Rapid, Easy, and Reliable Identification of Nocardia sp. by MALDI-TOF Mass Spectrometry, VITEK®-MS IVD V3.2 Database, Using Direct Deposit
Abstract
1. Introduction
2. Results
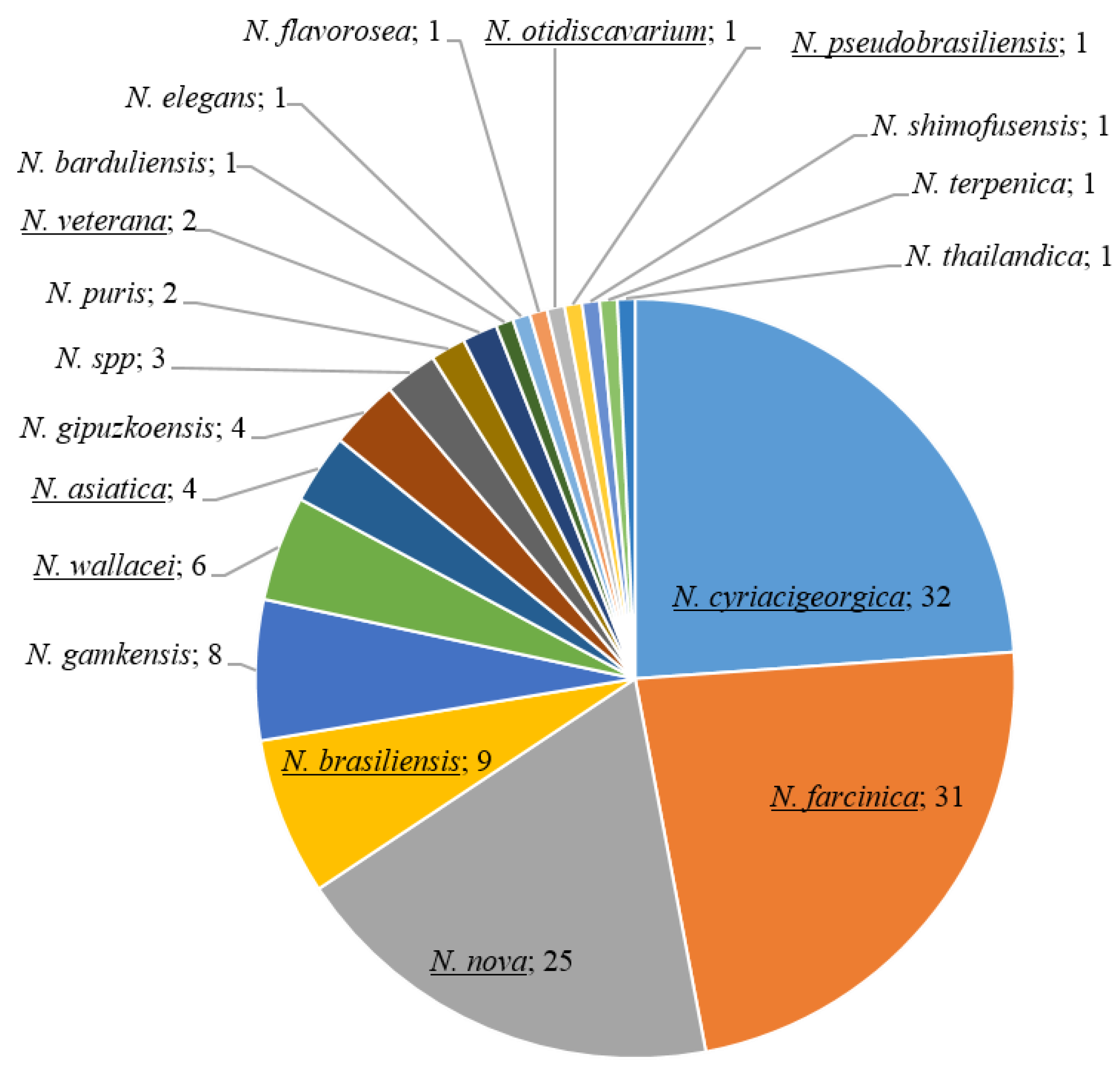
2.1. Nocardia Collection
2.2. Performance of VITEK®-MS for Nocardia Species Identification Using Direct Deposit
2.3. Discrepancy Analysis
3. Discussion
4. Materials and Methods
4.1. Nocardia Isolates
4.2. 16S rRNA/hsp65 Sequencing-Based Nocardia Identification
4.3. VITEK®-MS IVD v3.2 Database Nocardia Identification
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Margalit, I.; Lebeaux, D.; Tishler, O.; Goldberg, E.; Bishara, J.; Yahav, D.; Coussement, J. How do I manage nocardiosis? Clin. Microbiol. Infect. 2021, 27, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Soueges, S.; Bouiller, K.; Botelho-Nevers, E.; Gagneux-Brunon, A.; Chirouze, C.; Rodriguez-Nava, V.; Dumitrescu, O.; Triffault-Fillit, C.; Conrad, A.; Lebeaux, D.; et al. Prognosis and factors associated with disseminated nocardiosis: A ten-year multicenter study. J. Infect. 2022, 85, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. M62—Performance Standards for Susceptibility Testing of Mycobacetria, Nocardia spp., and Other Aerobic Actinomycetes, 1st ed.; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2018. [Google Scholar]
- Sánchez-Herrera, K.; Sandoval, H.; Mouniee, D.; Ramírez-Durán, N.; Bergeron, E.; Boiron, P.; Sánchez-Saucedo, N.; Rodríguez-Nava, V. Molecular identification of Nocardia species using the sodA gene: Identificación molecular de especies de Nocardia utilizando el gen sodA. New Microbes New Infect. 2017, 19, 96–116. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Wang, H.; Zhang, E.; Sintchenko, V.; Xiao, M.; Sorrell, T.C.; Chen, X.; Chen, S.C.-A. secA1 gene sequence polymorphisms for species identification of Nocardia species and recognition of intraspecies genetic diversity. J. Clin. Microbiol. 2010, 48, 3928–3934. [Google Scholar] [CrossRef] [PubMed]
- Conville, P.S.; Murray, P.R.; Zelazny, A.M. Evaluation of the integrated database network system (IDNS) SmartGene software for analysis of 16S rRNA gene sequences for identification of Nocardia species. J. Clin. Microbiol. 2010, 48, 2995–2998. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Nava, V.; Couble, A.; Devulder, G.; Flandrois, J.-P.; Boiron, P.; Laurent, F. Use of PCR-restriction enzyme pattern analysis and sequencing database for hsp65 gene-based identification of Nocardia species. J. Clin. Microbiol. 2006, 44, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Dingle, T.C.; Butler-Wu, S.M. Maldi-tof mass spectrometry for microorganism identification. Clin. Lab. Med. 2013, 33, 589–609. [Google Scholar] [CrossRef] [PubMed]
- Body, B.A.; Beard, M.A.; Slechta, E.S.; Hanson, K.E.; Barker, A.P.; Babady, N.E.; McMillen, T.; Tang, Y.-W.; Brown-Elliott, B.A.; Iakhiaeva, E.; et al. Evaluation of the Vitek MS v3.0 Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry System for Identification of Mycobacterium and Nocardia Species. J. Clin. Microbiol. 2018, 56, e00237-18. [Google Scholar] [CrossRef] [PubMed]
- Durand, T.; Vautrin, F.; Bergeron, E.; Girard, V.; Polsinelli, S.; Monnin, V.; Durand, G.; Dauwalder, O.; Dumitrescu, O.; Laurent, F.; et al. Assessment of VITEK® MS IVD database V3.0 for identification of Nocardia spp. using two culture media and comparing direct smear and protein extraction procedures. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Girard, V.; Mailler, S.; Polsinelli, S.; Jacob, D.; Saccomani, M.C.; Celliere, B.; Monnin, V.; van Belkum, A.; Hagen, F.; Meis, J.F.; et al. Routine identification of Nocardia species by MALDI-TOF mass spectrometry. Diagn. Microbiol. Infect. Dis. 2017, 87, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Temporal, D.; Zvezdánova, M.E.; Benedí, P.; Marín, M.; Blázquez-Sánchez, M.; Ruiz-Serrano, M.J.; Muñoz, P.; Rodríguez-Sánchez, B. Identification of Nocardia and non-tuberculous Mycobacterium species by MALDI-TOF MS using the VITEK MS coupled to IVD and RUO databases. Microb. Biotechnol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Haigh, J.; Degun, A.; Eydmann, M.; Millar, M.; Wilks, M. Improved performance of bacterium and yeast identification by a commercial matrix-assisted laser desorption ionization-time of flight mass spectrometry system in the clinical microbiology laboratory. J. Clin. Microbiol. 2011, 49, 3441. [Google Scholar] [CrossRef] [PubMed]
- Conville, P.S.; Brown-Elliott, B.A.; Smith, T.; Zelazny, A.M. The Complexities of Nocardia Taxonomy and Identification. J. Clin. Microbiol. 2018, 56, e01419-17. [Google Scholar] [CrossRef] [PubMed]
- Marín, M.; Ruiz, A.; Iglesias, C.; Quiroga, L.; Cercenado, E.; Martín-Rabadán, P.; Bouza, E.; Rodríguez-Sánchez, B. Identification of Nocardia species from clinical isolates using MALDI-TOF mass spectrometry. Clin. Microbiol. Infect. 2018, 24, 1342.e5–1342.e8. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Bergeron, E.; Berthet, J.; Djadi-Prat, J.; Mouniée, D.; Boiron, P.; Lortholary, O.; Rodriguez-Nava, V. Antibiotic susceptibility testing and species identification of Nocardia isolates: A retrospective analysis of data from a French expert laboratory, 2010–2015. Clin. Microbiol. Infect. 2019, 25, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Nouioui, I.; Cortés-Albayay, C.; Neumann-Schaal, M.; Vicente, D.; Cilla, G.; Klenk, H.-P.; Marimón, J.M.; Ercibengoa, M. Genomic Virulence Features of Two Novel Species Nocardia barduliensis sp. nov. and Nocardia gipuzkoensis sp. nov., Isolated from Patients with Chronic Pulmonary Diseases. Microorganisms 2020, 8, 1517. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, A.; Poonwan, N.; Yazawa, K.; Suzuki, S.; Kroppenstedt, R.; Mikami, Y. Nocardia vermiculata sp. nov. and Nocardia thailandica sp. nov. Isolated from Clinical Specimens. Actinomycetologica 2004, 18, 27–33. [Google Scholar] [CrossRef]
- le Roes, M.; Meyers, P.R. Nocardia gamkensis sp. nov. Antonie Van Leeuwenhoek 2006, 90, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. M24—Susceptibility Testing of Mycobacteria, Nocardia spp., and Other Aerobic Actinomycetes, 2nd ed.; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2012. [Google Scholar]
- Lafont, E.; Conan, P.-L.; Rodriguez-Nava, V.; Lebeaux, D. Invasive Nocardiosis: Disease Presentation, Diagnosis and Treatment—Old Questions, New Answers? Infect. Drug Resist. 2020, 13, 4601–4613. [Google Scholar] [CrossRef] [PubMed]
- Cloud, J.L.; Conville, P.S.; Croft, A.; Harmsen, D.; Witebsky, F.G.; Carroll, K.C. Evaluation of partial 16S ribosomal DNA sequencing for identification of nocardia species by using the MicroSeq 500 system with an expanded database. J. Clin. Microbiol. 2004, 42, 578–584. [Google Scholar] [CrossRef] [PubMed]
| Reference Identification | Number (%) of Isolates | |||||||
|---|---|---|---|---|---|---|---|---|
| Correct to Species Level | Correct to Complex Level | Incorrect | No. Identification | |||||
| 2 Wells/2 after the First Run | 1 Wells/2 after the First Run | >First Run (New Deposit) | Total | 1 Wells/2 after the First Run | Total | |||
| N. asiatica (n = 4) | 4 (100.0) | |||||||
| N. barduliensis N (n = 1) | 1 (100.0) | |||||||
| N. brasilliensis (n = 9) | 4 (44.4) | 5 (55.6) | 9 (100.0) | |||||
| N. cyriacigeorgica (n = 32) | 14 (43.7) | 10 (31.3) | 6 (18.7) | 30 (93.7) | 2 (6.3) | |||
| N. elegans N (n = 1) | 1 (100.0) | 1 (100.0) | ||||||
| N. farcinica (n = 31) | 16 (51.6) | 12 (38.7) | 2 (6.5) | 30 (96.8) | 1 (3.2) | |||
| N. flavorosea N (n = 1) | 1 (100.0) | |||||||
| N. gamkensis N (n = 8) | 1 (12.5) | 7 (87.5) | ||||||
| N. gipuzkoensis N (n = 4) | 2 (50.0) | 2 (50.0) | ||||||
| N. nova (n = 25) | 15 (60.0) | 7 (28.0) | 3 (12.0) | 25 (100.0) | ||||
| N. otitidiscavarium (n = 1) | 1 (100.0) | 1 (100.0) | ||||||
| N. pseudobrasiliensis (n = 1) | 1 (100.0) | 1 (100.0) | ||||||
| N. puris N (n = 2) | 2 (100.0) | |||||||
| N. shimofusensis N (n = 1) | 1 (100.0) | |||||||
| N. spp N* (n = 3) | 3 (100.0) | |||||||
| N. terpenica N (n = 1) | 1 (100.0) | |||||||
| N. thailandica N (n = 1) | 1 (100.0) | |||||||
| N. veterana (n = 2) | 1 (50.0) | 1 (50.0) | 2 (100.0) | |||||
| N. wallacei (n = 6) | 1 (16.7) | 3 (50.0) | 2 (33.3) | 6 (100.0) | ||||
| Total (n = 134) | 53 (39.6) | 37 (27.6) | 14 (10.4) | 104 (77.6) | 1 (0.7) | 1 (0.7) | 4 (3.0) | 25 (18.7) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hodille, E.; Prudhomme, C.; Dumitrescu, O.; Benito, Y.; Dauwalder, O.; Lina, G. Rapid, Easy, and Reliable Identification of Nocardia sp. by MALDI-TOF Mass Spectrometry, VITEK®-MS IVD V3.2 Database, Using Direct Deposit. Int. J. Mol. Sci. 2023, 24, 5469. https://doi.org/10.3390/ijms24065469
Hodille E, Prudhomme C, Dumitrescu O, Benito Y, Dauwalder O, Lina G. Rapid, Easy, and Reliable Identification of Nocardia sp. by MALDI-TOF Mass Spectrometry, VITEK®-MS IVD V3.2 Database, Using Direct Deposit. International Journal of Molecular Sciences. 2023; 24(6):5469. https://doi.org/10.3390/ijms24065469
Chicago/Turabian StyleHodille, Elisabeth, Clémence Prudhomme, Oana Dumitrescu, Yvonne Benito, Olivier Dauwalder, and Gérard Lina. 2023. "Rapid, Easy, and Reliable Identification of Nocardia sp. by MALDI-TOF Mass Spectrometry, VITEK®-MS IVD V3.2 Database, Using Direct Deposit" International Journal of Molecular Sciences 24, no. 6: 5469. https://doi.org/10.3390/ijms24065469
APA StyleHodille, E., Prudhomme, C., Dumitrescu, O., Benito, Y., Dauwalder, O., & Lina, G. (2023). Rapid, Easy, and Reliable Identification of Nocardia sp. by MALDI-TOF Mass Spectrometry, VITEK®-MS IVD V3.2 Database, Using Direct Deposit. International Journal of Molecular Sciences, 24(6), 5469. https://doi.org/10.3390/ijms24065469






