RIG-I Promotes Tumorigenesis and Confers Radioresistance of Esophageal Squamous Cell Carcinoma by Regulating DUSP6
Abstract
1. Introduction
2. Results
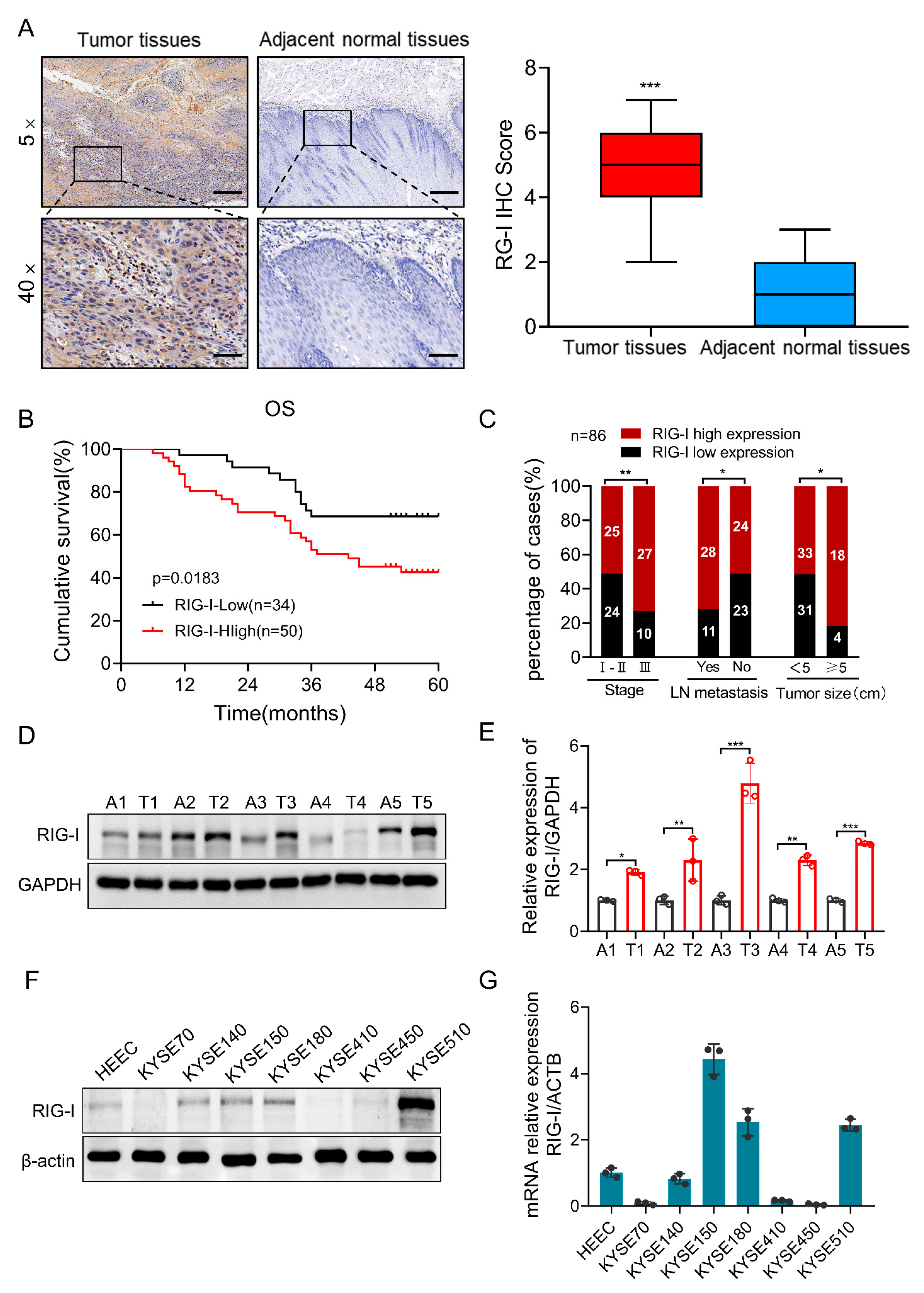
2.1. High Expression of RIG-I in Human ESCC Tissue Samples and Cell Lines
2.2. RIG-I Enhances Proliferation, Migration, and Invasion in ESCC Cells
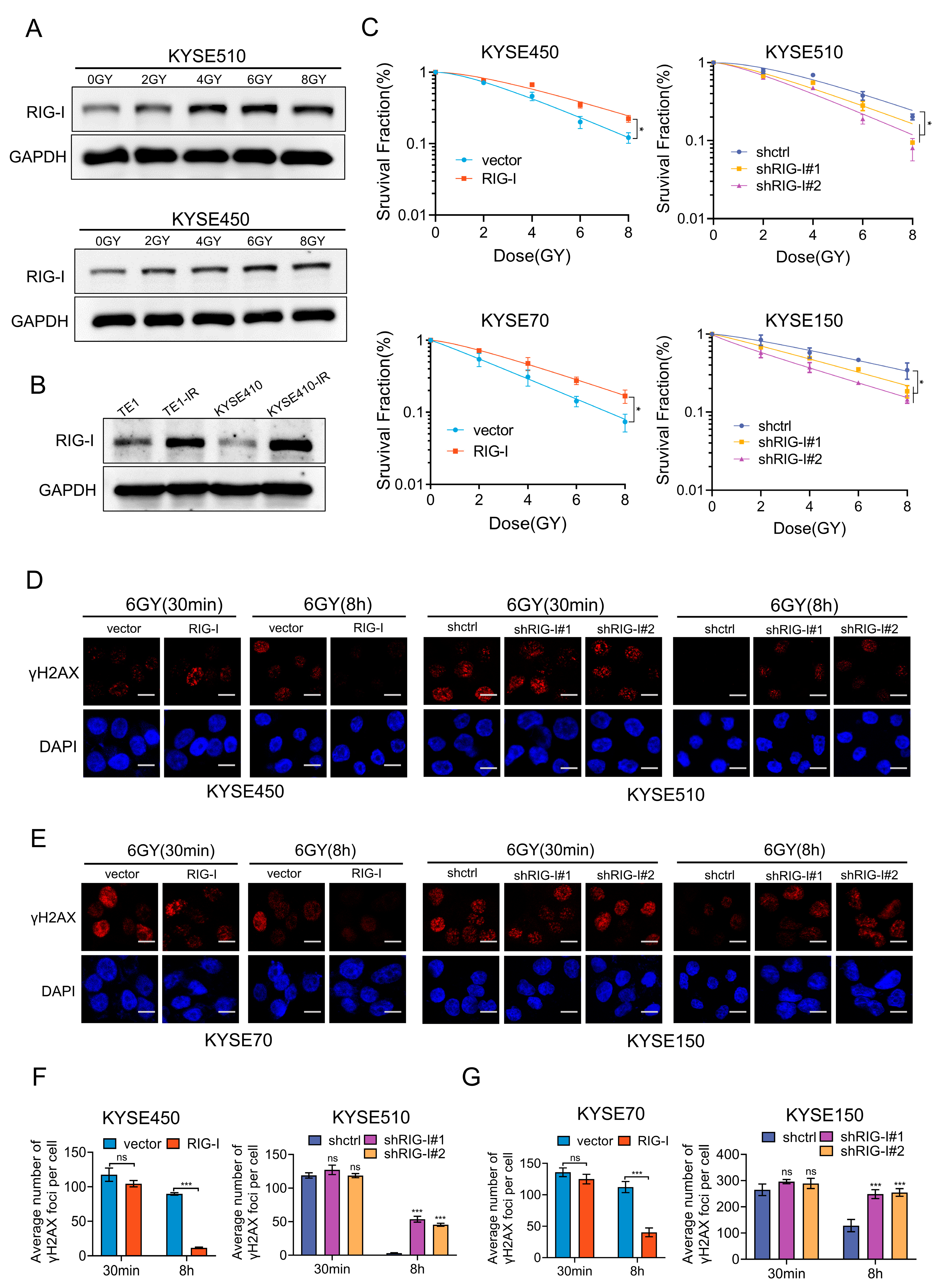
2.3. RIG-I Confers Radioresistance to ESCC Cells
2.4. DUSP6 Enhances the Radioresistance of ESCC Cells
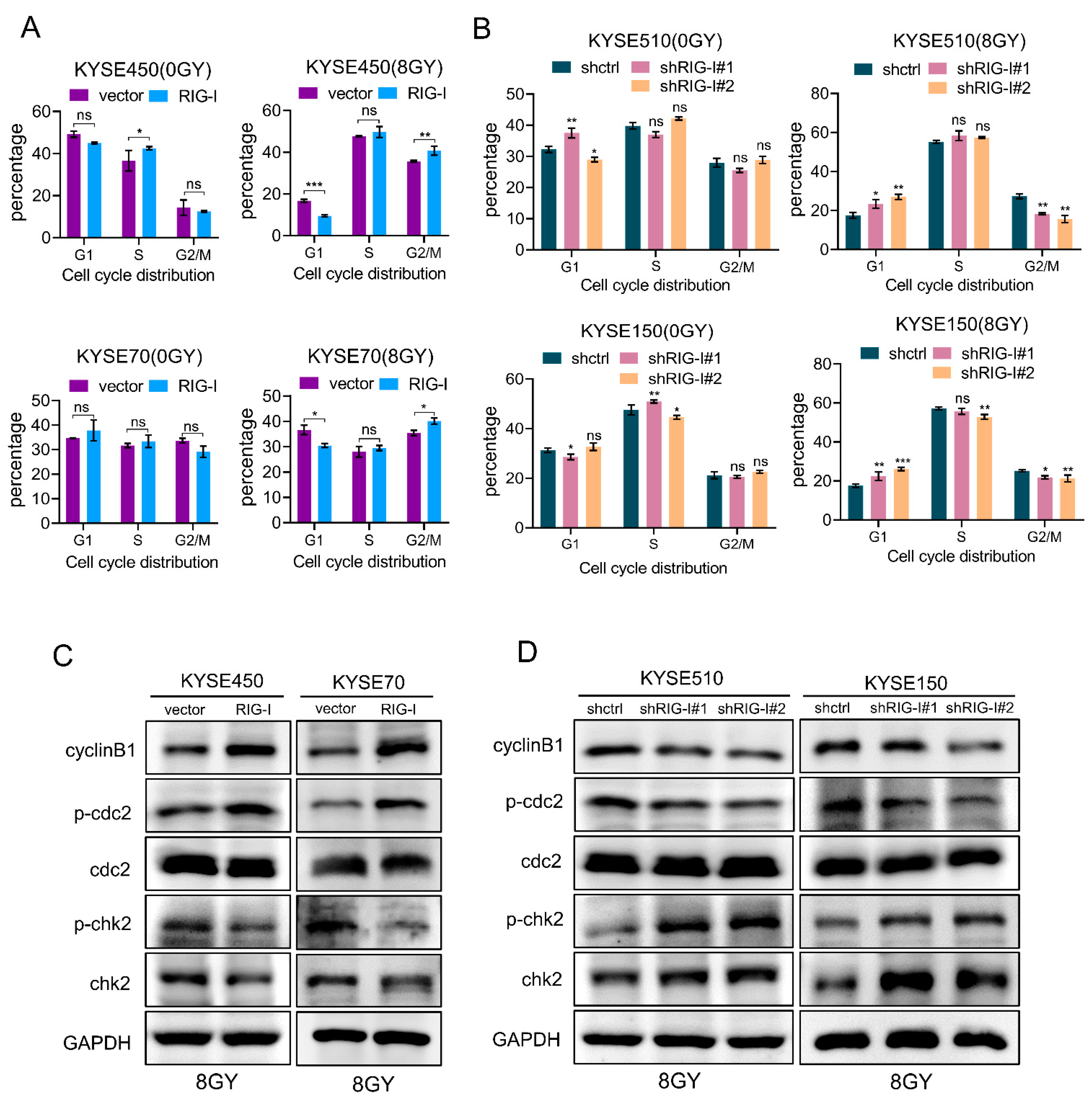
2.5. RIG-I Contributes to Radiation-Induced G2/M Phase Arrest in ESCC Cells
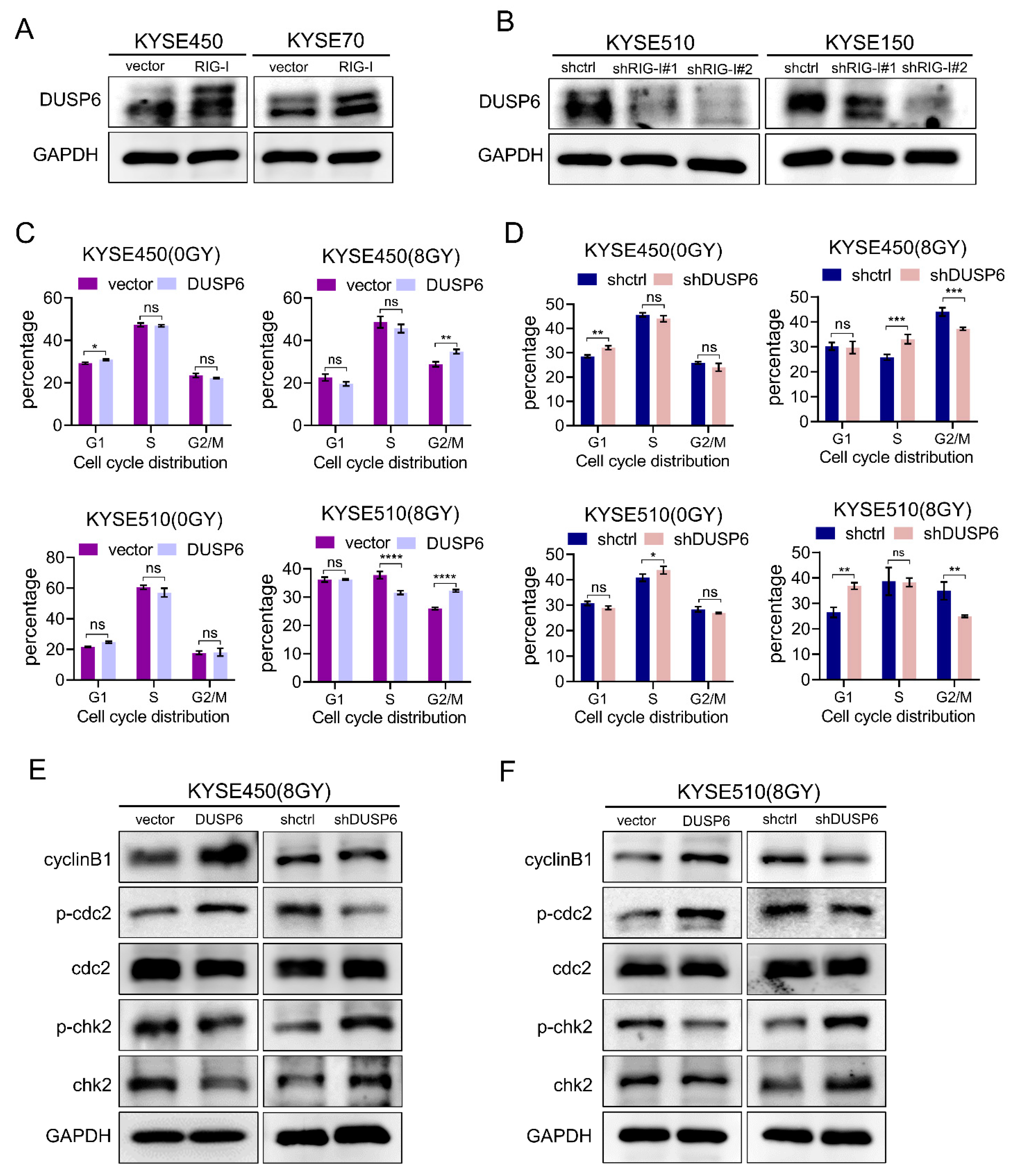
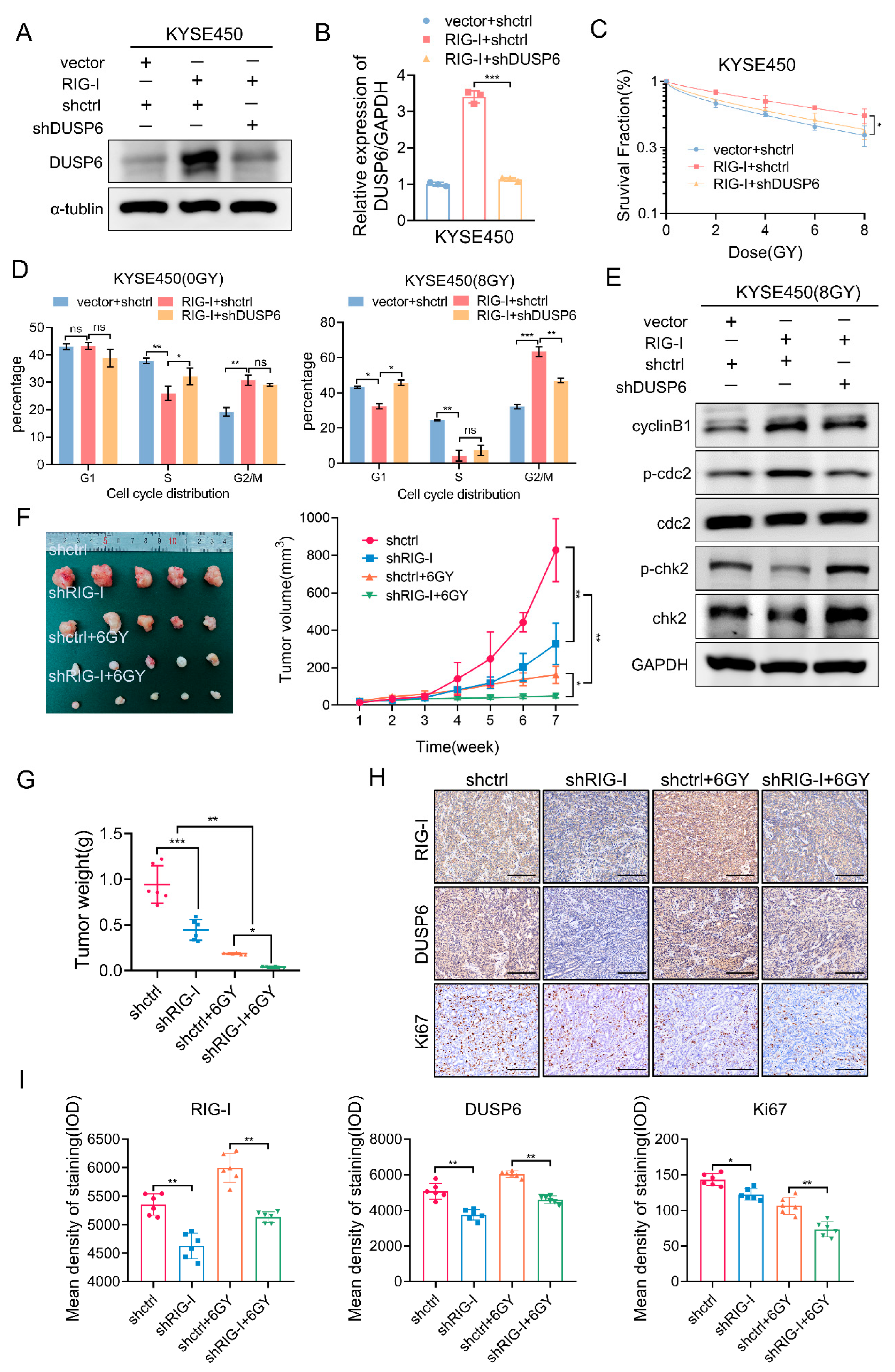
2.6. RIG-I-Induces Radioresistance and G2/M Phase Arrest by Regulating DUSP6 in ESCC Cells
2.7. RIG-I Promotes ESCC Tumorigenesis and Radioresistance In Vivo
3. Discussion
4. Materials and Methods
4.1. Collection of Tissue Specimens from Patients
4.2. Immunohistochemistry
4.3. Cell Lines and Culture
4.4. Lentivirus Packaging and Transduction
4.5. Real-Time RT-PCR
4.6. Western Blotting
4.7. Cell Counting Kit-8 Assay
4.8. Cell Migration and Invasion Assays
4.9. Wound-Healing Assay
4.10. Colony Formation Assay
4.11. Immunofluorescence Assay
4.12. Flow Cytometry
4.13. Xenograft Tumor Model
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smyth, E.C.; Lagergren, J.; Fitzgerald, R.C.; Lordick, F.; Shah, M.A.; Lagergren, P.; Cunningham, D. Oesophageal cancer. Nat. Rev. Dis. Primers 2017, 3, 17048. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Short, M.W.; Burgers, K.G.; Fry, V.T. Esophageal Cancer. Am. Fam. Physician 2017, 95, 22–28. [Google Scholar] [PubMed]
- Abnet, C.C.; Arnold, M.; Wei, W.Q. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology 2018, 154, 360–373. [Google Scholar] [CrossRef]
- Di Pietro, M.; Canto, M.I.; Fitzgerald, R.C. Endoscopic Management of Early Adeno- carcinoma and Squamous Cell Carcinoma of the Esophagus: Screening, Diagnosis, and Therapy. Gastroenterology 2018, 154, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.J. Emerging Multimodality Approaches to Treat Localized Esophageal Cancer. J. Natl. Compr. Cancer Netw. 2019, 17, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Ajani, J.A.; D’Amico, T.A.; Bentrem, D.J.; Chao, J.; Corvera, C.; Das, P.; Denlinger, C.S.; Enzinger, P.C.; Fanta, P.; Farjah, F.; et al. Esophageal and Esophagogastric Junction Cancers, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2019, 17, 855–883. [Google Scholar] [CrossRef]
- Eyck, B.; van der Wilk, B.; Lagarde, S.; Wijnhoven, B.; Valkema, R.; Spaander, M.; Nuyttens, J.; van der Gaast, A.; van Lanschot, J. Neoadjuvant chemoradiotherapy for resectable oesophageal cancer. Best Pract. Res. Clin. Gastroenterol 2018, 36–37, 37–44. [Google Scholar] [CrossRef]
- Leng, X.F.; Daiko, H.; Han, Y.T.; Mao, Y.S. Optimal preoperative neoadjuvant therapy for resectable locally advanced esophageal squamous cell carcinoma. Ann. N. Y. Acad. Sci. 2020, 1482, 213–224. [Google Scholar] [CrossRef]
- Reichenbach, Z.W.; Murray, M.G.; Saxena, R.; Farkas, D.; Karassik, E.G.; Klochkova, A.; Patel, K.; Tice, C.; Hall, T.M.; Gang, J.; et al. Clinical and translational advances in esophageal squamous cell carcinoma. Adv. Cancer Res. 2019, 144, 95–135. [Google Scholar] [CrossRef]
- Xu, X.X.; Wan, H.; Nie, L.; Shao, T.; Xiang, L.X.; Shao, J.Z. RIG-I: A multifunctional protein beyond a pattern recognition receptor. Protein Cell 2018, 9, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Thoresen, D.; Wang, W.; Galls, D.; Guo, R.; Xu, L.; Pyle, A.M. The molecular mechanism of RIG-I activation and signaling. Immunol. Rev. 2021, 304, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Brisse, M.; Ly, H. Comparative Structure and Function Analysis of the RIG-I-Like Receptors: RIG-I and MDA5. Front. Immunol. 2019, 10, 1586. [Google Scholar] [CrossRef]
- Koerner, J.; Horvath, D.; Herrmann, V.L.; MacKerracher, A.; Gander, B.; Yagita, H.; Rohayem, J.; Groettrup, M. PLGA-particle vaccine carrying TLR3/RIG-I ligand Riboxxim synergizes with immune checkpoint blockade for effective anti-cancer immunotherapy. Nat. Commun. 2021, 12, 2935. [Google Scholar] [CrossRef] [PubMed]
- Xian, H.; Yang, S.; Jin, S.; Zhang, Y.; Cui, J. LRRC59 modulates type I interferon signaling by restraining the SQSTM1/p62-mediated autophagic degradation of pattern recognition receptor DDX58/RIG-I. Autophagy 2020, 16, 408–418. [Google Scholar] [CrossRef]
- Elion, D.L.; Jacobson, M.E.; Hicks, D.J.; Rahman, B.; Sanchez, V.; Gonzales-Ericsson, P.I.; Fedorova, O.; Pyle, A.M.; Wilson, J.T.; Cook, R.S. Therapeutically Active RIG-I Agonist Induces Immunogenic Tumor Cell Killing in Breast Cancers. Cancer Res. 2018, 78, 6183–6195. [Google Scholar] [CrossRef]
- Das, M.; Shen, L.; Liu, Q.; Goodwin, T.J.; Huang, L. Nanoparticle Delivery of RIG-I Agonist Enables Effective and Safe Adjuvant Therapy in Pancreatic Cancer. Mol. Ther. 2019, 27, 507–517. [Google Scholar] [CrossRef]
- Jacobson, M.E.; Wang-Bishop, L.; Becker, K.W.; Wilson, J.T. Delivery of 5′-triphosphate RNA with endosomolytic nanoparticles potently activates RIG-I to improve cancer immunotherapy. Biomater. Sci. 2019, 7, 547–559. [Google Scholar] [CrossRef]
- Zhong, M.; Zhong, C.; Cui, W.; Wang, G.; Zheng, G.; Li, L.; Zhang, J.; Ren, R.; Gao, H.; Wang, T.; et al. Induction of tolerogenic dendritic cells by activated TGF-β/Akt/Smad2 signaling in RIG-I-deficient stemness-high human liver cancer cells. BMC Cancer 2019, 19, 439. [Google Scholar] [CrossRef]
- Li, Y.; Song, Y.; Li, P.; Li, M.; Wang, H.; Xu, T.; Yu, X.; Yu, Y.; Tai, Y.; Chen, P.; et al. Downregulation of RIG-I mediated by ITGB3/c-SRC/STAT3 signaling confers resistance to interferon-α-induced apoptosis in tumor-repopulating cells of melanoma. J. Immunother. Cancer 2020, 8, e000111. [Google Scholar] [CrossRef]
- Deng, Y.; Fu, H.; Han, X.; Li, Y.; Zhao, W.; Zhao, X.; Yu, C.; Guo, W.; Lei, K.; Wang, T. Activation of DDX58/RIG-I suppresses the growth of tumor cells by inhibiting STAT3/CSE signaling in colon cancer. Int. J. Oncol. 2022, 61, 120. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.; Fiegl, H.; Zeimet, A.G.; Wieser, V.; Marth, C.; Sprung, S.; Sopper, S.; Hartmann, G.; Reimer, D.; Boesch, M. High RIG-I expression in ovarian cancer associates with an immune-escape signature and poor clinical outcome. Int. J. Cancer 2020, 146, 2007–2018. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ye, S.; Zhu, Y.; Chen, L.; Zhang, Z. RIG-I Promotes Cell Viability, Colony Formation, and Glucose Metabolism and Inhibits Cell Apoptosis in Colorectal Cancer by NF-κB Signaling Pathway. Dis. Markers 2022, 2022, 1247007. [Google Scholar] [CrossRef] [PubMed]
- Beyer, S.; Müller, L.; Mitter, S.; Keilmann, L.; Meister, S.; Buschmann, C.; Kraus, F.; Topalov, N.E.; Czogalla, B.; Trillsch, F.; et al. High RIG-I and EFTUD2 expression predicts poor survival in endometrial cancer. J. Cancer Res. Clin. Oncol. 2022. [Google Scholar] [CrossRef]
- Guo, G.; Gao, M.; Gao, X.; Zhu, B.; Huang, J.; Tu, X.; Kim, W.; Zhao, F.; Zhou, Q.; Zhu, S.; et al. Reciprocal regulation of RIG-I and XRCC4 connects DNA repair with RIG-I immune signaling. Nat. Commun. 2021, 12, 2187. [Google Scholar] [CrossRef]
- Boelens, M.C.; Wu, T.J.; Nabet, B.Y.; Xu, B.; Qiu, Y.; Yoon, T.; Azzam, D.J.; Victor, C.T.-S.; Wiemann, B.Z.; Ishwaran, H.; et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell 2014, 159, 499–513. [Google Scholar] [CrossRef]
- Wu, Y.; Wu, X.; Wu, L.; Wang, X.; Liu, Z. The anticancer functions of RIG-I-like receptors, RIG-I and MDA5, and their applications in cancer therapy. Transl. Res. 2017, 190, 51–60. [Google Scholar] [CrossRef]
- Caunt, C.J.; Keyse, S.M. Dual-specificity MAP kinase phosphatases (MKPs): Shaping the outcome of MAP kinase signalling. FEBS J. 2013, 280, 489–504. [Google Scholar] [CrossRef]
- Ahmad, M.K.; Abdollah, N.A.; Shafie, N.H.; Yusof, N.M.; Razak, S.R.A. Dual-specificity phosphatase 6 (DUSP6): A review of its molecular characteristics and clinical relevance in cancer. Cancer Biol. Med. 2018, 15, 14–28. [Google Scholar] [CrossRef]
- Bagnyukova, T.V.; Restifo, D.; Beeharry, N.; Gabitova, L.; Li, T.; Serebriiskii, I.G.; A Golemis, E.; Astsaturov, I. DUSP6 regulates drug sensitivity by modulating DNA damage response. Br. J. Cancer 2013, 109, 1063–1071. [Google Scholar] [CrossRef]
- Nair, J.; Syed, S.B.; Mahaddalkar, T.; Ketkar, M.; Thorat, R.; Goda, J.S.; Dutt, S. DUSP6 regulates radiosensitivity in glioblastoma by modulating the recruitment of phosphorylated DNAPKcs at DNA double-strand breaks. J. Cell Sci. 2021, 134, jcs259520. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, S.; Hu, X.; Gao, F. Tumor-associated fibroblasts derived exosomes induce the proliferation and cisplatin resistance in esophageal squamous cell carcinoma cells through RIG-I/IFN-β signaling. Bioengineered 2022, 13, 12462–12474. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Li, Y.; Zhang, H.; Wang, F.; He, H.-L.; Bai, X.; Li, S.-S. Prognostic implications and oncogenic roles of MYBL2 protein expression in esophageal squamous-cell carcinoma. OncoTargets Ther. 2019, 12, 1917–1927. [Google Scholar] [CrossRef] [PubMed]
- Sugase, T.; Takahashi, T.; Serada, S.; Fujimoto, M.; Hiramatsu, K.; Ohkawara, T.; Tanaka, K.; Miyazaki, Y.; Makino, T.; Kurokawa, Y.; et al. SOCS1 Gene Therapy Improves Radiosensitivity and Enhances Irradiation-Induced DNA Damage in Esophageal Squamous Cell Carcinoma. Cancer Res. 2017, 77, 6975–6986. [Google Scholar] [CrossRef]
- Chen, G.Z.; Zhu, H.C.; Dai, W.S.; Zeng, X.N.; Luo, J.H.; Sun, X.C. The mechanisms of radioresistance in esophageal squamous cell carcinoma and current strategies in radiosensitivity. J. Thorac. Dis. 2017, 9, 849–859. [Google Scholar] [CrossRef]
- Murray, J.M.; Carr, A.M. Integrating DNA damage repair with the cell cycle. Curr. Opin. Cell Biol. 2018, 52, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Kraikivski, P.; Shafiekhani, S.; Terhune, S.S.; Dash, R.K. Crosstalk between Plk1, p53, cell cycle, and G2/M DNA damage checkpoint regulation in cancer: Computational modeling and analysis. NPJ Syst. Biol. Appl. 2021, 7, 46. [Google Scholar] [CrossRef]
- Yang, Y.M.; Hong, P.; Xu, W.W.; He, Q.Y.; Li, B. Advances in targeted therapy for esophageal cancer. Signal Transduct. Target. Ther. 2020, 5, 229. [Google Scholar] [CrossRef]
- Ghosh, R.; Roy, S.; Franco, S. PARP1 depletion induces RIG-I-dependent signaling in human cancer cells. PLoS ONE 2018, 13, e0194611. [Google Scholar] [CrossRef]
- Zhou, B.; Li, C.; Yang, Y.; Wang, Z. RIG-I Promotes Cell Death in Hepatocellular Carcinoma by Inducing M1 Polarization of Perineal Macrophages Through the RIG-I/MAVS/NF-κB Pathway. OncoTargets Ther. 2020, 13, 8783–8794. [Google Scholar] [CrossRef]
- Theodosiou, A.; Ashworth, A. MAP kinase phosphatases. Genome Biol. 2002, 3, REVIEWS3009. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.A.; Nisar, S.; Maacha, S.; Carneiro-Lobo, T.C.; Akhtar, S.; Siveen, K.S.; Wani, N.A.; Rizwan, A.; Bagga, P.; Singh, M.; et al. Cytokine-chemokine network driven metastasis in esophageal cancer; promising avenue for targeted therapy. Mol. Cancer 2021, 20, 2. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.X.; Zhou, P.K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target. Ther. 2020, 5, 60. [Google Scholar] [CrossRef] [PubMed]
- Withers, H.R. The four r’s of radiotherapy. In Advances in Radiation Biology; Lett, J.T., Adler, H., Eds.; Academic Press: New York, NY, USA, 1975. [Google Scholar]
- Qin, Q.; Cheng, H.; Lu, J.; Zhan, L.; Zheng, J.; Cai, J.; Yang, X.; Xu, L.; Zhu, H.; Zhang, C.; et al. Small-molecule survivin inhibitor YM155 enhances radiosensitization in esophageal squamous cell carcinoma by the abrogation of G2 checkpoint and suppression of homologous recombination repair. J. Hematol. Oncol. 2014, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.L.; Southgate, H.; Tweddle, D.A.; Curtin, N.J. DNA damage checkpoint kinases in cancer. Expert Rev. Mol. Med. 2020, 22, e2. [Google Scholar] [CrossRef]
- Li, J.; Guan, H.-Y.; Gong, L.-Y.; Song, L.-B.; Zhang, N.; Wu, J.; Yuan, J.; Zheng, Y.-J.; Huang, Z.-S.; Li, M. Clinical significance of sphingosine kinase-1 expression in human astrocytomas progression and overall patient survival. Clin. Cancer Res. 2008, 14, 6996–7003. [Google Scholar] [CrossRef]
- Shen, Y.T.; Huang, X.; Zhang, G.; Jiang, B.; Li, C.J.; Wu, Z.S. Pan-Cancer Prognostic Role and Targeting Potential of the Estrogen-Progesterone Axis. Front. Oncol. 2021, 11, 636365. [Google Scholar] [CrossRef]
- Sommer, C.A.; Stadtfeld, M.; Murphy, G.J.; Hochedlinger, K.; Kotton, D.N.; Mostoslavsky, G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 2009, 27, 543–549. [Google Scholar] [CrossRef]
- Skvortsova, E.V.; Nazarov, I.B.; Tomilin, A.N.; Sinenko, S.A. Dual Mode of Mitochondri- al ROS Action during Reprogramming to Pluripotency. Int. J. Mol. Sci. 2022, 23, 10924. [Google Scholar] [CrossRef]
- Gu, C.; Luo, J.; Lu, X.; Tang, Y.; Ma, Y.; Yun, Y.; Cao, J.; Cao, J.; Huang, Z.; Zhou, X.; et al. REV7 confers radioresistance of esophagus squamous cell carcinoma by recruiting PRDX2. Cancer Sci. 2019, 110, 962–972. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, S.; Lin, C.; Li, Y.; Ye, L.; Wu, X.; Jian, Y.; Dai, Y.; Ouyang, Y.; Zhao, L.; et al. TRIB3 confers radiotherapy resistance in esophageal squamous cell carcinoma by stabilizing TAZ. Oncogene 2020, 39, 3710–3725. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Lv, L.; Xu, J.-C.; He, Q.; Chang, N.; Cui, Y.-Y.; Tao, Z.-C.; Zhu, T.; Qian, L.-T. RIG-I Promotes Tumorigenesis and Confers Radioresistance of Esophageal Squamous Cell Carcinoma by Regulating DUSP6. Int. J. Mol. Sci. 2023, 24, 5586. https://doi.org/10.3390/ijms24065586
Li L, Lv L, Xu J-C, He Q, Chang N, Cui Y-Y, Tao Z-C, Zhu T, Qian L-T. RIG-I Promotes Tumorigenesis and Confers Radioresistance of Esophageal Squamous Cell Carcinoma by Regulating DUSP6. International Journal of Molecular Sciences. 2023; 24(6):5586. https://doi.org/10.3390/ijms24065586
Chicago/Turabian StyleLi, Lu, Lei Lv, Jun-Chao Xu, Qing He, Na Chang, Ya-Yun Cui, Zhen-Chao Tao, Tao Zhu, and Li-Ting Qian. 2023. "RIG-I Promotes Tumorigenesis and Confers Radioresistance of Esophageal Squamous Cell Carcinoma by Regulating DUSP6" International Journal of Molecular Sciences 24, no. 6: 5586. https://doi.org/10.3390/ijms24065586
APA StyleLi, L., Lv, L., Xu, J.-C., He, Q., Chang, N., Cui, Y.-Y., Tao, Z.-C., Zhu, T., & Qian, L.-T. (2023). RIG-I Promotes Tumorigenesis and Confers Radioresistance of Esophageal Squamous Cell Carcinoma by Regulating DUSP6. International Journal of Molecular Sciences, 24(6), 5586. https://doi.org/10.3390/ijms24065586





