A Liquid–Liquid Phase Separation-Related Index Associate with Biochemical Recurrence and Tumor Immune Environment of Prostate Cancer Patients
Abstract
1. Introduction
2. Results
2.1. Identification of Differentially Expressed LRGs and Functional Enrichment
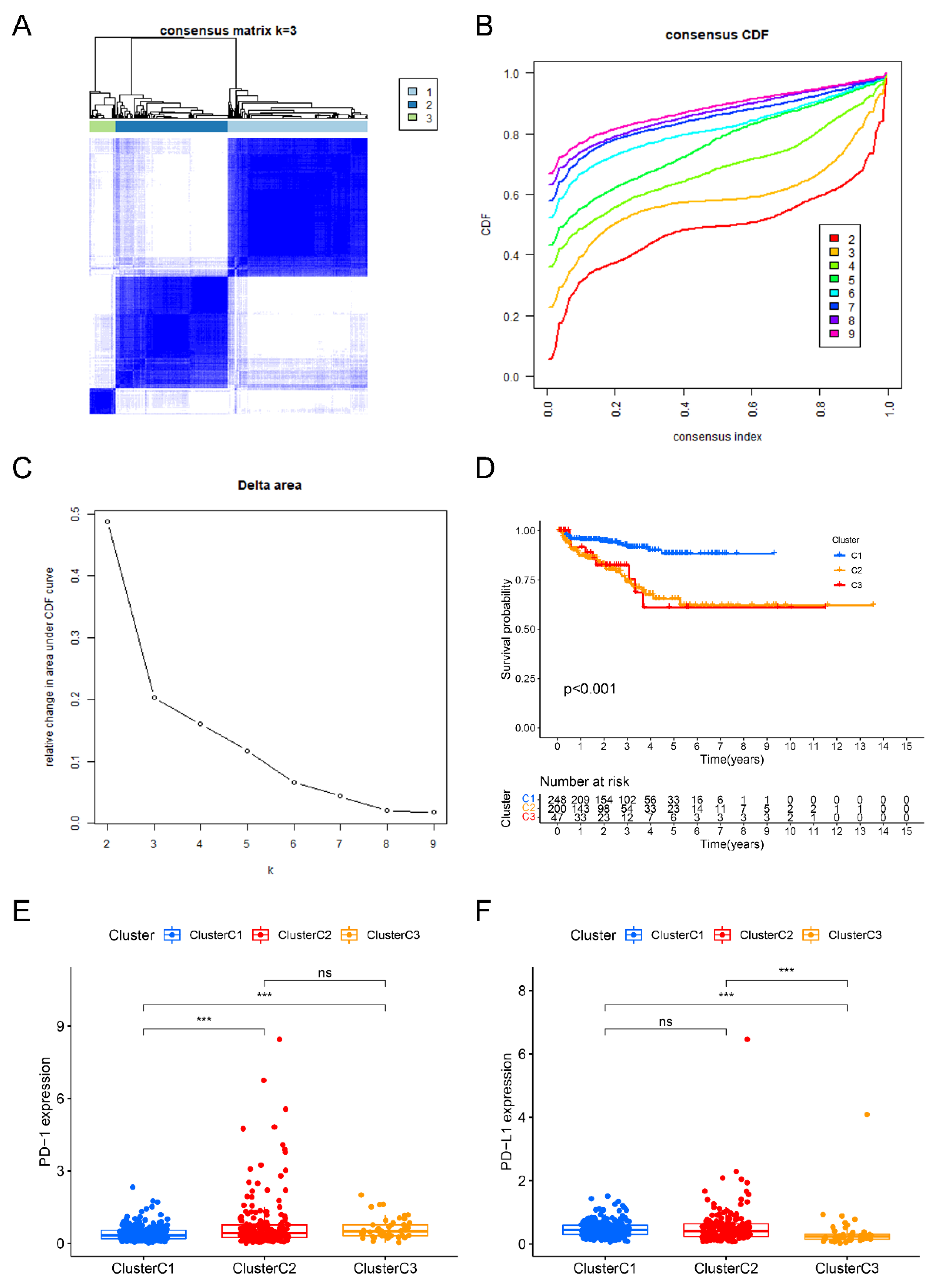
2.2. Identification of Three Liquid–Liquid Phase Separation-Based Molecular Clusters
2.3. Development and Verification of a Novel Index Related to Liquid–Liquid Phase Separation for Predicting BCRFS
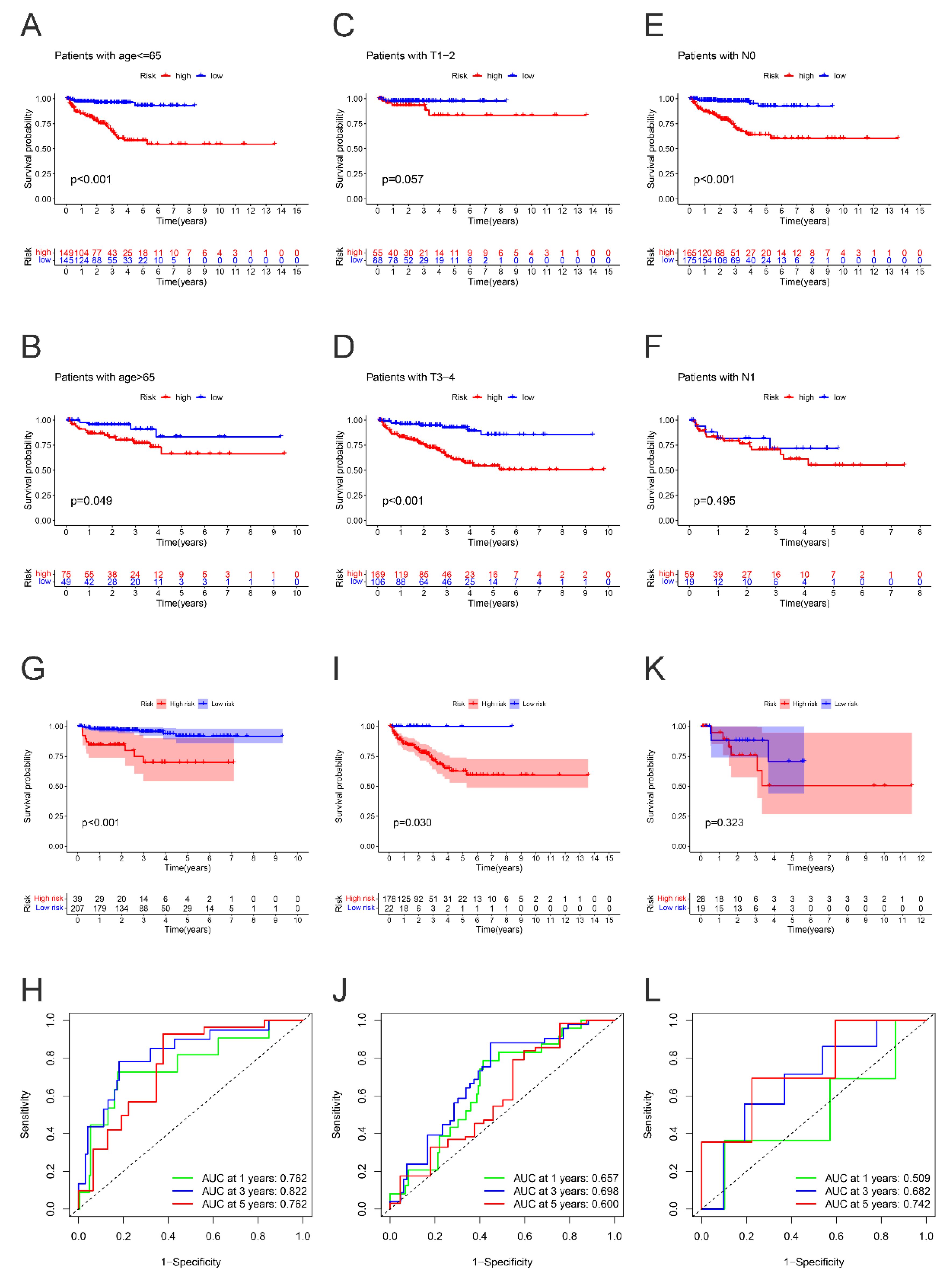
2.4. Subgroup Survival Analysis
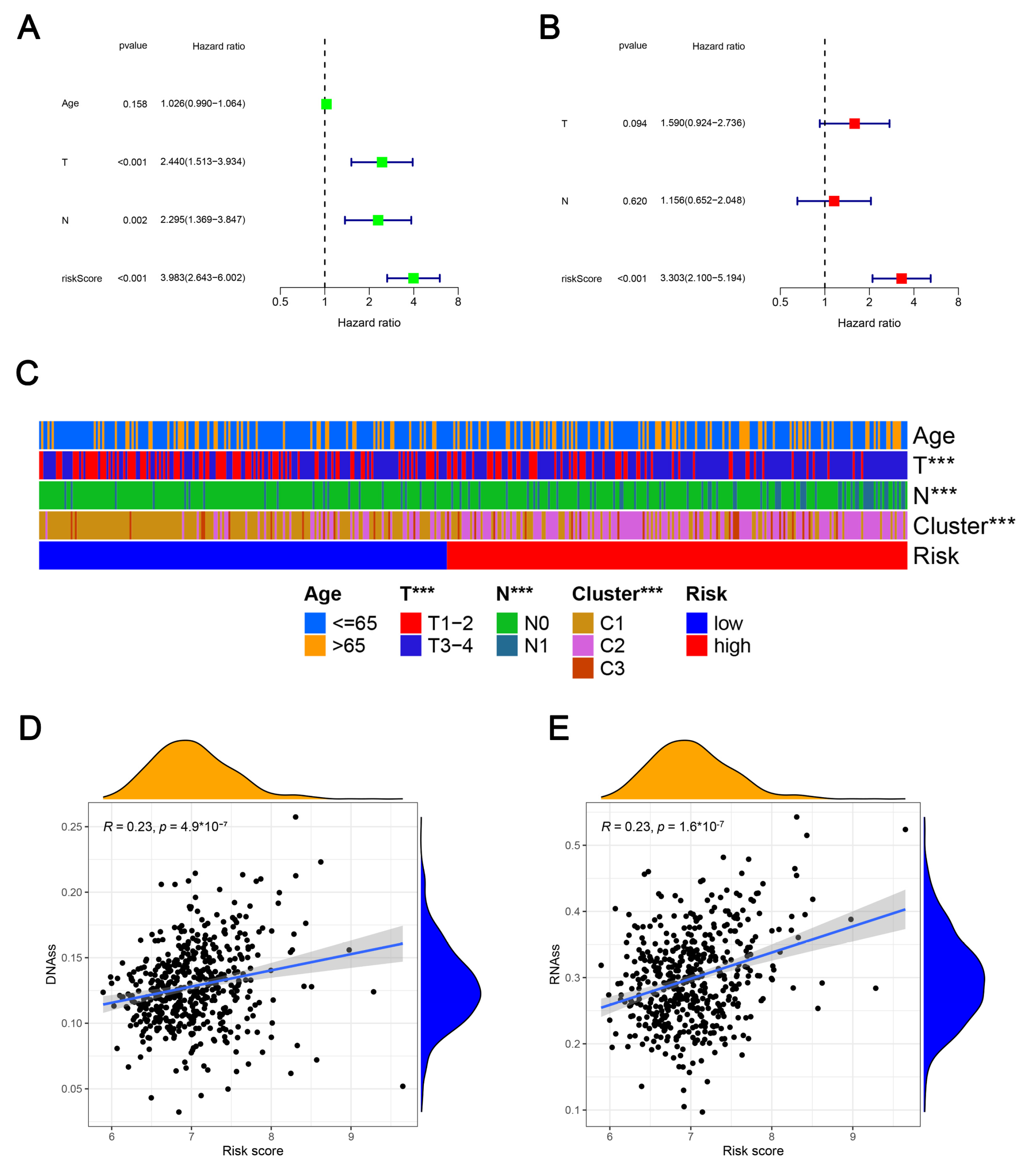
2.5. Clinicopathologic Characteristics, Caner Stemness Functional Enrichment, Cancer Stemness and Tumor Immune Microenvironment
2.6. Anti-Cancer Drug Sensitivity Prediction
2.7. Validation mRNA Expression Levels of Risk Genes Using UALCAN Database
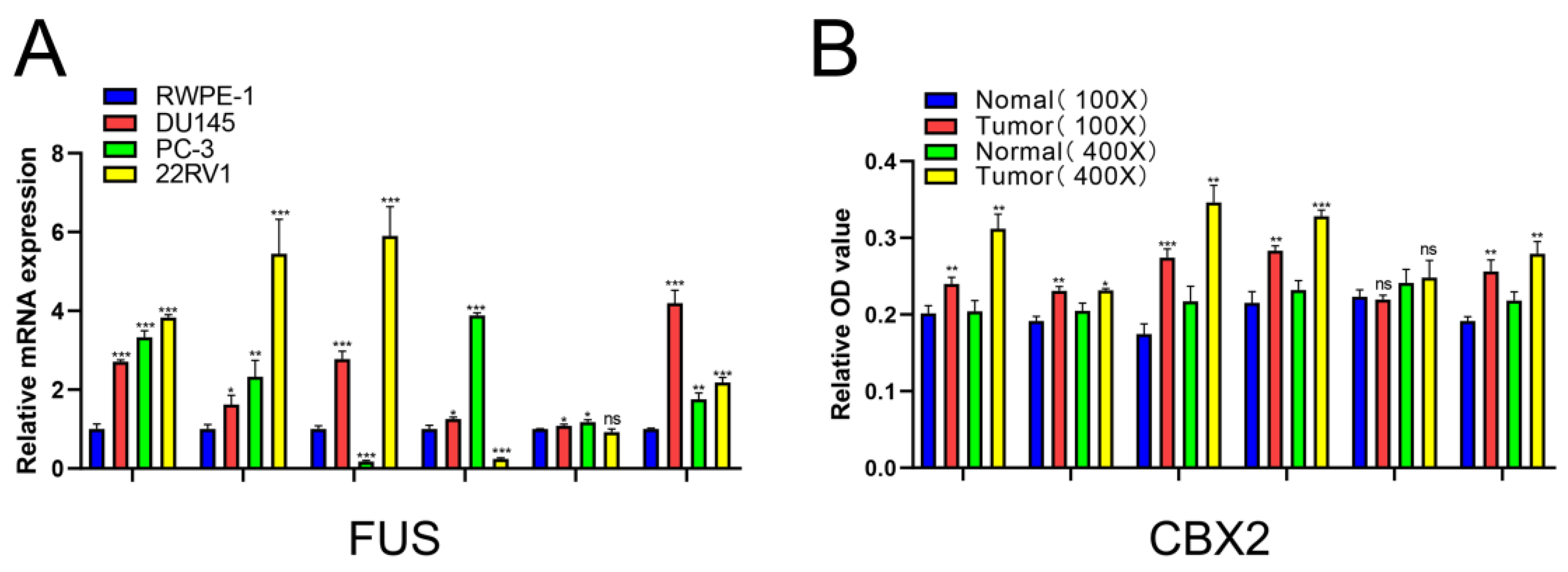
2.8. Verification of Relative mRNA Expression Levels of Risk DELRGs in RWPE-1 and PCa Cell Lines by Conducting qRT-PCR
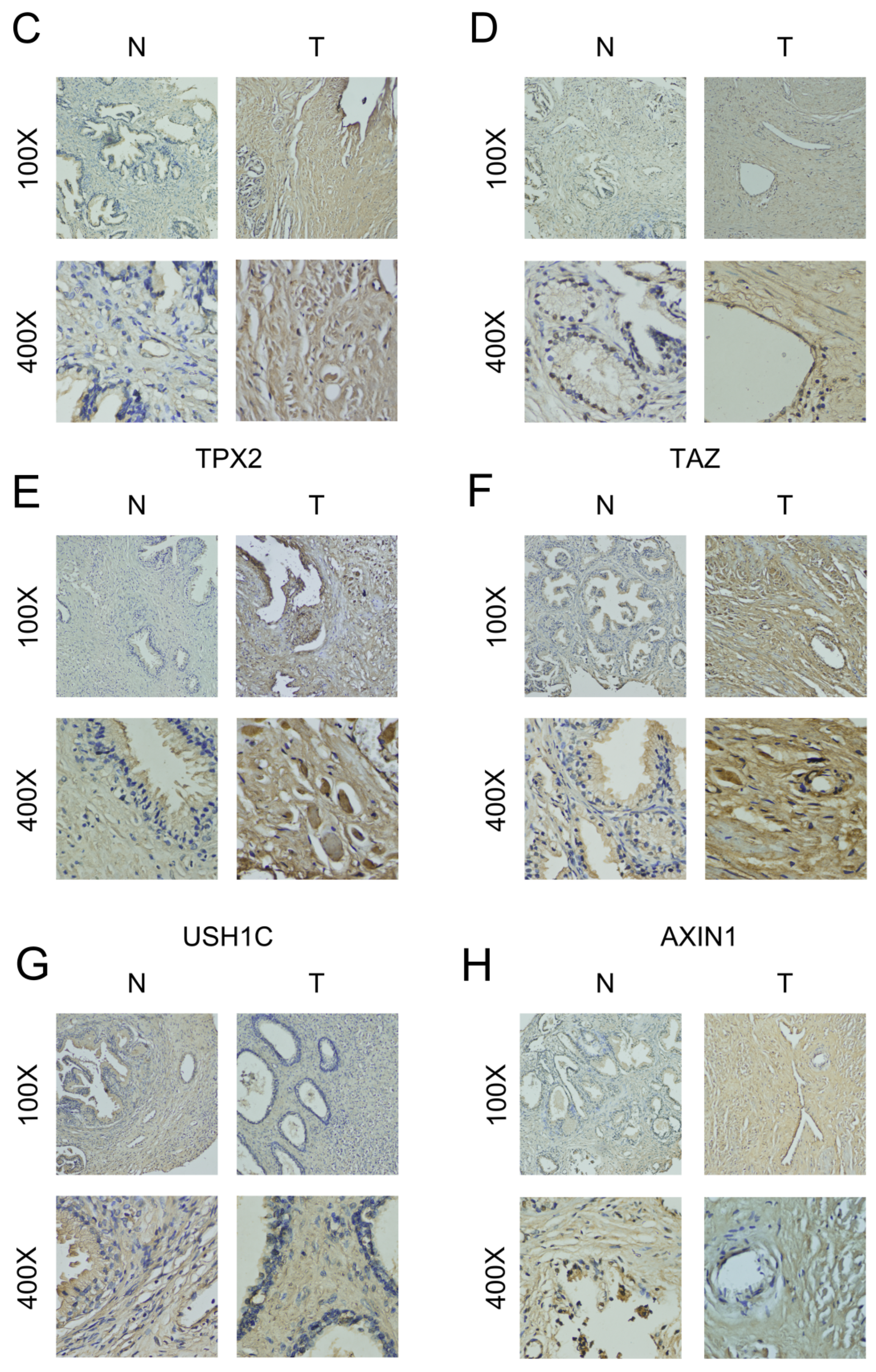
2.9. Verification of the Relative Protein Expression Levels of Six DELRGs in PCa Tissue through IHC Staining
2.10. Inhibition of FUS Could Reduce Proliferation, Migration and Invasion, and Promote Apoptosis of PCa Cells
3. Discussion
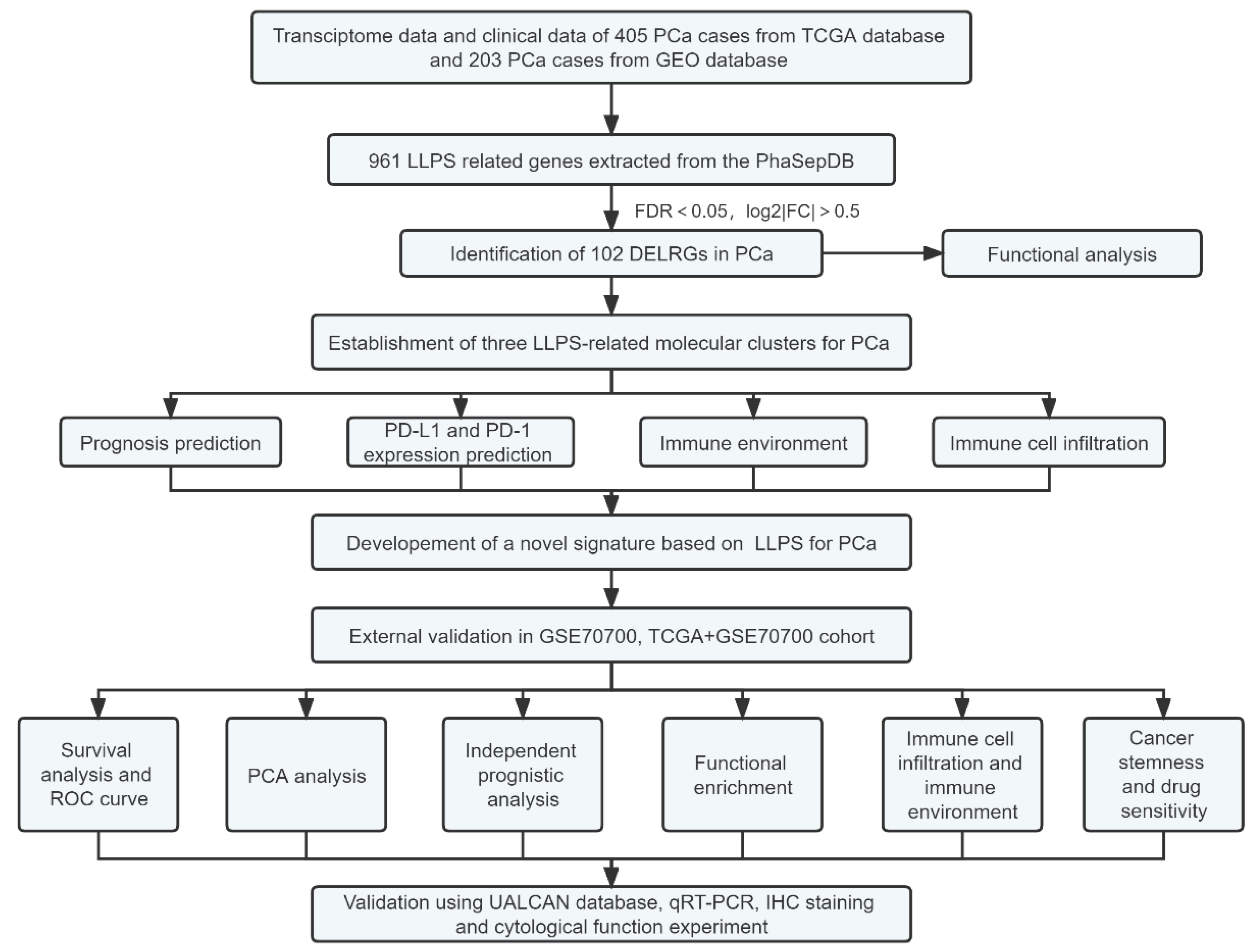
4. Materials and Methods
4.1. Data Collection and Preprocessing
4.2. Identification of Differentially Expressed LRGs (DELRGs) and Functional Enrichment
4.3. Establishment of LLPS-Related Molecular Clusters by Consensus Clustering Analysis
4.4. Establishment and Verification of a Novel LLPS-Related Prognostic Index for Prostate Cancer
4.5. Association of the LLPS-Related Signature with Clinicopathologic Features, Tumor Stemness Scores, Immune Microenvironment, and Functional Enrichment
4.6. Drug Sensitivity of Risk DELRGs
4.7. Validation of Risk Genes Using UALCAN
4.8. Cell Culture
4.9. Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
4.10. Immunohistochemical (IHC) Staining
4.11. RNA Interference
4.12. Western Blotting
4.13. CCK-8 Viability Assays
4.14. Transwell Migration and Invasion Assays
4.15. Flow Cytometry
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Rebello, R.J.; Oing, C.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Gillessen, S.; Van der Kwast, T.; Bristow, R.G. Prostate cancer. Nat. Rev. Dis. Prim. 2021, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef]
- Onderdonk, B.E.; Dorn, P.L.; Martinez, C.; Arif, F.; Cloutier, D.; Antic, T.; Golden, D.W.; Karrison, T.; Pitroda, S.P.; Szmulewitz, R.Z.; et al. A prospective clinical and transcriptomic feasibility study of oral-only hormonal therapy with radiation for unfavorable prostate cancer in men 70 years of age and older or with comorbidity. Cancer 2021, 127, 2631–2640. [Google Scholar] [CrossRef] [PubMed]
- Busetto, G.M.; Giovannone, R.; Antonini, G.; Rossi, A.; Del Giudice, F.; Tricarico, S.; Ragonesi, G.; Gentile, V.; De Berardinis, E. Short-term pretreatment with a dual 5α-reductase inhibitor before bipolar transurethral resection of the prostate (B-TURP): Evaluation of prostate vascularity and decreased surgical blood loss in large prostates. BJU Int. 2015, 116, 117–123. [Google Scholar] [CrossRef]
- Björnebo, L.; Nordström, T.; Discacciati, A.; Palsdottir, T.; Aly, M.; Grönberg, H.; Eklund, M.; Lantz, A. Association of 5α-Reductase Inhibitors With Prostate Cancer Mortality. JAMA Oncol. 2022, 8, 1019–1026. [Google Scholar] [CrossRef]
- Tan, Y.G.; Fang, A.H.S.; Lim, J.K.S.; Khalid, F.; Chen, K.; Ho, H.S.S.; Yuen, J.S.P.; Huang, H.H.; Tay, K.J. Incorporating artificial intelligence in urology: Supervised machine learning algorithms demonstrate comparative advantage over nomograms in predicting biochemical recurrence after prostatectomy. Prostate 2022, 82, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Knipper, S.; Ascalone, L.; Ziegler, B.; Hohenhorst, J.L.; Simon, R.; Berliner, C.; van Leeuwen, F.W.B.; van der Poel, H.; Giesel, F.; Graefen, M.; et al. Salvage Surgery in Patients with Local Recurrence After Radical Prostatectomy. Eur. Urol. 2021, 79, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Ke, Z.B.; You, Q.; Sun, J.B.; Zhu, J.M.; Li, X.D.; Chen, D.N.; Su, L.; Zheng, Q.S.; Wei, Y.; Xue, X.Y.; et al. A Novel Ferroptosis-Based Molecular Signature Associated with Biochemical Recurrence-Free Survival and Tumor Immune Microenvironment of Prostate Cancer. Front. Cell Dev. Biol. 2021, 9, 774625. [Google Scholar] [CrossRef]
- Qiu, Y.; Pan, M.; Chen, X. A Liquid-Liquid Phase Separation-Related Gene Signature as Prognostic Biomarker for Epithelial Ovarian Cancer. Front. Oncol. 2021, 11, 671892. [Google Scholar] [CrossRef]
- Kakiuchi, N.; Ogawa, S. Clonal expansion in non-cancer tissues. Nat. Rev. Cancer 2021, 21, 239–256. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, J.; Zhang, W.; Xiao, C.; Zhang, S.; Nian, C.; Li, J.; Su, D.; Chen, L.; Zhao, Q.; et al. Glycogen accumulation and phase separation drives liver tumor initiation. Cell 2021, 184, 5559–5576.e5519. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Qian, J.; Xu, Z.; Yin, S.; Zhou, L.; Zheng, S.; Zhang, W. Emerging Roles of Liquid-Liquid Phase Separation in Cancer: From Protein Aggregation to Immune-Associated Signaling. Front. Cell Dev. Biol. 2021, 9, 631486. [Google Scholar] [CrossRef] [PubMed]
- Zhuge, L.; Zhang, K.; Zhang, Z.; Guo, W.; Li, Y.; Bao, Q. A novel model based on liquid-liquid phase separation-Related genes correlates immune microenvironment profiles and predicts prognosis of lung squamous cell carcinoma. J. Clin. Lab. Anal. 2022, 36, e24135. [Google Scholar] [CrossRef]
- Takayama, K.-I.; Inoue, S. Targeting phase separation on enhancers induced by transcription factor complex formations as a new strategy for treating drug-resistant cancers. Front. Oncol. 2022, 12, 1024600. [Google Scholar] [CrossRef]
- Xie, J.; He, H.; Kong, W.; Li, Z.; Gao, Z.; Xie, D.; Sun, L.; Fan, X.; Jiang, X.; Zheng, Q.; et al. Targeting androgen receptor phase separation to overcome antiandrogen resistance. Nat. Chem. Biol. 2022, 18, 1341–1350. [Google Scholar] [CrossRef]
- Ahmed, J.; Meszaros, A.; Lazar, T.; Tompa, P. DNA-binding domain as the minimal region driving RNA-dependent liquid-liquid phase separation of androgen receptor. Protein Sci. 2021, 30, 1380–1392. [Google Scholar] [CrossRef]
- Takayama, K.-I.; Kosaka, T.; Suzuki, T.; Hongo, H.; Oya, M.; Fujimura, T.; Suzuki, Y.; Inoue, S. Subtype-specific collaborative transcription factor networks are promoted by OCT4 in the progression of prostate cancer. Nat. Commun. 2021, 12, 3766. [Google Scholar] [CrossRef]
- Roggero, C.M.; Esser, V.; Duan, L.; Rice, A.M.; Ma, S.; Raj, G.V.; Rosen, M.K.; Liu, Z.P.; Rizo, J. Poly-glutamine-dependent self-association as a potential mechanism for regulation of androgen receptor activity. PLoS ONE 2022, 17, e0258876. [Google Scholar] [CrossRef]
- Ahn, J.H.; Davis, E.S.; Daugird, T.A.; Zhao, S.; Quiroga, I.Y.; Uryu, H.; Li, J.; Storey, A.J.; Tsai, Y.H.; Keeley, D.P.; et al. Phase separation drives aberrant chromatin looping and cancer development. Nature 2021, 595, 591–595. [Google Scholar] [CrossRef]
- Nozawa, R.S.; Yamamoto, T.; Takahashi, M.; Tachiwana, H.; Maruyama, R.; Hirota, T.; Saitoh, N. Nuclear microenvironment in cancer: Control through liquid-liquid phase separation. Cancer Sci. 2020, 111, 3155–3163. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, L.; Dai, T.; Qin, Z.; Lu, H.; Zhang, L.; Zhou, F. Liquid-liquid phase separation in human health and diseases. Signal Transduct. Target Ther. 2021, 6, 290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ji, X.; Li, P.; Liu, C.; Lou, J.; Wang, Z.; Wen, W.; Xiao, Y.; Zhang, M.; Zhu, X. Liquid-liquid phase separation in biology: Mechanisms, physiological functions and human diseases. Sci. China Life Sci. 2020, 63, 953–985. [Google Scholar] [CrossRef] [PubMed]
- Sperger, J.M.; Emamekhoo, H.; McKay, R.R.; Stahlfeld, C.N.; Singh, A.; Chen, X.E.; Kwak, L.; Gilsdorf, C.S.; Wolfe, S.K.; Wei, X.X.; et al. Prospective Evaluation of Clinical Outcomes Using a Multiplex Liquid Biopsy Targeting Diverse Resistance Mechanisms in Metastatic Prostate Cancer. J. Clin. Oncol. 2021, 39, 2926–2937. [Google Scholar] [CrossRef]
- Takayama, K.-I.; Suzuki, T.; Fujimura, T.; Yamada, Y.; Takahashi, S.; Homma, Y.; Suzuki, Y.; Inoue, S. Dysregulation of spliceosome gene expression in advanced prostate cancer by RNA-binding protein PSF. Proc. Natl. Acad. Sci. USA 2017, 114, 10461–10466. [Google Scholar] [CrossRef]
- Salciccia, S.; Capriotti, A.L.; Laganà, A.; Fais, S.; Logozzi, M.; De Berardinis, E.; Busetto, G.M.; Di Pierro, G.B.; Ricciuti, G.P.; Del Giudice, F.; et al. Biomarkers in Prostate Cancer Diagnosis: From Current Knowledge to the Role of Metabolomics and Exosomes. Int. J. Mol. Sci. 2021, 22, 4367. [Google Scholar] [CrossRef]
- Logozzi, M.; Angelini, D.F.; Giuliani, A.; Mizzoni, D.; Di Raimo, R.; Maggi, M.; Gentilucci, A.; Marzio, V.; Salciccia, S.; Borsellino, G.; et al. Increased Plasmatic Levels of PSA-Expressing Exosomes Distinguish Prostate Cancer Patients from Benign Prostatic Hyperplasia: A Prospective Study. Cancers 2019, 11, 1449. [Google Scholar] [CrossRef]
- He, Z.; Ruan, X.; Liu, X.; Zheng, J.; Liu, Y.; Liu, L.; Ma, J.; Shao, L.; Wang, D.; Shen, S.; et al. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in Glioma. J. Exp. Clin. Cancer Res. 2019, 38, 65. [Google Scholar] [CrossRef]
- Chen, T.; Wang, X.; Li, C.; Zhang, H.; Liu, Y.; Han, D.; Li, Y.; Li, Z.; Luo, D.; Zhang, N.; et al. CircHIF1A regulated by FUS accelerates triple-negative breast cancer progression by modulating NFIB expression and translocation. Oncogene 2021, 40, 2756–2771. [Google Scholar] [CrossRef]
- Zeng, M.; Li, B.; Yang, L.; Guan, Q. CBX2 depletion inhibits the proliferation, invasion and migration of gastric cancer cells by inactivating the YAP/β-catenin pathway. Mol. Med. Rep. 2021, 23, 137. [Google Scholar] [CrossRef]
- Kahl, I.; Mense, J.; Finke, C.; Boller, A.L.; Lorber, C.; Győrffy, B.; Greve, B.; Götte, M.; Espinoza-Sánchez, N.A. The cell cycle-related genes RHAMM, AURKA, TPX2, PLK1, and PLK4 are associated with the poor prognosis of breast cancer patients. J. Cell. Biochem. 2022, 123, 581–600. [Google Scholar] [CrossRef] [PubMed]
- Matson, D.R.; Denu, R.A.; Zasadil, L.M.; Burkard, M.E.; Weaver, B.A.; Flynn, C.; Stukenberg, P.T. High nuclear TPX2 expression correlates with TP53 mutation and poor clinical behavior in a large breast cancer cohort, but is not an independent predictor of chromosomal instability. BMC Cancer 2021, 21, 186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhang, M.; Li, Q.; Yang, Y.; Shang, Z.; Luo, J. TPX2 mediates prostate cancer epithelial-mesenchymal transition through CDK1 regulated phosphorylation of ERK/GSK3β/SNAIL pathway. Biochem. Biophys. Res. Commun. 2021, 546, 1–6. [Google Scholar] [CrossRef]
- Li, D.; Chen, L.; Zhang, X.; Wang, Y.; Huang, C.; Li, J.; He, F.; He, W. miR-125a-5p reverses epithelial-mesenchymal transition and restores drug sensitivity by negatively regulating TAFAZZIN signaling in breast cancer. Mol. Med. Rep. 2021, 24, 1–11. [Google Scholar] [CrossRef]
- Chen, S.C.; Chen, F.W.; Hsu, Y.L.; Kuo, P.L. Systematic Analysis of Transcriptomic Profile of Renal Cell Carcinoma under Long-Term Hypoxia Using Next-Generation Sequencing and Bioinformatics. Int. J. Mol. Sci. 2017, 18, 2657. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sheng, H.; Ma, F.; Wu, Q.; Huang, J.; Chen, Q.; Sheng, L.; Zhu, X.; Zhu, X.; Xu, M. RNA m(6)A reader YTHDF2 facilitates lung adenocarcinoma cell proliferation and metastasis by targeting the AXIN1/Wnt/β-catenin signaling. Cell Death Dis. 2021, 12, 479. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Fu, H.; Wei, Z.; Yang, D.; Yan, R. UBE3C promotes proliferation and inhibits apoptosis by activating the β-catenin signaling via degradation of AXIN1 in gastric cancer. Carcinogenesis 2021, 42, 285–293. [Google Scholar] [CrossRef]
- Nair, S.; Weil, R.; Dovey, Z.; Davis, A.; Tewari, A. The Tumor Microenvironment and Immunotherapy in Prostate and Bladder Cancer. Urol. Clin. North Am. 2020, 47, e17–e54. [Google Scholar] [CrossRef]
- Majidpoor, J.; Mortezaee, K. The efficacy of PD-1/PD-L1 blockade in cold cancers and future perspectives. Clin. Immunol. 2021, 226, 108707. [Google Scholar] [CrossRef]
- Cha, H.; Lee, J.; Ponnazhagan, S. Revisiting Immunotherapy: A Focus on Prostate Cancer. Cancer Res. 2020, 80, 1615–1623. [Google Scholar] [CrossRef]
- Xu, Y.; Wan, B.; Chen, X.; Zhan, P.; Zhao, Y.; Zhang, T.; Liu, H.; Afzal, M.Z.; Dermime, S.; Hochwald, S.N.; et al. The association of PD-L1 expression with the efficacy of anti-PD-1/PD-L1 immunotherapy and survival of non-small cell lung cancer patients: A meta-analysis of randomized controlled trials. Transl. Lung Cancer Res. 2019, 8, 413–428. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Peng, Z.; Qin, M.; Liu, Y.; Wang, J.; Zhang, C.; Lin, J.; Dong, T.; Wang, L.; Li, S.; et al. Interferon-γ induces tumor resistance to anti-PD-1 immunotherapy by promoting YAP phase separation. Mol. Cell 2021, 81, 1216–1230.e1219. [Google Scholar] [CrossRef]
- Xu, Y.; Song, G.; Xie, S.; Jiang, W.; Chen, X.; Chu, M.; Hu, X.; Wang, Z.-W. The roles of PD-1/PD-L1 in the prognosis and immunotherapy of prostate cancer. Mol. Ther. 2021, 29, 1958–1969. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, X.; Dou, Z.; Yang, W.; Huang, B.; Lou, J.; Zhang, Z. Protein Databases Related to Liquid-Liquid Phase Separation. Int. J. Mol. Sci. 2020, 21, 6796. [Google Scholar] [CrossRef] [PubMed]
- Alivand, M.R.; Najafi, S.; Esmaeili, S.; Rahmanpour, D.; Zhaleh, H.; Rahmati, Y. Integrative analysis of DNA methylation and gene expression profiles to identify biomarkers of glioblastoma. Cancer Genet. 2021, 258–259, 135–150. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Cheng, L.; Yuan, M.; Li, S.; Lian, Z.; Chen, J.; Lin, W.; Zhang, J.; Zhong, S. Identification of an IFN-β-associated gene signature for the prediction of overall survival among glioblastoma patients. Ann. Transl. Med. 2021, 9, 925. [Google Scholar] [CrossRef]




| Variables | Training Cohort | Testing Cohort | p-Value |
|---|---|---|---|
| Age | 60.87 ± 6.59 | 60.98 ± 6.86 | 0.869 |
| T stage | — | — | 0.349 |
| T2a | 1 (0.5%) | 5 (2.5%) | — |
| T2b | 4 (2.0%) | 5 (2.5%) | — |
| T2c | 62 (30.4%) | 73 (36.3%) | — |
| T3a | 69 (33.7%) | 62 (30.8%) | — |
| T3b | 62 (30.4%) | 52 (25.9%) | — |
| T4 | 2 (1.0%) | 3 (1.5%) | — |
| Unknown | 4 (2.0%) | 1 (0.5%) | — |
| N stage | — | — | 0.252 |
| N0 | 148 (72.6%) | 142 (70.7%) | — |
| N1 | 34 (16.7%) | 27 (13.4%) | — |
| Unknown | 32 (15.7%) | 32 (15.9%) | — |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, Q.; Chen, J.-Y.; Wu, X.-H.; Xue, Y.-T.; Sun, J.-B.; Wei, Y.; Zheng, Q.-S.; Xue, X.-Y.; Chen, D.-N.; Xu, N. A Liquid–Liquid Phase Separation-Related Index Associate with Biochemical Recurrence and Tumor Immune Environment of Prostate Cancer Patients. Int. J. Mol. Sci. 2023, 24, 5515. https://doi.org/10.3390/ijms24065515
You Q, Chen J-Y, Wu X-H, Xue Y-T, Sun J-B, Wei Y, Zheng Q-S, Xue X-Y, Chen D-N, Xu N. A Liquid–Liquid Phase Separation-Related Index Associate with Biochemical Recurrence and Tumor Immune Environment of Prostate Cancer Patients. International Journal of Molecular Sciences. 2023; 24(6):5515. https://doi.org/10.3390/ijms24065515
Chicago/Turabian StyleYou, Qi, Jia-Yin Chen, Xiao-Hui Wu, Yu-Ting Xue, Jiang-Bo Sun, Yong Wei, Qing-Shui Zheng, Xue-Yi Xue, Dong-Ning Chen, and Ning Xu. 2023. "A Liquid–Liquid Phase Separation-Related Index Associate with Biochemical Recurrence and Tumor Immune Environment of Prostate Cancer Patients" International Journal of Molecular Sciences 24, no. 6: 5515. https://doi.org/10.3390/ijms24065515
APA StyleYou, Q., Chen, J.-Y., Wu, X.-H., Xue, Y.-T., Sun, J.-B., Wei, Y., Zheng, Q.-S., Xue, X.-Y., Chen, D.-N., & Xu, N. (2023). A Liquid–Liquid Phase Separation-Related Index Associate with Biochemical Recurrence and Tumor Immune Environment of Prostate Cancer Patients. International Journal of Molecular Sciences, 24(6), 5515. https://doi.org/10.3390/ijms24065515






