Investigating Circular RNAs Using qRT-PCR; Roundup of Optimization and Processing Steps
Abstract
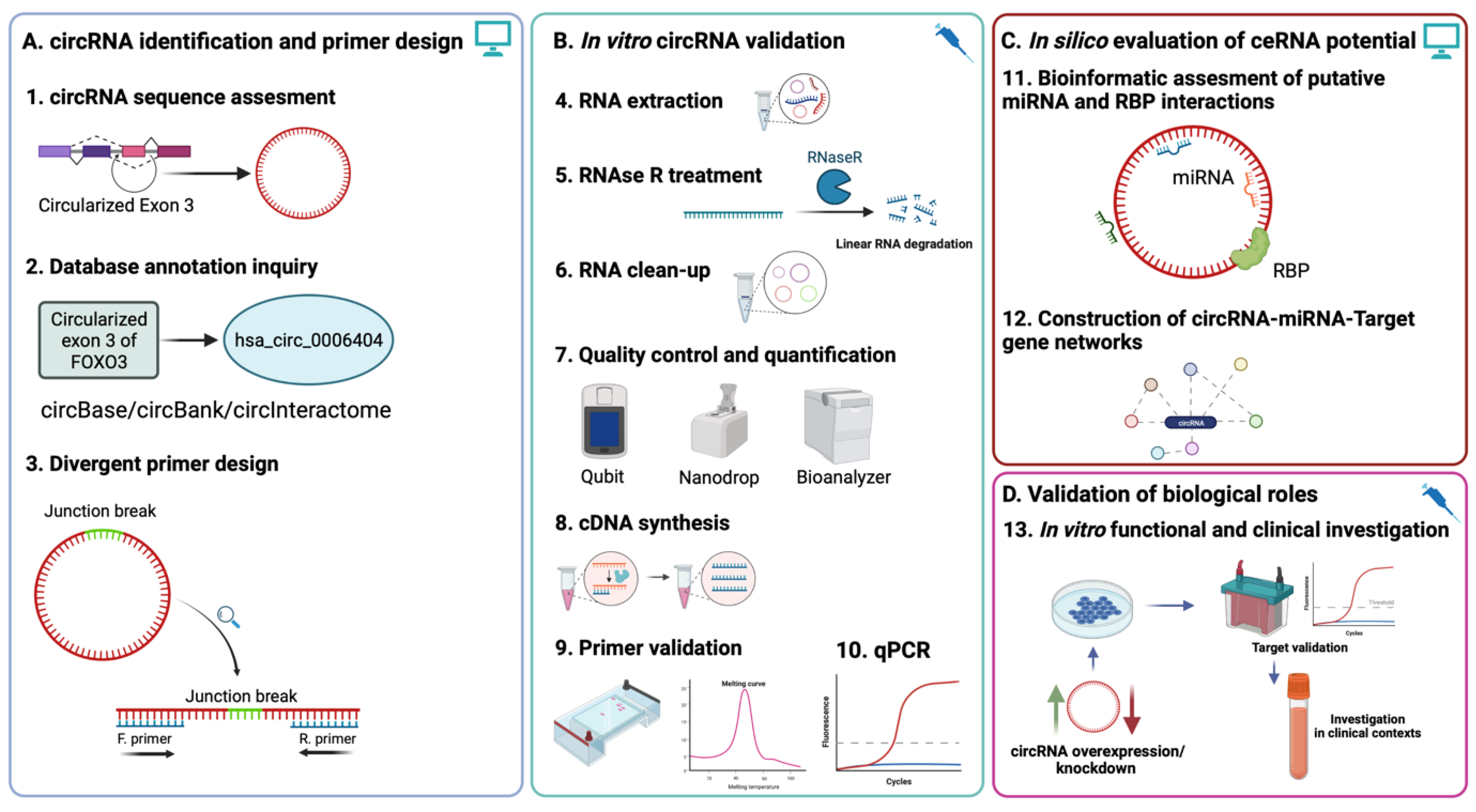
1. Introduction
2. Results
2.1. Database Annotation, Composition, and Sequence Characterization of circRNAs
2.2. Junction Sequence Mapping and Primer Design
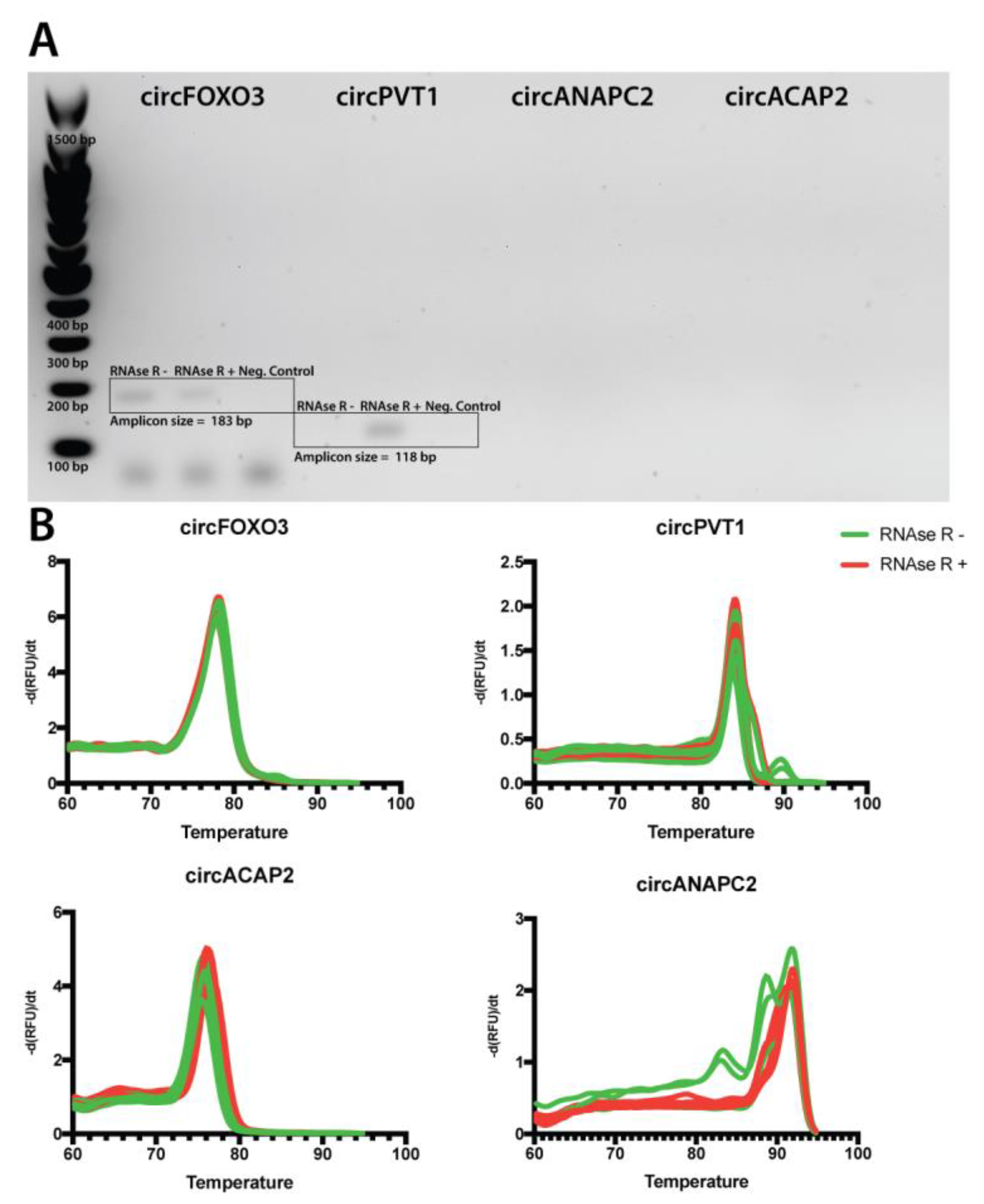
2.3. Primer Validation
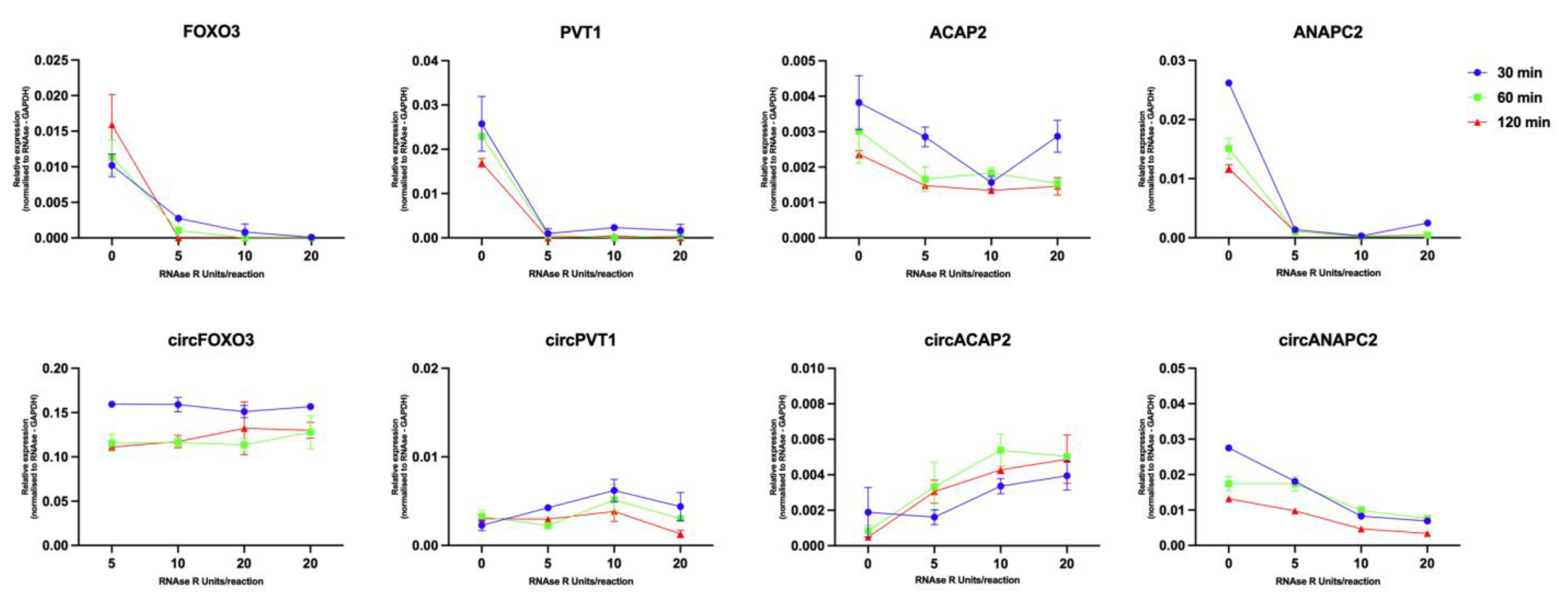
2.4. RNAse R Treatment Optimization and RNA Quality Control
2.5. Assessing Enrichment of circRNA following RNAse R Treatment
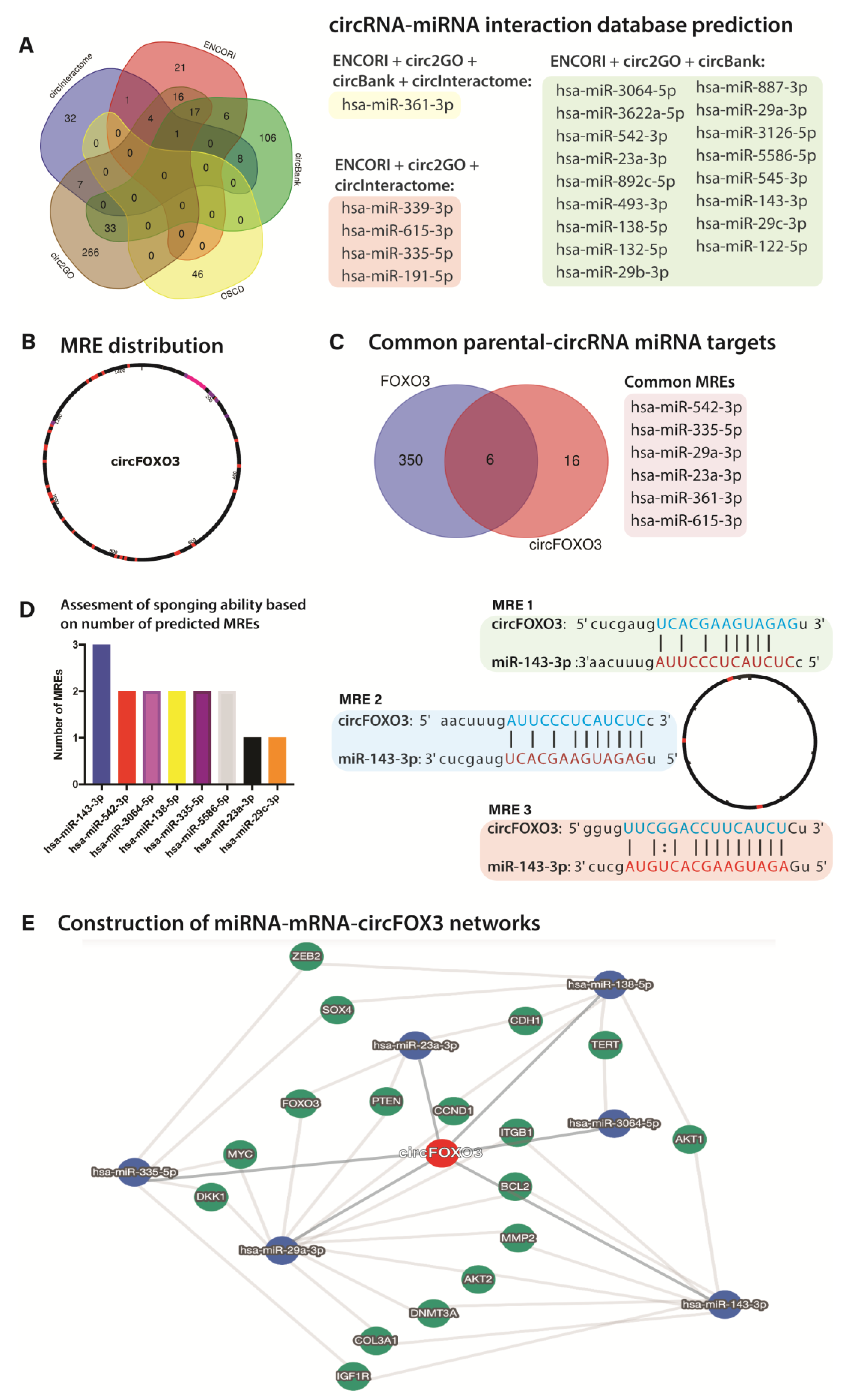
2.6. Database Prediction of Interacting miRNAs and Construction of circRNA-miRNA-mRNA Networks
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. RNA Extraction
4.3. RNAse R Treatment
4.4. RNA Clean-Up and Quantification
4.5. cDNA Synthesis
4.6. Primer Validation PCR and Agarose Gel Electrophoresis
4.7. qRT-PCR
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schneider, T.; Bindereif, A. Circular RNAs: Coding or Noncoding? Cell Res. 2017, 27, 724–725. [Google Scholar] [CrossRef]
- Kong, S.; Tao, M.; Shen, X.; Ju, S. Translatable CircRNAs and LncRNAs: Driving Mechanisms and Functions of Their Translation Products. Cancer Lett. 2020, 483, 59–65. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. CircRNA Biogenesis Competes with Pre-MRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids Are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-like Structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-Type Specific Features of Circular RNA Expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, Present, and Future of CircRNAs. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-O.; Dong, R.; Zhang, Y.; Zhang, J.-L.; Luo, Z.; Zhang, J.; Chen, L.-L.; Yang, L. Diverse Alternative Back-Splicing and Alternative Splicing Landscape of Circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Zhang, X.-O.; Wang, H.-B.; Zhang, Y.; Lu, X.; Chen, L.-L.; Yang, L. Complementary Sequence-Mediated Exon Circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-Intron Circular RNAs Regulate Transcription in the Nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular Intronic Long Noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of Intron Sequences Reveals Hallmarks of Circular RNA Biogenesis in Animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.-H.; Bindereif, A. Exon Circularization Requires Canonical Splice Signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L. The Expanding Regulatory Mechanisms and Cellular Functions of Circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Lu, T.; Cui, L.; Zhou, Y.; Zhu, C.; Fan, D.; Gong, H.; Zhao, Q.; Zhou, C.; Zhao, Y.; Lu, D.; et al. Transcriptome-Wide Investigation of Circular RNAs in Rice. RNA 2015, 21, 2076–2087. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, F. Computational Strategies for Exploring Circular RNAs. Trends Genet. 2018, 34, 389–400. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, S.; Yang, J.; Zhao, F. Accurate Quantification of Circular RNAs Identifies Extensive Circular Isoform Switching Events. Nat. Commun. 2020, 11, 90. [Google Scholar] [CrossRef]
- Vromman, M.; Vandesompele, J.; Volders, P.-J. Closing the Circle: Current State and Perspectives of Circular RNA Databases. Brief. Bioinform. 2021, 22, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, C.; Sun, H.; Wang, J.; Liang, Y.; Wang, Y.; Wong, G. The Bioinformatics Toolbox for CircRNA Discovery and Analysis. Brief. Bioinform. 2021, 22, 1706–1728. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Chen, M. Circular RNA Databases. Methods Mol. Biol. 2021, 2362, 109–118. [Google Scholar] [CrossRef]
- Hu, D.; Zhang, P.; Chen, M. Database Resources for Functional Circular RNAs. Methods Mol. Biol. 2021, 2284, 457–466. [Google Scholar] [CrossRef]
- Rahimi, K.; Venø, M.T.; Dupont, D.M.; Kjems, J. Nanopore Sequencing of Brain-Derived Full-Length CircRNAs Reveals CircRNA-Specific Exon Usage, Intron Retention and Microexons. Nat. Commun. 2021, 12, 4825. [Google Scholar] [CrossRef]
- Zhou, R.-M.; Shi, L.-J.; Shan, K.; Sun, Y.-N.; Wang, S.-S.; Zhang, S.-J.; Li, X.-M.; Jiang, Q.; Yan, B.; Zhao, C. Circular RNA-ZBTB44 Regulates the Development of Choroidal Neovascularization. Theranostics 2020, 10, 3293–3307. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shan, G. CircRNA in Cancer: Fundamental Mechanism and Clinical Potential. Cancer Lett. 2021, 505, 49–57. [Google Scholar] [CrossRef]
- Drula, R.; Braicu, C.; Harangus, A.; Nabavi, S.M.; Trif, M.; Slaby, O.; Ionescu, C.; Irimie, A.; Berindan-Neagoe, I. Critical Function of Circular RNAs in Lung Cancer. Wiley Interdiscip. Rev. RNA 2020, 11, e1592. [Google Scholar] [CrossRef]
- Pandey, P.R.; Munk, R.; Kundu, G.; De, S.; Abdelmohsen, K.; Gorospe, M. Methods for Analysis of Circular RNAs. Wiley Interdiscip. Rev. RNA 2020, 11, e1566. [Google Scholar] [CrossRef]
- Drula, R.; Pirlog, R.; Trif, M.; Slaby, O.; Braicu, C.; Berindan-Neagoe, I. CircFOXO3: Going around the Mechanistic Networks in Cancer by Interfering with MiRNAs Regulatory Networks. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2021, 1867, 166045. [Google Scholar] [CrossRef]
- Xiang, T.; Jiang, H.-S.; Zhang, B.-T.; Liu, G. CircFOXO3 Functions as a Molecular Sponge for MiR-143-3p to Promote the Progression of Gastric Carcinoma via Upregulating USP44. Gene 2020, 753, 144798. [Google Scholar] [CrossRef]
- Kong, Z.; Wan, X.; Lu, Y.; Zhang, Y.; Huang, Y.; Xu, Y.; Liu, Y.; Zhao, P.; Xiang, X.; Li, L.; et al. Circular RNA CircFOXO3 Promotes Prostate Cancer Progression through Sponging MiR-29a-3p. J. Cell. Mol. Med. 2020, 24, 799–813. [Google Scholar] [CrossRef]
- Glažar, P.; Papavasileiou, P.; Rajewsky, N. CircBase: A Database for Circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Q.; Shen, J.; Yang, B.B.; Ding, X. Circbank: A Comprehensive Database for CircRNA with Standard Nomenclature. RNA Biol. 2019, 16, 899–905. [Google Scholar] [CrossRef]
- Liu, X.; Du, Z.; Yi, X.; Sheng, T.; Yuan, J.; Jia, J. Circular RNA CircANAPC2 Mediates the Impairment of Endochondral Ossification by MiR-874-3p/SMAD3 Signalling Pathway in Idiopathic Short Stature. J. Cell. Mol. Med. 2021, 25, 3408–3426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, Z.; Zhong, J.; Lin, L. Circ-ACAP2 Facilitates the Progression of Colorectal Cancer through Mediating MiR-143-3p/FZD4 Axis. Eur. J. Clin. Investig. 2021, 51, e13607. [Google Scholar] [CrossRef]
- Zhu, J.; Xiang, X.-L.; Cai, P.; Jiang, Y.-L.; Zhu, Z.-W.; Hu, F.-L.; Wang, J. CircRNA-ACAP2 Contributes to the Invasion, Migration, and Anti-Apoptosis of Neuroblastoma Cells through Targeting the MiRNA-143-3p-Hexokinase 2 Axis. Transl. Pediatr. 2021, 10, 3237–3247. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, L.; Yang, Z.; Wen, D.; Hu, Z. Circular RNA CircACAP2 Suppresses Ferroptosis of Cervical Cancer during Malignant Progression by MiR-193a-5p/GPX4. J. Oncol. 2022, 2022, 5228874. [Google Scholar] [CrossRef]
- Zhao, B.; Song, X.; Guan, H. CircACAP2 Promotes Breast Cancer Proliferation and Metastasis by Targeting MiR-29a/b-3p-COL5A1 Axis. Life Sci. 2020, 244, 117179. [Google Scholar] [CrossRef]
- Zuker, M. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.-S.; Wilusz, J.E. An Improved Method for Circular RNA Purification Using RNAse R That Efficiently Removes Linear RNAs Containing G-Quadruplexes or Structured 3′ Ends. Nucleic Acids Res. 2019, 47, 8755–8769. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.C.; Gorospe, M. Detection and Analysis of Circular RNAs by RT-PCR. Bio-Protoc. J. 2018, 8, e2775. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The Ultimate QPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. StarBase v2.0: Decoding MiRNA-CeRNA, MiRNA-NcRNA and Protein-RNA Interaction Networks from Large-Scale CLIP-Seq Data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- CSCD: A Database for Cancer-Specific Circular RNAs—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/29036403/ (accessed on 23 August 2022).
- Lyu, Y.; Caudron-Herger, M.; Diederichs, S. Circ2GO: A Database Linking Circular RNAs to Gene Function. Cancers 2020, 12, E2975. [Google Scholar] [CrossRef]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A Web Tool for Exploring Circular RNAs and Their Interacting Proteins and MicroRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Targeting Circular RNAs as a Therapeutic Approach: Current Strategies and Challenges|Signal Transduction and Targeted Therapy. Available online: https://www.nature.com/articles/s41392-021-00569-5 (accessed on 4 September 2022).
- Danac, J.M.C.; Garcia, R.L. CircPVT1 Attenuates Negative Regulation of NRAS by Let-7 and Drives Cancer Cells towards Oncogenicity. Sci. Rep. 2021, 11, 9021. [Google Scholar] [CrossRef]
- Shi, J.; Lv, X.; Zeng, L.; Li, W.; Zhong, Y.; Yuan, J.; Deng, S.; Liu, B.; Yuan, B.; Chen, Y.; et al. CircPVT1 Promotes Proliferation of Lung Squamous Cell Carcinoma by Binding to MiR-30d/e. J. Exp. Clin. Cancer Res. 2021, 40, 193. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Khoshbakht, T.; Taheri, M.; Jamali, E. A Concise Review on the Role of CircPVT1 in Tumorigenesis, Drug Sensitivity, and Cancer Prognosis. Front. Oncol. 2021, 11, 762960. [Google Scholar] [CrossRef]
- Vromman, M.; Yigit, N.; Verniers, K.; Lefever, S.; Vandesompele, J.; Volders, P.-J. Validation of Circular RNAs Using RT-QPCR After Effective Removal of Linear RNAs by Ribonuclease R. Curr. Protoc. 2021, 1, e181. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.R.; Rout, P.K.; Das, A.; Gorospe, M.; Panda, A.C. RPAD (RNAse R Treatment, Polyadenylation, and Poly(A)+ RNA Depletion) Method to Isolate Highly Pure Circular RNA. Methods 2019, 155, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, B.; Huang, S. Identification of CircRNAs for MiRNA Targets by Argonaute2 RNA Immunoprecipitation and Luciferase Screening Assays. Methods Mol. Biol. 2018, 1724, 209–218. [Google Scholar] [CrossRef]
- Das, A.; Das, D.; Panda, A.C. Validation of Circular RNAs by PCR. Methods Mol. Biol. 2022, 2392, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Schreiner, S.; Preußer, C.; Bindereif, A.; Rossbach, O. Northern Blot Analysis of Circular RNAs. Methods Mol. Biol. 2018, 1724, 119–133. [Google Scholar] [CrossRef] [PubMed]
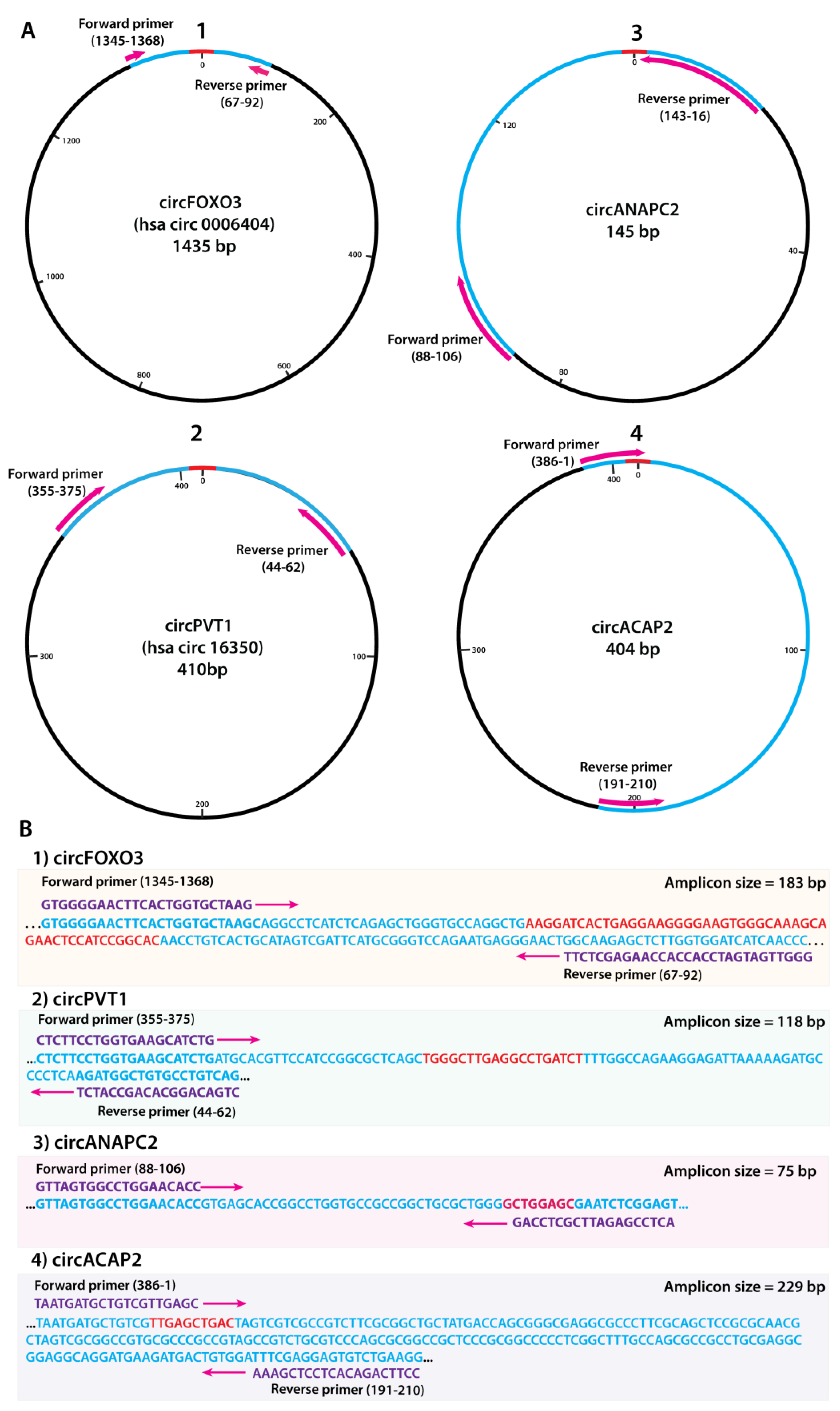

| CircRNA | Parental Gene | CircBase ID | Structure | Genomic Location | Length |
|---|---|---|---|---|---|
| circFOXO3 | FOXO3 | hsa_circ_0006404 | Exon 2 | chr6:108984657-108986092 | 1435 bp |
| circPVT1 | PVT1 lncRNA | hsa_circ_0009143 | Exon 2 | chr8:128867400-128903244 | 410 bp |
| circANAPC2 | ANAPC2 | Unannotated variant | Exon 1 | chr9:137188416-137188560 | 145 bp |
| circACAP2 | ACAP2 | Unannotated variant | Exon 3-1 | Chr3:195442795-195381903 | 404 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drula, R.; Braicu, C.; Chira, S.; Berindan-Neagoe, I. Investigating Circular RNAs Using qRT-PCR; Roundup of Optimization and Processing Steps. Int. J. Mol. Sci. 2023, 24, 5721. https://doi.org/10.3390/ijms24065721
Drula R, Braicu C, Chira S, Berindan-Neagoe I. Investigating Circular RNAs Using qRT-PCR; Roundup of Optimization and Processing Steps. International Journal of Molecular Sciences. 2023; 24(6):5721. https://doi.org/10.3390/ijms24065721
Chicago/Turabian StyleDrula, Rares, Cornelia Braicu, Sergiu Chira, and Ioana Berindan-Neagoe. 2023. "Investigating Circular RNAs Using qRT-PCR; Roundup of Optimization and Processing Steps" International Journal of Molecular Sciences 24, no. 6: 5721. https://doi.org/10.3390/ijms24065721
APA StyleDrula, R., Braicu, C., Chira, S., & Berindan-Neagoe, I. (2023). Investigating Circular RNAs Using qRT-PCR; Roundup of Optimization and Processing Steps. International Journal of Molecular Sciences, 24(6), 5721. https://doi.org/10.3390/ijms24065721








