Effect of Ultraviolet Radiation and Benzo[a]pyrene Co-Exposure on Skin Biology: Autophagy as a Potential Target
Abstract
1. Introduction
2. Results
2.1. Identification of Dysregulated Proteins following Skin Exposure to UV Alone or UV + BaP
2.2. Identification of Biological Processes Dysregulated Following Skin Exposure to UV + BaP Compared to Exposition to UV Alone
2.3. Autophagy Is Downregulated after Co-Exposure of Skin Cells to UV + BaP
3. Discussion
4. Materials and Methods
4.1. Human Skin Explant Samples’ Exposition
4.2. Proteomic Samples’ Processing
4.3. LC-MS/MS Data Acquisition and Analysis
4.4. Bioinformatic Analysis
4.5. Immunohistochemistry Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wickett, R.R.; Visscher, M.O. Structure and function of the epidermal barrier. Am. J. Infect. Control 2006, 34, S98–S110. [Google Scholar] [CrossRef]
- Slominski, A.; Zmijewski, A.; Plonka, A.; Szaflarski, J.; Paus, R. How UV Light Touches the Brain and Endocrine System Through Skin, and Why. Endocr. J. 2018, 159, 1992–2007. [Google Scholar] [CrossRef]
- Slominski, A.; Slominski, R.; Raman, C.; Chen, J.; Athar, M.; Elmets, C. Neuroendocrine signaling in the skin with a special focus on the epidermal neuropeptides. Am. J. Physiol. Cell Physiol. 2022, 323, C1757–C1776. [Google Scholar] [CrossRef]
- Baudouin, C.; Charveron, M.; Tarroux, R.; Gall, Y. Environmental pollutants and skin cancer. Cell Biol. Toxicol. 2002, 18, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Drakaki, E.; Dessinioti, C.; Antoniou, C.V. Air pollution and the skin. Front. Environ. Sci. 2014, 2, 11. [Google Scholar] [CrossRef]
- Katsouyanni, K. Ambient air pollution and health. Br. Med. Bull. 2003, 68, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Dessinioti, C.; Antoniou, C.; Katsambas, A.; Stratigos, A.J. Basal Cell Carcinoma: What’s New Under the Sun. Photochem. Photobiol. 2010, 86, 481–491. [Google Scholar] [CrossRef]
- Fisher, G.J.; Kang, S.; Varani, J.; Bata-Csorgo, Z.; Wan, Y.; Datta, S.; Voorhees, J.J. Mechanisms of photoaging and chronological skin aging. Arch. Dermatol. 2002, 138, 1462–1470. [Google Scholar] [CrossRef]
- Valacchi, G.; Sticozzi, C.; Pecorelli, A.; Cervellati, F.; Cervellati, C.; Maioli, E. Cutaneous responses to environmental stressors. Ann. N. Y. Acad. Sci. 2021, 1271, 75–81. [Google Scholar] [CrossRef]
- Puri, P.; Nandar, S.; Kathuria, S.; Ramesh, V. Effects of air pollution on the skin: A review. Indian J. Dermatol. Venereol. Leprol. 2017, 83, 415. [Google Scholar] [CrossRef]
- Burke, K.; Wei, H. Synergistic damage by UVA radiation and pollutants. Toxicol. Ind. Health. 2009, 25, 219–224. [Google Scholar] [CrossRef]
- Siddique, R.; Zahoor, A.F.; Ahmad, H.; Zahid, F.M.; Karrar, E. Impact of different cooking methods on polycyclic aromatic hydrocarbons in rabbit meat. Food Sci. Nutr. 2021, 9, 3219–3227. [Google Scholar] [CrossRef] [PubMed]
- Migdał, W.; Walczycka, M.; Migdał, L. The Levels of Polycyclic Aromatic Hydrocarbons in Traditionally Smoked Cheeses in Poland. Polycycl. Aromat. Compd. 2022, 42, 1391–1403. [Google Scholar] [CrossRef]
- Ekström, G.; von Bahr, C.; Glaumann, H.; Ingelman-Sundberg, M. Interindividual variation in benzo(a)pyrene metabolism and composition of isoenzymes of cytochrome P-450 as revealed by SDS-gel electrophoresis of human liver microsomal fractions. Acta Pharmacol. Toxicol. Copenh 1982, 50, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Perera, F. Carcinogenicity of airborne fine particulate benzo(a)pyrene: An appraisal of the evidence and the need for control. Environ. Health Perspect. 1981, 42, 163. [Google Scholar] [CrossRef]
- Amadou, A.; Praud, D.; Coudon, T.; Deygas, F.; Grassot, L.; Faure, E.; Couvidat, F.; Caudeville, J.; Bessagnet, B.; Salizzoni, P.; et al. Risk of breast cancer associated with long-term exposure to benzo[a]pyrene (BaP) air pollution: Evidence from the French E3N cohort study. Environ. Int. 2021, 149, 106399. [Google Scholar] [CrossRef]
- Sample, A.; He, Y.Y. Autophagy in UV Damage Response. Photochem. Photobiol. 2017, 93, 943–955. [Google Scholar] [CrossRef]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Runwal, G.; Stamatakou, E.; Siddiqi, F.H.; Puri, C.; Zhu, Y.; Rubinsztein, D.C. LC3-positive structures are prominent in autophagy-deficient cells. Sci. Rep. 2019, 9, 10147. [Google Scholar] [CrossRef]
- Birch-Machin, M.A.; Bowman, A. Oxidative stress and ageing. Br. J. Dermatol. 2016, 175, 26–29. [Google Scholar] [CrossRef]
- Krutmann, J.; Bouloc, A.; Sore, G.; Bernard, B.A.; Passeron, T. The skin aging exposome. J. Dermatol. Sci. 2017, 85, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Matsui, M.S.; Wang, N.; Deleo, V.A. Ultraviolet radiation B induces differentiation and protein kinase C in normal human epidermal keratinocytes. Photodermatol. Photoimmunol. Photomed. 1996, 12, 103–108. [Google Scholar] [CrossRef]
- Syed, D.N.; Afaq, F.; Mukhtar, H. Differential Activation of Signaling Pathways by UVA and UVB Radiation in Normal Human Epidermal Keratinocytes. Photochem. Photobiol. 2012, 88, 1184–1190. [Google Scholar] [CrossRef]
- He, Y.Y.; Huang, J.L.; Sik, R.H.; Liu, J.; Waalkes, M.P.; Chignell, C.F. Expression Profiling of Human Keratinocyte Response to Ultraviolet A: Implications in Apoptosis. J. Investig. Dermatol. 2004, 122, 533–543. [Google Scholar] [CrossRef]
- Perez, D.S.; Handa, R.J.; Yang, R.S.H.; Campain, J.A. Gene expression changes associated with altered growth and differentiation in benzo[a]pyrene or arsenic exposed normal human epidermal keratinocytes. J. Appl. Toxicol. 2008, 28, 491–508. [Google Scholar] [CrossRef] [PubMed]
- Perez, D.S.; Armstrong-Lea, L.; Fox, M.H.; Yang, R.S.H.; Campain, J.A. Arsenic and Benzo[a]pyrene Differentially Alter the Capacity for Differentiation and Growth Properties of Primary Human Epidermal Keratinocytes. Toxicol. Sci. 2003, 76, 280–290. [Google Scholar] [CrossRef]
- Zorina, A.; Zorin, V.; Kudlay, D.; Kopnin, P. Molecular Mechanisms of Changes in Homeostasis of the Dermal Extracellular Matrix: Both Involutional and Mediated by Ultraviolet Radiation. Int. J. Mol. Sci. 2022, 23, 6655. [Google Scholar] [CrossRef] [PubMed]
- Schalka, S.; Silva, M.S.; Lopes, L.F.; de Freitas, L.M.; Baptista, M.S. The skin redoxome. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Wagener, F.A.D.T.G.; Carels, C.E.; Lundvig, D.M.S. Targeting the Redox Balance in Inflammatory Skin Conditions. Int. J. Mol. Sci. 2013, 14, 9126. [Google Scholar] [CrossRef] [PubMed]
- Fisher, G.J.; Choi, H.C.; Bata-Csorgo, Z.; Shao, Y.; Datta, S.; Wang, Z.Q.; Kang, S.; Voorhees, J.J. Ultraviolet Irradiation Increases Matrix Metalloproteinase-8 Protein in Human Skin In Vivo. J. Investig. Dermatol. 2001, 117, 219–226. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, C.F.; Rossiter, H.; Eckhart, L.; König, U.; Karner, S.; Mildner, M.; Bochkov, V.N.; Tschachler, E.; Gruber, F. Autophagy Is Induced by UVA and Promotes Removal of Oxidized Phospholipids and Protein Aggregates in Epidermal Keratinocytes. J. Investig. Dermatol. 2013, 133, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Charareh, P.; Lei, X.; Zhong, J.L. Autophagy: Multiple Mechanisms to Protect Skin from Ultraviolet Radiation-Driven Photoaging. Oxid. Med. Cell. Longev. 2019, 2019, 8135985. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.; Qomaladewi, N.P.; Lee, J.; Park, S.H.; Cho, J.Y. The Role of Autophagy in Skin Fibroblasts, Keratinocytes, Melanocytes, and Epidermal Stem Cells. J. Investig. Dermatol. 2020, 140, 1691–1697. [Google Scholar] [CrossRef] [PubMed]
- Vogeley, C.; Esser, C.; Tüting, T.; Krutmann, J.; Haarmann-Stemmann, T. Role of the Aryl Hydrocarbon Receptor in Environmentally Induced Skin Aging and Skin Carcinogenesis. Int. J. Mol. Sci. 2019, 20, 6005. [Google Scholar] [CrossRef]
- Burke, K.E. Mechanisms of aging and development—A new understanding of environmental damage to the skin and prevention with topical antioxidants. Mech. Ageing Dev. 2018, 172, 123–130. [Google Scholar] [CrossRef]
- Umar, S.A.; Shahid, N.H.; Nazir, L.A.; Tanveer, M.A.; Divya, G.; Archoo, S.; Raghu, S.R.; Tasduq, S.A. Pharmacological Activation of Autophagy Restores Cellular Homeostasis in Ultraviolet-(B)-Induced Skin Photodamage. Front. Oncol. 2021, 11, 726066. [Google Scholar] [CrossRef]
- Mildner, M.; Weninger, W.; Trautinger, F.; Ban, J.; Tschachler, E. UVA and UVB radiation differentially regulate vascular endothelial growth factor expression in keratinocyte-derived cell lines and in human keratinocytes. J. Photochem. Photobiol. B Biol. 1999, 70, 674–679. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Huang, D.; Sherman, B.; Lempicki, R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
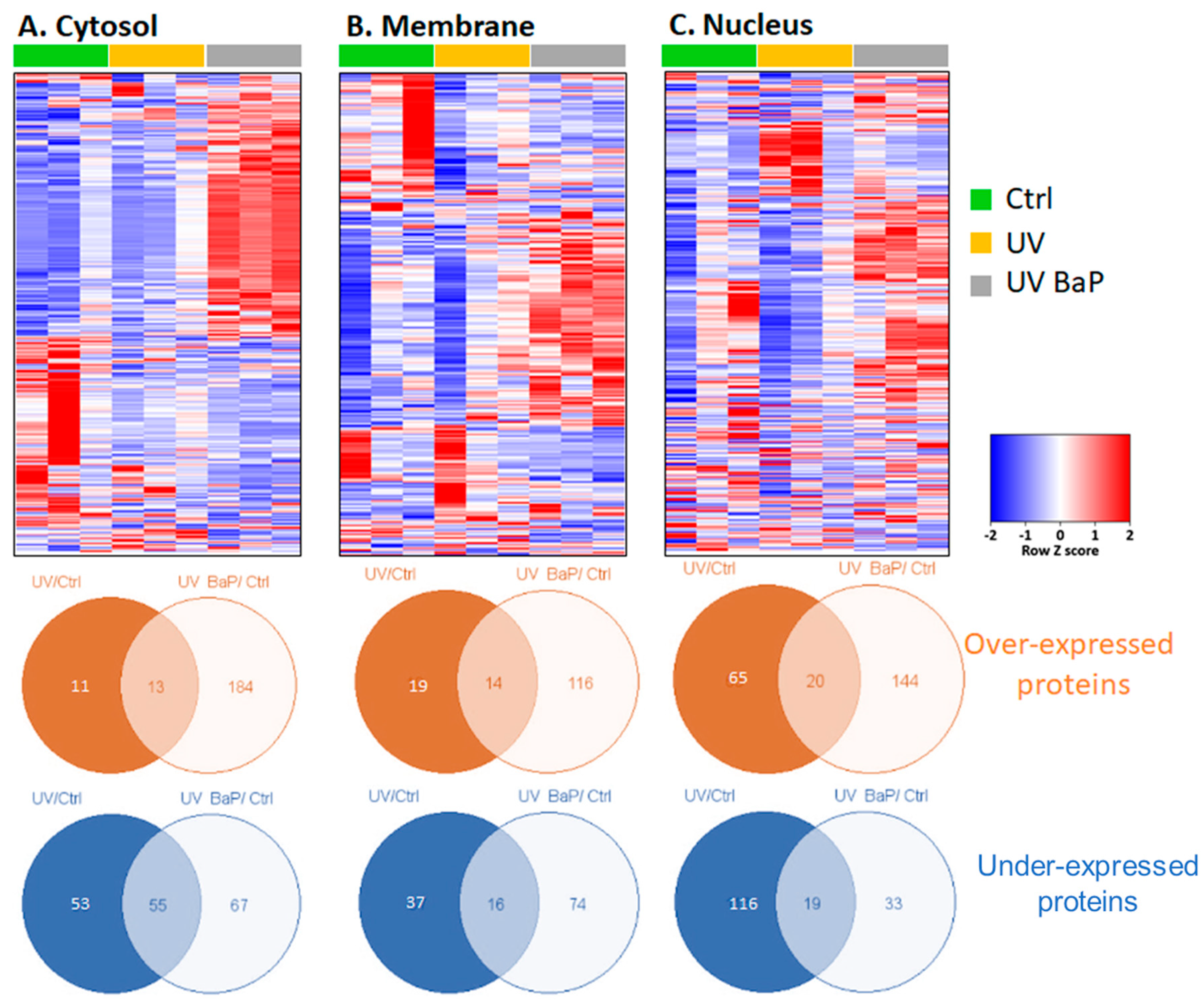
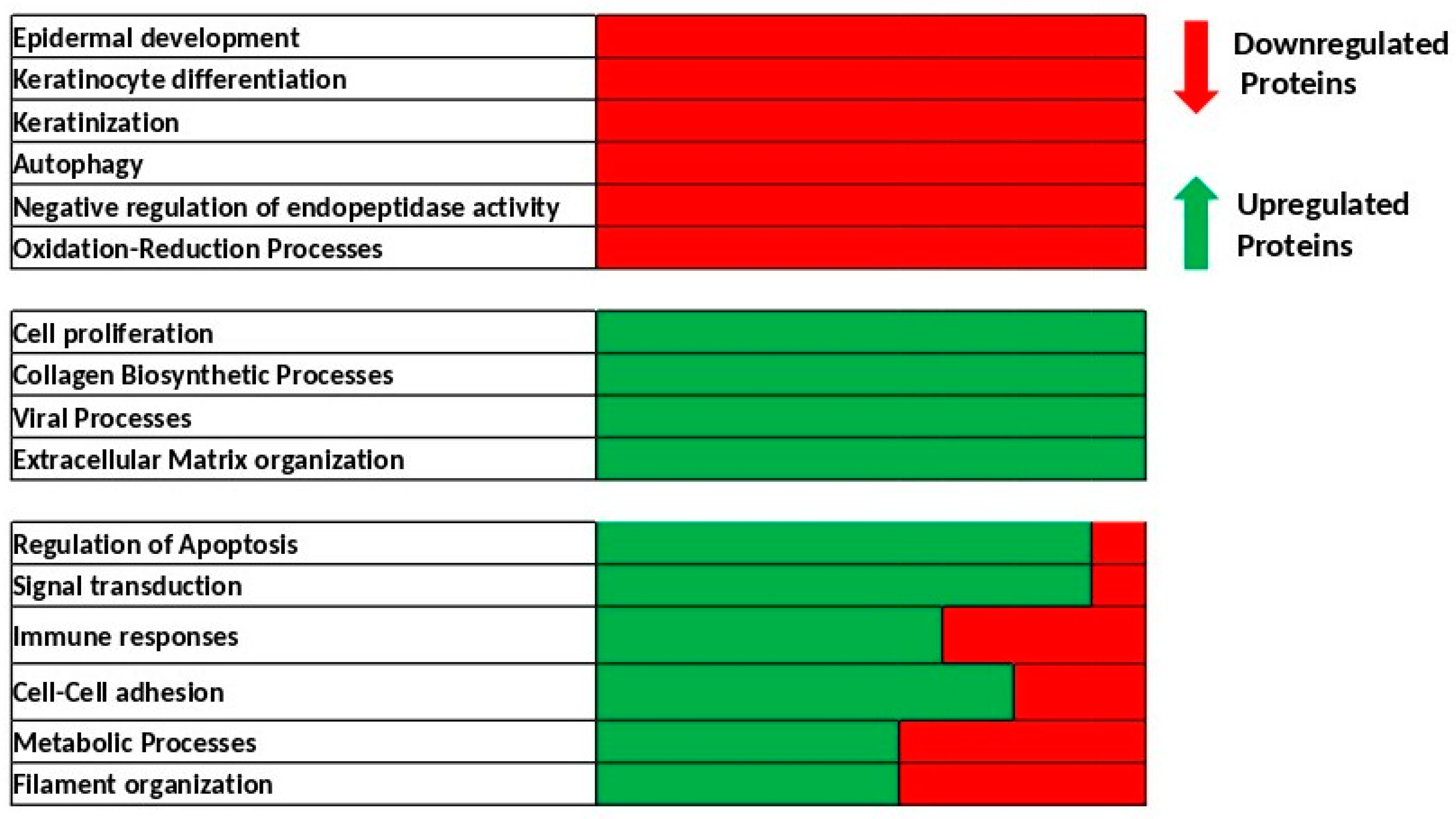

| Terms | Protein Count | p-Value |
|---|---|---|
| Biological processes of cytosolic upregulated proteins specific to UV + BaP condition | ||
| SRP-dependent cotranslational protein targeting to membrane | 66 | 5.80 × 10−111 |
| viral transcription | 66 | 3.20 × 10−103 |
| nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 66 | 9.50 × 10−101 |
| translational initiation | 67 | 2.10 × 10−97 |
| rRNA processing | 68 | 2.70 × 10−83 |
| translation | 69 | 2.10 × 10−79 |
| cytoplasmic translation | 14 | 5.90 × 10−20 |
| ribosomal large subunit assembly | 8 | 1.30 × 10−09 |
| ribosomal small subunit biogenesis | 7 | 9.20 × 10−09 |
| ribosomal small subunit assembly | 7 | 3.10 × 10−08 |
| Biological processes of cytosolic downregulated proteins specific to UV + BaP condition | ||
| negative regulation of endopeptidase activity | 7 | 7.20 × 10−06 |
| epidermis development | 6 | 1.90 × 10−05 |
| negative regulation of peptidase activity | 4 | 3.60 × 10−05 |
| keratinocyte differentiation | 5 | 2.10 × 10−04 |
| negative regulation of cytokine-mediated signaling pathway | 3 | 5.20 × 10−04 |
| keratinization | 4 | 8.50 × 10−04 |
| peptide cross-linking | 4 | 9.50 × 10−04 |
| innate immune response | 8 | 1.30 × 10−03 |
| negative regulation of catalytic activity | 4 | 3.10 × 10−03 |
| cell–cell adhesion | 6 | 4.00 × 10−03 |
| Biological processes of cell membrane upregulated proteins specific to UV + BaP condition | ||
| viral entry into host cell | 6 | 1.30 × 10−04 |
| protein transport | 9 | 3.00 × 10−03 |
| substantia nigra development | 4 | 3.40 × 10−03 |
| small GTPase-mediated signal transduction | 7 | 4.10 × 10−03 |
| leukocyte migration | 5 | 6.70 × 10−03 |
| extracellular matrix organization | 6 | 7.20 × 10−03 |
| vesicle-mediated transport | 5 | 1.40 × 10−02 |
| antigen processing and presentation of peptide antigen via MHC class I | 3 | 1.40 × 10−02 |
| membrane organization | 3 | 1.60 × 10−02 |
| regulation of oxidative-stress-induced intrinsic apoptotic signaling pathway | 2 | 1.80 × 10−02 |
| Biological processes of cell membrane downregulated proteins specific to UV + BaP condition | ||
| tricarboxylic acid cycle | 5 | 6.80 × 10−06 |
| keratinocyte differentiation | 5 | 3.20 × 10−04 |
| keratinization | 4 | 1.10 × 10−03 |
| peptide cross-linking | 4 | 1.30 × 10−03 |
| 2-oxoglutarate metabolic process | 3 | 2.70 × 10−03 |
| epidermis development | 4 | 5.80 × 10−03 |
| neutrophil aggregation | 2 | 8.60 × 10−03 |
| chemokine production | 2 | 1.30 × 10−02 |
| sequestering of zinc ion | 2 | 1.70 × 10−02 |
| ketone body catabolic process | 2 | 1.70 × 10−02 |
| Biological processes of nucleus upregulated proteins specific to UV + BaP condition | ||
| positive regulation of telomerase RNA localization to Cajal body | 8 | 1.60 × 10−11 |
| positive regulation of establishment of protein localization to telomere | 7 | 2.70 × 10−11 |
| positive regulation of protein localization to Cajal body | 6 | 2.20 × 10−09 |
| positive regulation of telomere maintenance via telomerase | 7 | 2.50 × 10−07 |
| response to drug | 12 | 5.60 × 10−05 |
| cell–cell adhesion | 11 | 1.00 × 10−04 |
| protein folding | 9 | 1.50 × 10−04 |
| protein stabilization | 8 | 1.60 × 10−04 |
| binding of sperm to zona pellucida | 5 | 2.10 × 10−04 |
| toxin transport | 5 | 2.30 × 10−04 |
| Biological processes of nucleus downregulated proteins specific to UV + BaP condition | ||
| autophagy | 5 | 6.60 × 10−05 |
| activation of cysteine-type endopeptidase activity involved in apoptotic process | 4 | 3.50 × 10−04 |
| neutrophil aggregation | 2 | 3.30 × 10−03 |
| chemokine production | 2 | 5.00 × 10−03 |
| sequestering of zinc ion | 2 | 6.70 × 10−03 |
| positive regulation of peptide secretion | 2 | 6.70 × 10−03 |
| leukocyte migration involved in inflammatory response | 2 | 1.80 × 10−02 |
| positive regulation of NF-kappaB transcription factor activity | 3 | 2.10 × 10−02 |
| astrocyte development | 2 | 2.60 × 10−02 |
| regulation of cytoskeleton organization | 2 | 3.10 × 10−02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fayyad-Kazan, M.; Kobaisi, F.; Nasrallah, A.; Matarrese, P.; Fitoussi, R.; Bourgoin-Voillard, S.; Seve, M.; Rachidi, W. Effect of Ultraviolet Radiation and Benzo[a]pyrene Co-Exposure on Skin Biology: Autophagy as a Potential Target. Int. J. Mol. Sci. 2023, 24, 5863. https://doi.org/10.3390/ijms24065863
Fayyad-Kazan M, Kobaisi F, Nasrallah A, Matarrese P, Fitoussi R, Bourgoin-Voillard S, Seve M, Rachidi W. Effect of Ultraviolet Radiation and Benzo[a]pyrene Co-Exposure on Skin Biology: Autophagy as a Potential Target. International Journal of Molecular Sciences. 2023; 24(6):5863. https://doi.org/10.3390/ijms24065863
Chicago/Turabian StyleFayyad-Kazan, Mohammad, Farah Kobaisi, Ali Nasrallah, Patrick Matarrese, Richard Fitoussi, Sandrine Bourgoin-Voillard, Michel Seve, and Walid Rachidi. 2023. "Effect of Ultraviolet Radiation and Benzo[a]pyrene Co-Exposure on Skin Biology: Autophagy as a Potential Target" International Journal of Molecular Sciences 24, no. 6: 5863. https://doi.org/10.3390/ijms24065863
APA StyleFayyad-Kazan, M., Kobaisi, F., Nasrallah, A., Matarrese, P., Fitoussi, R., Bourgoin-Voillard, S., Seve, M., & Rachidi, W. (2023). Effect of Ultraviolet Radiation and Benzo[a]pyrene Co-Exposure on Skin Biology: Autophagy as a Potential Target. International Journal of Molecular Sciences, 24(6), 5863. https://doi.org/10.3390/ijms24065863










