10233–10241. [Google Scholar] [CrossRef]


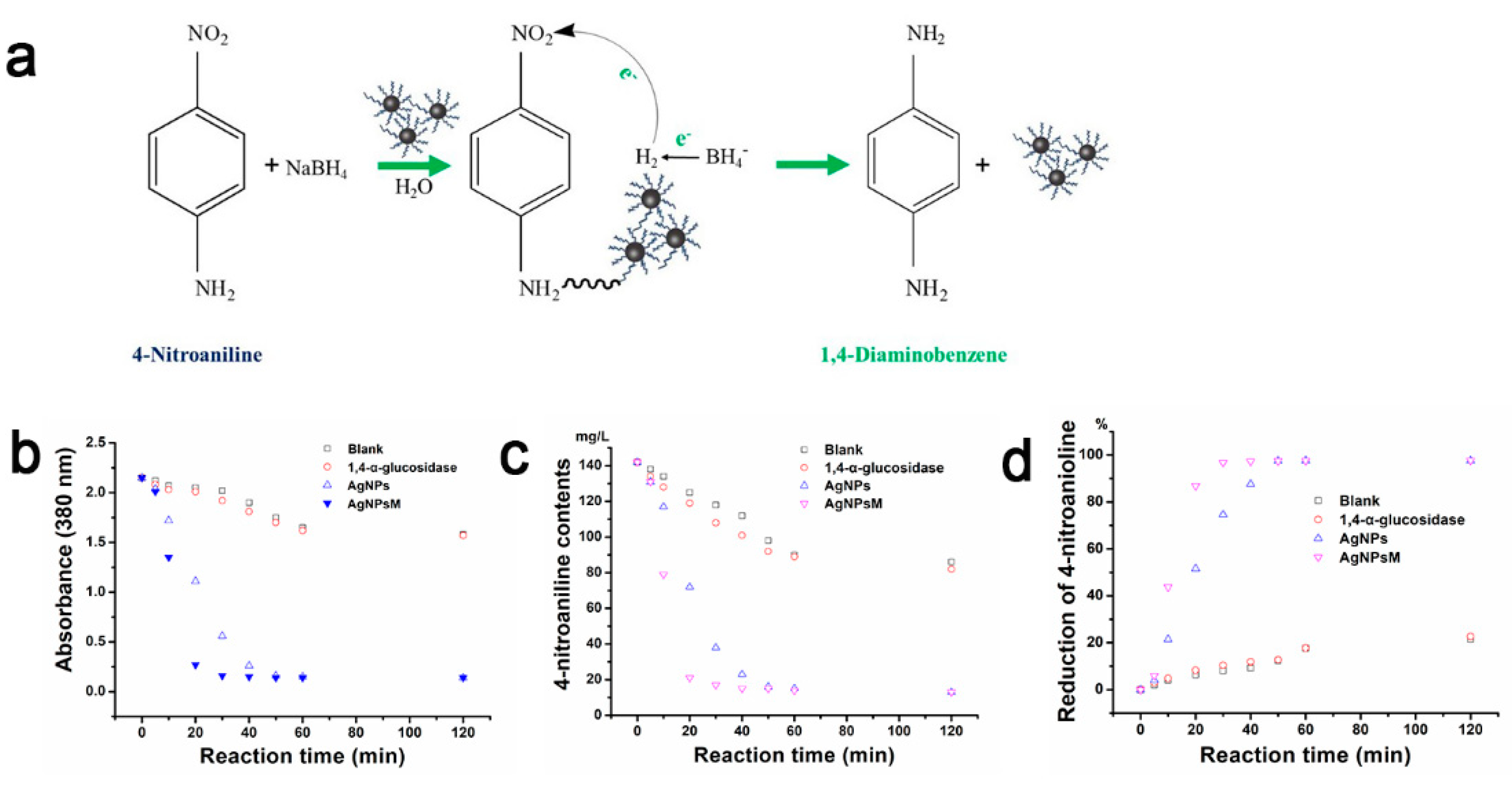
| Samples | Rate Constant (k) (10−3 min−1) | t1/2 (min) |
|---|---|---|
| Blank | 2.3 | 263.7 |
| 1,4-α-glucosidase | 2.8 | 227.8 |
| AgNPs | 12.4 | 56.3 |
| mAgNPs | 35.6 | 19.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, Y.-J.; Wu, X.-L.; Yang, H.-R.; Zong, M.-H.; Lou, W.-Y. 1,4-α-Glucosidase from Fusarium solani for Controllable Biosynthesis of Silver Nanoparticles and Their Multifunctional Applications. Int. J. Mol. Sci. 2023, 24, 5865. https://doi.org/10.3390/ijms24065865
Zeng Y-J, Wu X-L, Yang H-R, Zong M-H, Lou W-Y. 1,4-α-Glucosidase from Fusarium solani for Controllable Biosynthesis of Silver Nanoparticles and Their Multifunctional Applications. International Journal of Molecular Sciences. 2023; 24(6):5865. https://doi.org/10.3390/ijms24065865
Chicago/Turabian StyleZeng, Ying-Jie, Xiao-Ling Wu, Hui-Rong Yang, Min-Hua Zong, and Wen-Yong Lou. 2023. "1,4-α-Glucosidase from Fusarium solani for Controllable Biosynthesis of Silver Nanoparticles and Their Multifunctional Applications" International Journal of Molecular Sciences 24, no. 6: 5865. https://doi.org/10.3390/ijms24065865
APA StyleZeng, Y.-J., Wu, X.-L., Yang, H.-R., Zong, M.-H., & Lou, W.-Y. (2023). 1,4-α-Glucosidase from Fusarium solani for Controllable Biosynthesis of Silver Nanoparticles and Their Multifunctional Applications. International Journal of Molecular Sciences, 24(6), 5865. https://doi.org/10.3390/ijms24065865







