Shared Genes of PPARG and NOS2 in Alzheimer’s Disease and Ulcerative Colitis Drive Macrophages and Microglia Polarization: Evidence from Bioinformatics Analysis and Following Validation
Abstract
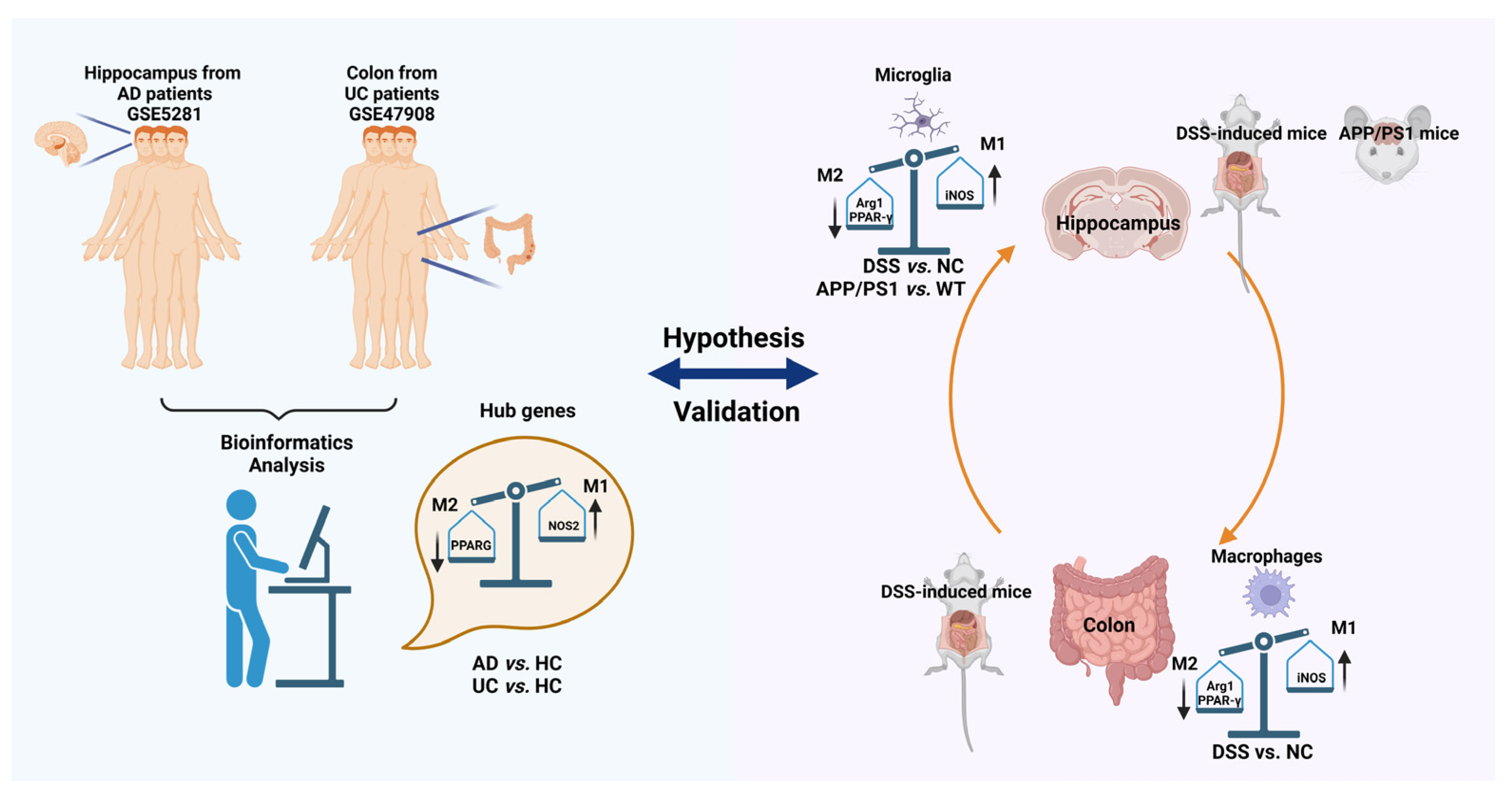
1. Introduction
2. Results
2.1. PPARG, NOS2, SELE, CXCL1, and HSP90AB1 Are Hub Genes of Both AD and UC
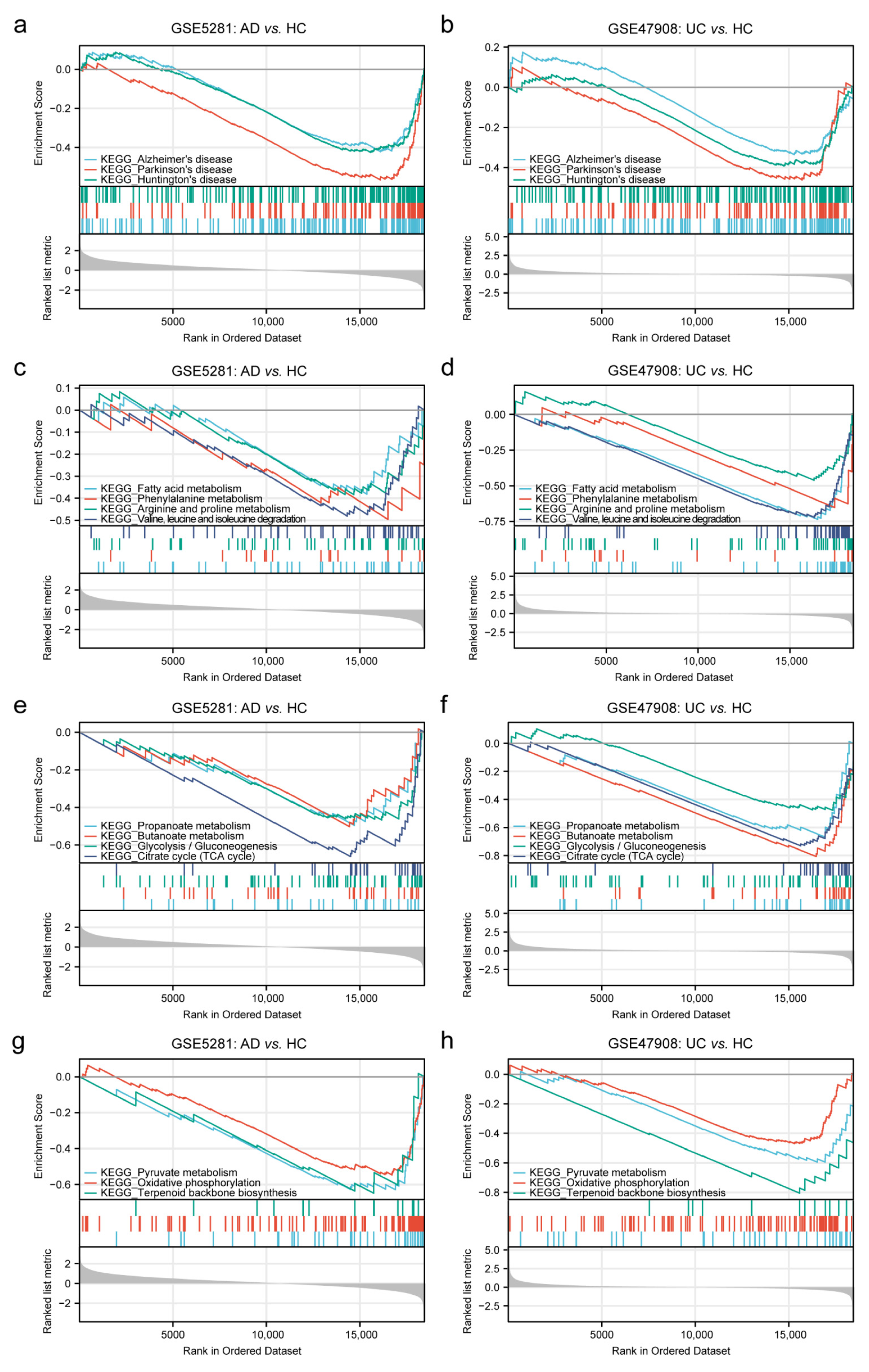
2.1.1. GSEA Analysis in AD and UC
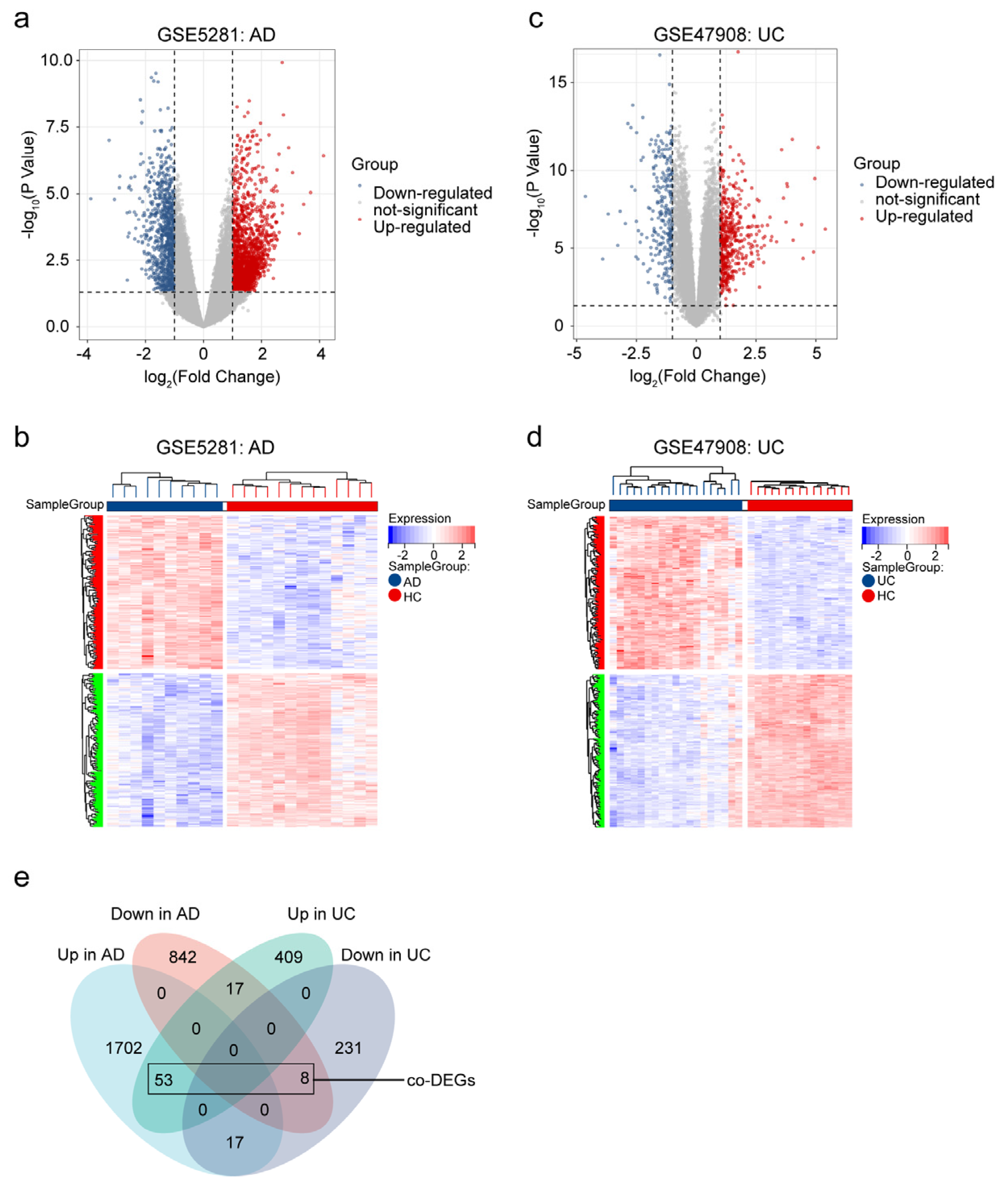
2.1.2. Identified Co-DEGs in AD and UC
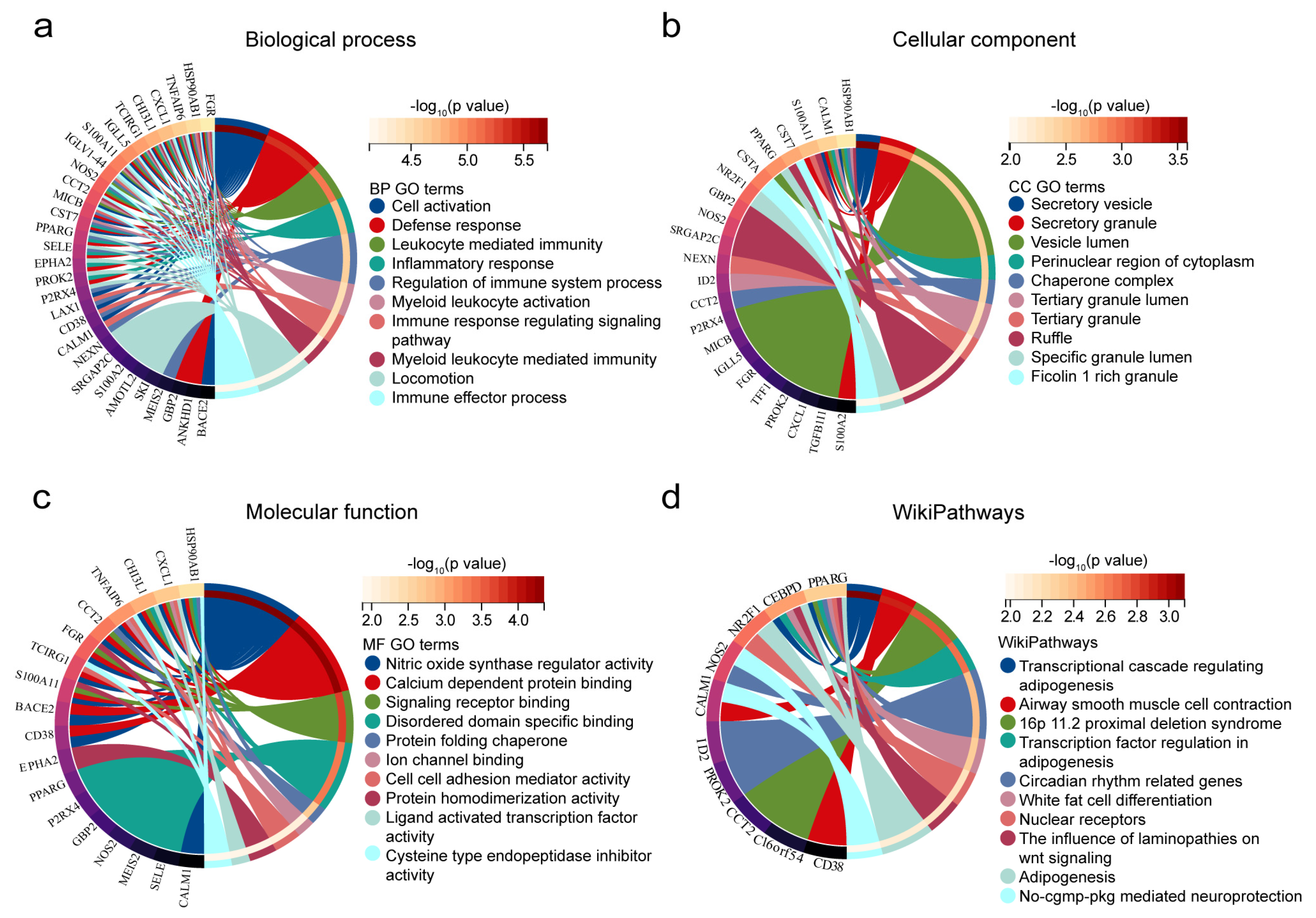
2.1.3. Enrichment Analysis in Co-DEGs
2.1.4. Constructed PPI Network, Determined Hub Genes, and ROC Analysis
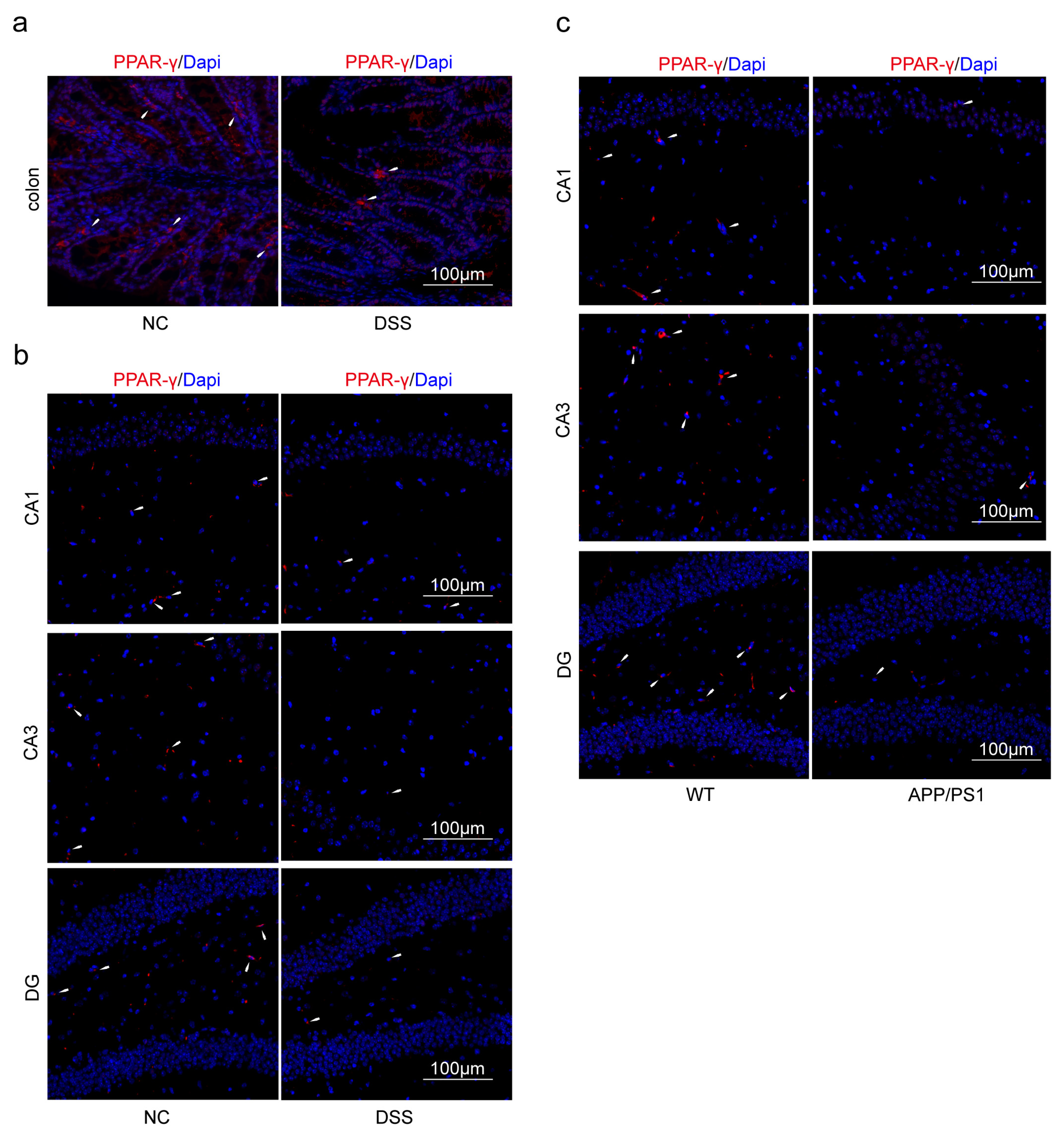
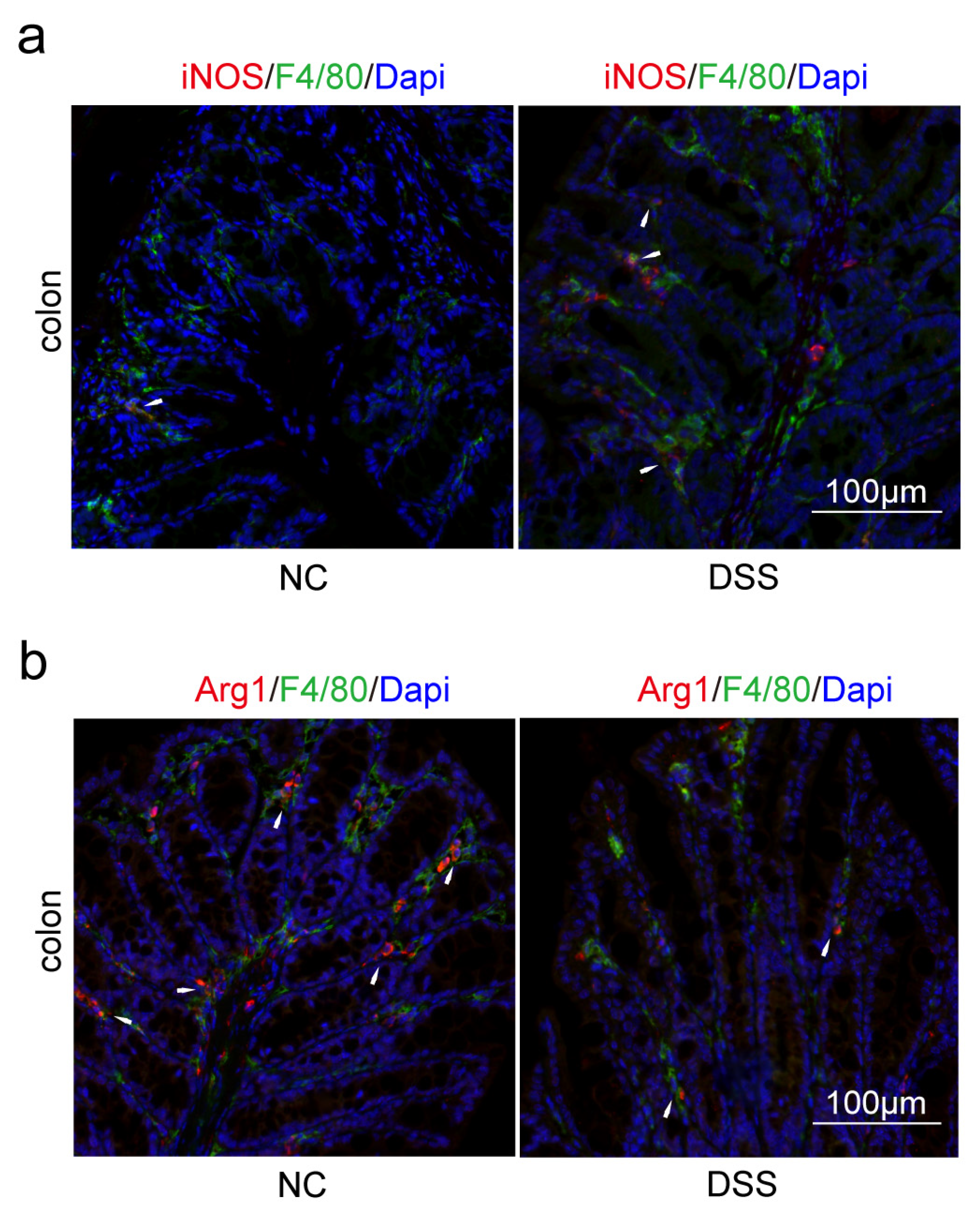
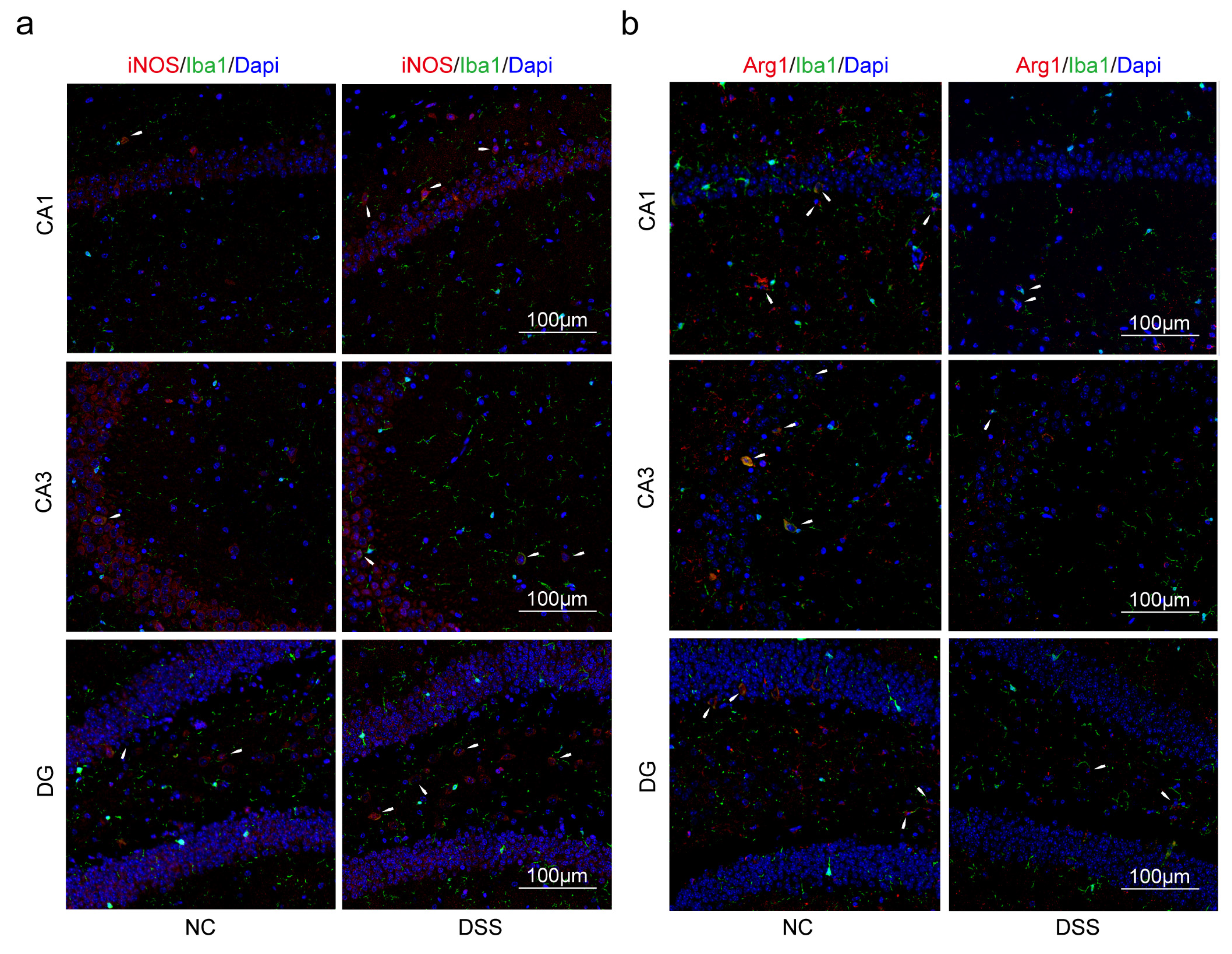
2.2. Decreasing of PPARG and HSP90AB1, and Increasing of NOS2, SELE and CXCL1 Were Found Both in AD and DSS Mice
2.2.1. Induced of the Mouse Model and qRT-PCR Validation of RNA-Seq Data
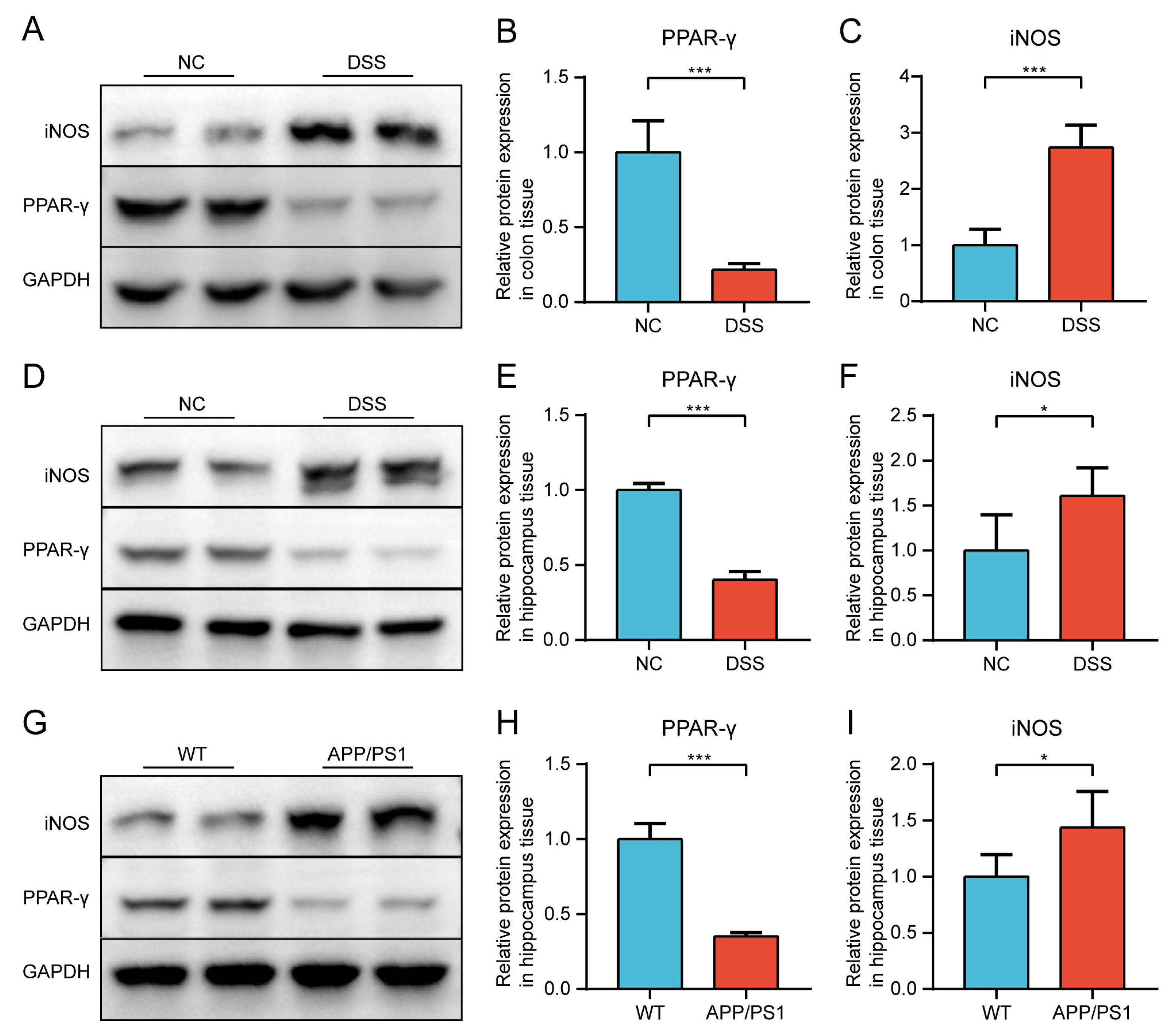
2.2.2. Protein Expression of PPAR-γ and iNOS in Both Diseases Was Confirmed by Western Blot (WB)
2.3. Macrophages and Microglia Tended to Be Inflammatory Polarization Both in AD and UC
3. Discussion
4. Materials and Methods
4.1. GEO Dataset Selection
4.2. Gene Set Enrichment Analysis (GSEA)
4.3. Identification and Enrichment Analyses of DEGs, PPI Network Construction, and Selection of Hub Genes
4.4. Animals
4.5. Morris Water Maze (MWM) Test
4.6. Disease Activity Index (DAI)
4.7. Hematoxylin and Eosin (HE) Staining and Periodic Acid Schiff (PAS) Staining
4.8. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.9. Western Blot (WB)
4.10. Immunofluorescence (IF)
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marizzoni, M.; Cattaneo, A.; Mirabelli, P.; Festari, C.; Lopizzo, N.; Nicolosi, V.; Mombelli, E.; Mazzelli, M.; Luongo, D.; Naviglio, D.; et al. Short-Chain Fatty Acids and Lipopolysaccharide as Mediators Between Gut Dysbiosis and Amyloid Pathology in Alzheimer’s Disease. J. Alzheimer’s Dis. 2020, 78, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Osadchiy, V.; Martin, C.R.; Mayer, E.A. The Gut-Brain Axis and the Microbiome: Mechanisms and Clinical Implications. Clin. Gastroenterol. Hepatol. 2019, 17, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Lobbestael, E.; Vermeire, S.; Sabino, J.; Cleynen, I. Inflammatory bowel disease and Parkinson’s disease: Common pathophysiological links. Gut 2021, 70, 408–417. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Jiang, L.; Zhang, J.; Tong, X.; Chen, D.; Le, W. Intestinal Inflammation and Parkinson’s Disease. Aging Dis. 2021, 12, 2052–2068. [Google Scholar] [CrossRef]
- Liao, P.H.; Chiang, H.L.; Shun, C.T.; Hang, J.F.; Chiu, H.M.; Wu, M.S.; Lin, C.H. Colonic Leucine-Rich Repeat Kinase 2 Expression Is Increased and Associated With Disease Severity in Patients With Parkinson’s Disease. Front. Aging Neurosci. 2021, 13, 819373. [Google Scholar] [CrossRef] [PubMed]
- Gampierakis, I.A.; Koutmani, Y.; Semitekolou, M.; Morianos, I.; Polissidis, A.; Katsouda, A.; Charalampopoulos, I.; Xanthou, G.; Gravanis, A.; Karalis, K.P. Hippocampal neural stem cells and microglia response to experimental inflammatory bowel disease (IBD). Mol. Psychiatr. 2021, 26, 1248–1263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, H.E.; Bai, Y.M.; Tsai, S.J.; Su, T.P.; Chen, T.J.; Wang, Y.P.; Chen, M.H. Inflammatory bowel disease is associated with higher dementia risk: A nationwide longitudinal study. Gut 2021, 70, 85–91. [Google Scholar] [CrossRef]
- Ronnow, S.J.; Troelsen, F.S.; Horvath-Puho, E.; Henderson, V.W.; Sorensen, H.T.; Erichsen, R. Risk of dementia in patients with inflammatory bowel disease: A Danish population-based study. Aliment. Pharm. Ther. 2022, 56, 831–843. [Google Scholar] [CrossRef]
- Kim, G.H.; Lee, Y.C.; Kim, T.J.; Kim, E.R.; Hong, S.N.; Chang, D.K.; Kim, Y.H. Risk of Neurodegenerative Diseases in Patients with Inflammatory Bowel Disease: A Nationwide Population-based Cohort Study. J. Crohn’s Colitis 2022, 16, 436–443. [Google Scholar] [CrossRef]
- Arranz, A.M.; De Strooper, B. The role of astroglia in Alzheimer’s disease: Pathophysiology and clinical implications. Lancet Neurol. 2019, 18, 406–414. [Google Scholar] [CrossRef]
- De Strooper, B.; Karran, E. The Cellular Phase of Alzheimer’s Disease. Cell 2016, 164, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Scheltens, P.; De Strooper, B.; Kivipelto, M.; Holstege, H.; Chetelat, G.; Teunissen, C.E.; Cummings, J.; van der Flier, W.M. Alzheimer’s disease. Lancet 2021, 397, 1577–1590. [Google Scholar] [CrossRef]
- Singh, N.A.; Bhardwaj, V.; Ravi, C.; Ramesh, N.; Mandal, A.; Khan, Z.A. EGCG Nanoparticles Attenuate Aluminum Chloride Induced Neurobehavioral Deficits, Beta Amyloid and Tau Pathology in a Rat Model of Alzheimer’s Disease. Front. Aging Neurosci. 2018, 10, 244. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, J.S.; Wang, Y.; Schork, A.J.; Thompson, W.K.; Karch, C.M.; Cruchaga, C.; McEvoy, L.K.; Witoelar, A.; Chen, C.H.; Holland, D.; et al. Association Between Genetic Traits for Immune-Mediated Diseases and Alzheimer Disease. JAMA Neurol. 2016, 73, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Petralia, M.C.; Battaglia, G.; Bruno, V.; Pennisi, M.; Mangano, K.; Lombardo, S.D.; Fagone, P.; Cavalli, E.; Saraceno, A.; Nicoletti, F.; et al. The Role of Macrophage Migration Inhibitory Factor in Alzheimer’s Disease: Conventionally Pathogenetic or Unconventionally Protective? Molecules 2020, 25, 291. [Google Scholar] [CrossRef] [PubMed]
- Plavec, T.V.; Kuchar, M.; Benko, A.; Liskova, V.; Cerny, J.; Berlec, A.; Maly, P. Engineered Lactococcus lactis Secreting IL-23 Receptor-Targeted REX Protein Blockers for Modulation of IL-23/Th17-Mediated Inflammation. Microorganisms 2019, 7, 152. [Google Scholar] [CrossRef]
- Cosnes, J.; Gower-Rousseau, C.; Seksik, P.; Cortot, A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology 2011, 140, 1785–1794. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, Q.; Xie, L.; Li, C.; Zhuang, Z.; Lin, S.; Lv, P.; Liu, Y.; Wu, Q.; Yu, S. Electroacupuncture and Moxibustion Regulate Hippocampus Glia and Mitochondria Activation in DSS-Induced Colitis Mice. Evid. -Based Complement. Altern. Med. 2020, 2020, 2530253. [Google Scholar] [CrossRef]
- Wei, D.; Zhao, N.; Xie, L.; Huang, B.; Zhuang, Z.; Tang, Y.; Yu, S.; Wu, Q. Electroacupuncture and Moxibustion Improved Anxiety Behavior in DSS-Induced Colitis Mice. Gastroent. Res. Pract. 2019, 2019, 2345890. [Google Scholar] [CrossRef]
- Yao, M.; Zhang, C.; Gao, C.; Wang, Q.; Dai, M.; Yue, R.; Sun, W.; Liang, W.; Zheng, Z. Exploration of the Shared Gene Signatures and Molecular Mechanisms Between Systemic Lupus Erythematosus and Pulmonary Arterial Hypertension: Evidence From Transcriptome Data. Front. Immunol. 2021, 12, 658341. [Google Scholar] [CrossRef]
- Aggarwal, M.; Alkhayyat, M.; Abou, S.M.; Sarmini, M.T.; Singh, A.; Garg, R.; Garg, P.; Mansoor, E.; Padival, R.; Cohen, B.L. Alzheimer Disease Occurs More Frequently In Patients With Inflammatory Bowel Disease: Insight From a Nationwide Study. J. Clin. Gastroenterol. 2022. [Google Scholar] [CrossRef]
- D’Avila, J.C.; Siqueira, L.D.; Mazeraud, A.; Azevedo, E.P.; Foguel, D.; Castro-Faria-Neto, H.C.; Sharshar, T.; Chretien, F.; Bozza, F.A. Age-related cognitive impairment is associated with long-term neuroinflammation and oxidative stress in a mouse model of episodic systemic inflammation. J. Neuroinflamm. 2018, 15, 28. [Google Scholar] [CrossRef] [PubMed]
- Kinney, J.W.; Bemiller, S.M.; Murtishaw, A.S.; Leisgang, A.M.; Salazar, A.M.; Lamb, B.T. Inflammation as a central mechanism in Alzheimer’s disease. Alzh. Dement-Trci. 2018, 4, 575–590. [Google Scholar] [CrossRef]
- Cavalli, E.; Battaglia, G.; Basile, M.S.; Bruno, V.; Petralia, M.C.; Lombardo, S.D.; Pennisi, M.; Kalfin, R.; Tancheva, L.; Fagone, P.; et al. Exploratory Analysis of iPSCS-Derived Neuronal Cells as Predictors of Diagnosis and Treatment of Alzheimer Disease. Brain Sci. 2020, 10, 166. [Google Scholar] [CrossRef]
- Xin, Z.; Zhai, Z.; Long, H.; Zhang, F.; Ni, X.; Deng, J.; Yi, L.; Deng, B. Metabolic Profiling by UPLC-Orbitrap-MS/MS of Liver from C57BL/6 Mice with DSS-Induced Inflammatory Bowel Disease. Mediat. Inflamm. 2020, 2020, 6020247. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, Y.M.; Borcherding, D.; Kanthasamy, A.; Kim, H.J.; Willette, A.A.; Jergens, A.; Allenspach, K.; Mochel, J.P. The Gut-Brain Axis in Neurodegenerative Diseases and Relevance of the Canine Model: A Review. Front. Aging Neurosci. 2019, 11, 130. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.A. The tantalizing links between gut microbes and the brain. Nature 2015, 526, 312–314. [Google Scholar] [CrossRef]
- Robinson, L.; Tang, E.; Taylor, J.P. Dementia: Timely diagnosis and early intervention. BMJ-Brit. Med. J. 2015, 350, h3029. [Google Scholar] [CrossRef]
- Mandrekar, J.N. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef]
- Zhao, Q.; Yang, D.; Gao, L.; Zhao, M.; He, X.; Zhu, M.; Tian, C.; Liu, G.; Li, L.; Hu, C. Downregulation of peroxisome proliferator-activated receptor gamma in the placenta correlates to hyperglycemia in offspring at young adulthood after exposure to gestational diabetes mellitus. J. Diabetes Investig. 2019, 10, 499–512. [Google Scholar] [CrossRef]
- Janani, C.; Ranjitha, K.B. PPAR gamma gene—A review. Diabetes Metab. Synd. 2015, 9, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; He, J.; Koh, S.P.; Zhong, Y.; Liu, Z.; Wang, Z.; Zhang, Y.; Li, Z.; Tam, B.T.; Lin, P.; et al. Reprogrammed marrow adipocytes contribute to myeloma-induced bone disease. Sci. Transl. Med. 2019, 11, eaau9087. [Google Scholar] [CrossRef] [PubMed]
- Lopes, G.; Daletos, G.; Proksch, P.; Andrade, P.B.; Valentao, P. Anti-inflammatory potential of monogalactosyl diacylglycerols and a monoacylglycerol from the edible brown seaweed Fucus spiralis Linnaeus. Mar. Drugs 2014, 12, 1406–1418. [Google Scholar] [CrossRef]
- Anavi, S.; Tirosh, O. iNOS as a metabolic enzyme under stress conditions. Free. Radic. Biol. Med. 2020, 146, 16–35. [Google Scholar] [CrossRef] [PubMed]
- Dharmasiri, S.; Garrido-Martin, E.M.; Harris, R.J.; Bateman, A.C.; Collins, J.E.; Cummings, J.; Sanchez-Elsner, T. Human Intestinal Macrophages Are Involved in the Pathology of Both Ulcerative Colitis and Crohn Disease. Inflamm. Bowel. Dis. 2021, 27, 1641–1652. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.J.; Fisher, E.A.; Greaves, D.R. Macrophage differentiation and function in atherosclerosis: Opportunities for therapeutic intervention? J. Innate Immun. 2012, 4, 498–508. [Google Scholar] [CrossRef]
- Ma, S.; Zhang, J.; Liu, H.; Li, S.; Wang, Q. The Role of Tissue-Resident Macrophages in the Development and Treatment of Inflammatory Bowel Disease. Front. Cell Dev. Biol. 2022, 10, 896591. [Google Scholar] [CrossRef]
- Kashfi, K.; Kannikal, J.; Nath, N. Macrophage Reprogramming and Cancer Therapeutics: Role of iNOS-Derived NO. Cells 2021, 10, 3194. [Google Scholar] [CrossRef]
- Han, X.; Ding, S.; Jiang, H.; Liu, G. Roles of Macrophages in the Development and Treatment of Gut Inflammation. Front. Cell Dev. Biol. 2021, 9, 625423. [Google Scholar] [CrossRef]
- Croasdell, A.; Duffney, P.F.; Kim, N.; Lacy, S.H.; Sime, P.J.; Phipps, R.P. PPARgamma and the Innate Immune System Mediate the Resolution of Inflammation. PPAR Res. 2015, 2015, 549691. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Ricardo-Gonzalez, R.R.; Goforth, M.H.; Morel, C.R.; Subramanian, V.; Mukundan, L.; Red, E.A.; Vats, D.; Brombacher, F.; Ferrante, A.W.; et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 2007, 447, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Kieler, M.; Hofmann, M.; Schabbauer, G. More than just protein building blocks: How amino acids and related metabolic pathways fuel macrophage polarization. FEBS J. 2021, 288, 3694–3714. [Google Scholar] [CrossRef]
- Prinz, M.; Masuda, T.; Wheeler, M.A.; Quintana, F.J. Microglia and Central Nervous System-Associated Macrophages-From Origin to Disease Modulation. Annu. Rev. Immunol. 2021, 39, 251–277. [Google Scholar] [CrossRef] [PubMed]
- Borst, K.; Dumas, A.A.; Prinz, M. Microglia: Immune and non-immune functions. Immunity 2021, 54, 2194–2208. [Google Scholar] [CrossRef]
- Devanney, N.A.; Stewart, A.N.; Gensel, J.C. Microglia and macrophage metabolism in CNS injury and disease: The role of immunometabolism in neurodegeneration and neurotrauma. Exp. Neurol. 2020, 329, 113310. [Google Scholar] [CrossRef] [PubMed]
- El-Din, S.S.; Abd, E.S.; Rashed, L.; Fayez, S.; Aboulhoda, B.E.; Heikal, O.A.; Galal, A.F.; Nour, Z.A. Possible role of rice bran extract in microglial modulation through PPAR-gamma receptors in alzheimer’s disease mice model. Metab. Brain Dis. 2021, 36, 1903–1915. [Google Scholar] [CrossRef]
- Medrano-Jimenez, E.; Jimenez-Ferrer, C.I.; Pedraza-Escalona, M.; Ramirez-Serrano, C.E.; Alvarez-Arellano, L.; Cortes-Mendoza, J.; Herrera-Ruiz, M.; Jimenez-Ferrer, E.; Zamilpa, A.; Tortoriello, J.; et al. Malva parviflora extract ameliorates the deleterious effects of a high fat diet on the cognitive deficit in a mouse model of Alzheimer’s disease by restoring microglial function via a PPAR-gamma-dependent mechanism. J. Neuroinflamm. 2019, 16, 143. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, T.; Wang, M.; Zhang, F.; Zhang, G.; Zhao, J.; Zhang, Y.; Wu, E.; Li, X. Icariin Attenuates M1 Activation of Microglia and Abeta Plaque Accumulation in the Hippocampus and Prefrontal Cortex by Up-Regulating PPARgamma in Restraint/Isolation-Stressed APP/PS1 Mice. Front. Neurosci. 2019, 13, 291. [Google Scholar] [CrossRef]
- Cui, W.; Sun, C.; Ma, Y.; Wang, S.; Wang, X.; Zhang, Y. Inhibition of TLR4 Induces M2 Microglial Polarization and Provides Neuroprotection via the NLRP3 Inflammasome in Alzheimer’s Disease. Front. Neurosci. 2020, 14, 444. [Google Scholar] [CrossRef]
- Wang, Z.; Vilekar, P.; Huang, J.; Weaver, D.F. Furosemide as a Probe Molecule for the Treatment of Neuroinflammation in Alzheimer’s Disease. ACS Chem. Neurosci. 2020, 11, 4152–4168. [Google Scholar] [CrossRef]
- Teter, B.; Morihara, T.; Lim, G.P.; Chu, T.; Jones, M.R.; Zuo, X.; Paul, R.M.; Frautschy, S.A.; Cole, G.M. Curcumin restores innate immune Alzheimer’s disease risk gene expression to ameliorate Alzheimer pathogenesis. Neurobiol. Dis. 2019, 127, 432–448. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Gao, P.; Shi, L.; Chen, L.; Liu, J.; Long, J. Central and Peripheral Metabolic Defects Contribute to the Pathogenesis of Alzheimer’s Disease: Targeting Mitochondria for Diagnosis and Prevention. Antioxid. Redox Sign. 2020, 32, 1188–1236. [Google Scholar] [CrossRef] [PubMed]
- Mapstone, M.; Cheema, A.K.; Fiandaca, M.S.; Zhong, X.; Mhyre, T.R.; MacArthur, L.H.; Hall, W.J.; Fisher, S.G.; Peterson, D.R.; Haley, J.M.; et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat. Med. 2014, 20, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gong, Z.; Zhang, X.; Zhu, F.; Liu, Y.; Jin, C.; Du, X.; Xu, C.; Chen, Y.; Cai, W.; et al. Gut microbial bile acid metabolite skews macrophage polarization and contributes to high-fat diet-induced colonic inflammation. Gut Microbes 2020, 12, 1819155. [Google Scholar] [CrossRef]
- Yoo, W.; Zieba, J.K.; Foegeding, N.J.; Torres, T.P.; Shelton, C.D.; Shealy, N.G.; Byndloss, A.J.; Cevallos, S.A.; Gertz, E.; Tiffany, C.R.; et al. High-fat diet-induced colonocyte dysfunction escalates microbiota-derived trimethylamine N-oxide. Science 2021, 373, 813–818. [Google Scholar] [CrossRef]
- Yan, J.; Horng, T. Lipid Metabolism in Regulation of Macrophage Functions. Trends Cell Biol. 2020, 30, 979–989. [Google Scholar] [CrossRef]
- Victor, M.B.; Leary, N.; Luna, X.; Meharena, H.S.; Scannail, A.N.; Bozzelli, P.L.; Samaan, G.; Murdock, M.H.; von Maydell, D.; Effenberger, A.H.; et al. Lipid accumulation induced by APOE4 impairs microglial surveillance of neuronal-network activity. Cell Stem Cell 2022, 29, 1197–1212. [Google Scholar] [CrossRef]
- Marschallinger, J.; Iram, T.; Zardeneta, M.; Lee, S.E.; Lehallier, B.; Haney, M.S.; Pluvinage, J.V.; Mathur, V.; Hahn, O.; Morgens, D.W.; et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 2020, 23, 194–208. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Rahman, M.R.; Islam, T.; Nicoletti, F.; Petralia, M.C.; Ciurleo, R.; Fisicaro, F.; Pennisi, M.; Bramanti, A.; Demirtas, T.Y.; Gov, E.; et al. Identification of Common Pathogenetic Processes between Schizophrenia and Diabetes Mellitus by Systems Biology Analysis. Genes 2021, 12, 237. [Google Scholar] [CrossRef]
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Melius, J.; Cirillo, E.; Coort, S.L.; Digles, D.; et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018, 46, D661–D667. [Google Scholar] [CrossRef]
- The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Otasek, D.; Morris, J.H.; Boucas, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Aitken, J.D.; Malleshappa, M.; Vijay-Kumar, M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 2014, 104, 15–25. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, H.; Wang, C.; Yang, M.; Chang, S.; Geng, Y.; Yang, H.; Zhuang, Z.; Wang, X.; Xie, L.; et al. Regulating the Balance of Th17/Treg via Electroacupuncture and Moxibustion: An Ulcerative Colitis Mice Model Based Study. Evid. -Based Complement. Altern. Med. 2017, 2017, 7296353. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, L.; Shen, Y.; Li, H.; Zhang, R.; Yu, S.; Wu, Q. Shared Genes of PPARG and NOS2 in Alzheimer’s Disease and Ulcerative Colitis Drive Macrophages and Microglia Polarization: Evidence from Bioinformatics Analysis and Following Validation. Int. J. Mol. Sci. 2023, 24, 5651. https://doi.org/10.3390/ijms24065651
Dong L, Shen Y, Li H, Zhang R, Yu S, Wu Q. Shared Genes of PPARG and NOS2 in Alzheimer’s Disease and Ulcerative Colitis Drive Macrophages and Microglia Polarization: Evidence from Bioinformatics Analysis and Following Validation. International Journal of Molecular Sciences. 2023; 24(6):5651. https://doi.org/10.3390/ijms24065651
Chicago/Turabian StyleDong, Longcong, Yuan Shen, Hongying Li, Ruibin Zhang, Shuguang Yu, and Qiaofeng Wu. 2023. "Shared Genes of PPARG and NOS2 in Alzheimer’s Disease and Ulcerative Colitis Drive Macrophages and Microglia Polarization: Evidence from Bioinformatics Analysis and Following Validation" International Journal of Molecular Sciences 24, no. 6: 5651. https://doi.org/10.3390/ijms24065651
APA StyleDong, L., Shen, Y., Li, H., Zhang, R., Yu, S., & Wu, Q. (2023). Shared Genes of PPARG and NOS2 in Alzheimer’s Disease and Ulcerative Colitis Drive Macrophages and Microglia Polarization: Evidence from Bioinformatics Analysis and Following Validation. International Journal of Molecular Sciences, 24(6), 5651. https://doi.org/10.3390/ijms24065651



