Selective Targeting of Class I HDAC Reduces Microglial Inflammation in the Entorhinal Cortex of Young APP/PS1 Mice
Abstract
1. Introduction
2. Results
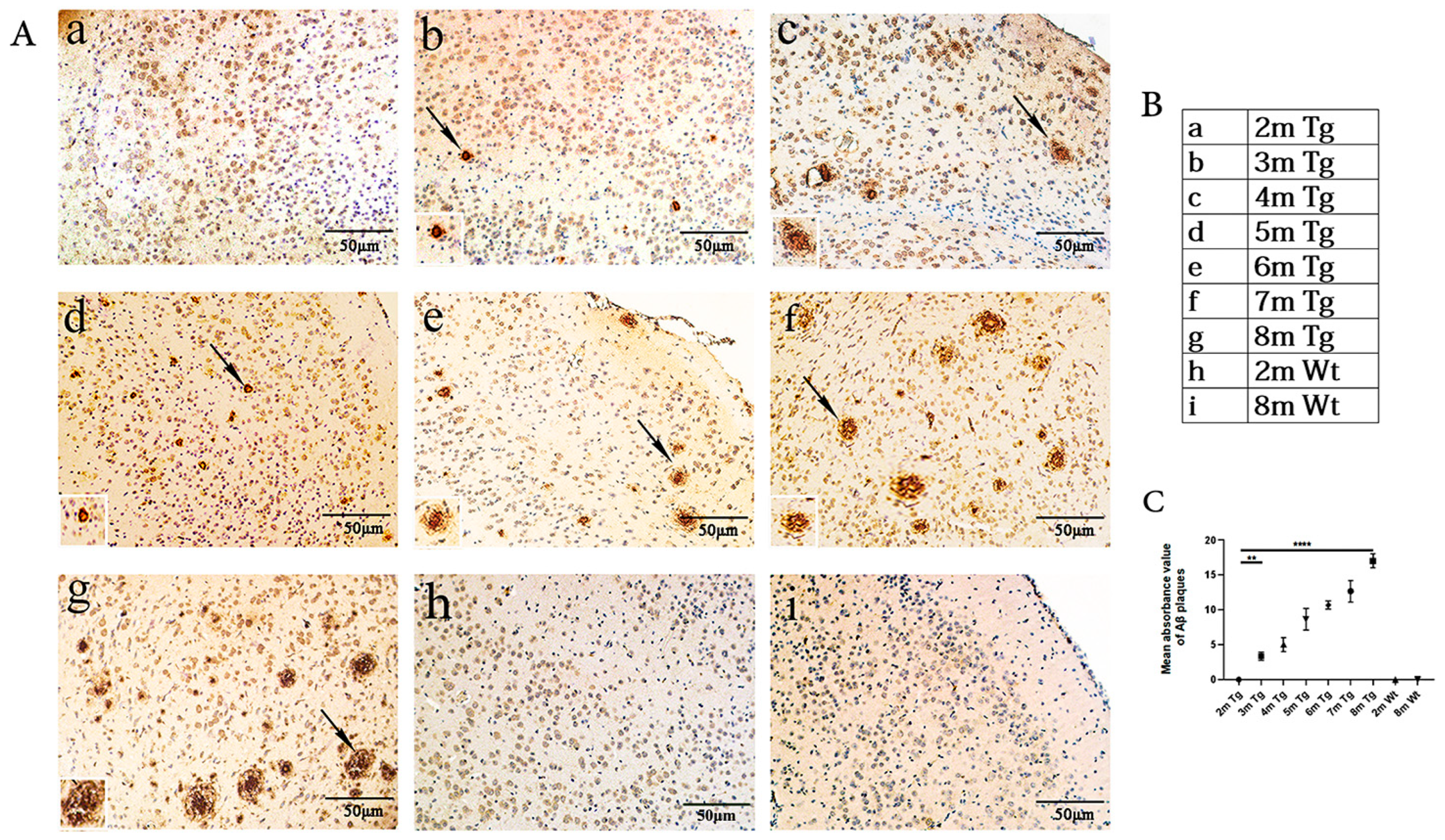
2.1. Age-Related Changes in Aβ in the Entorhinal Cortexes of the APP/PS1 and Wt Mice
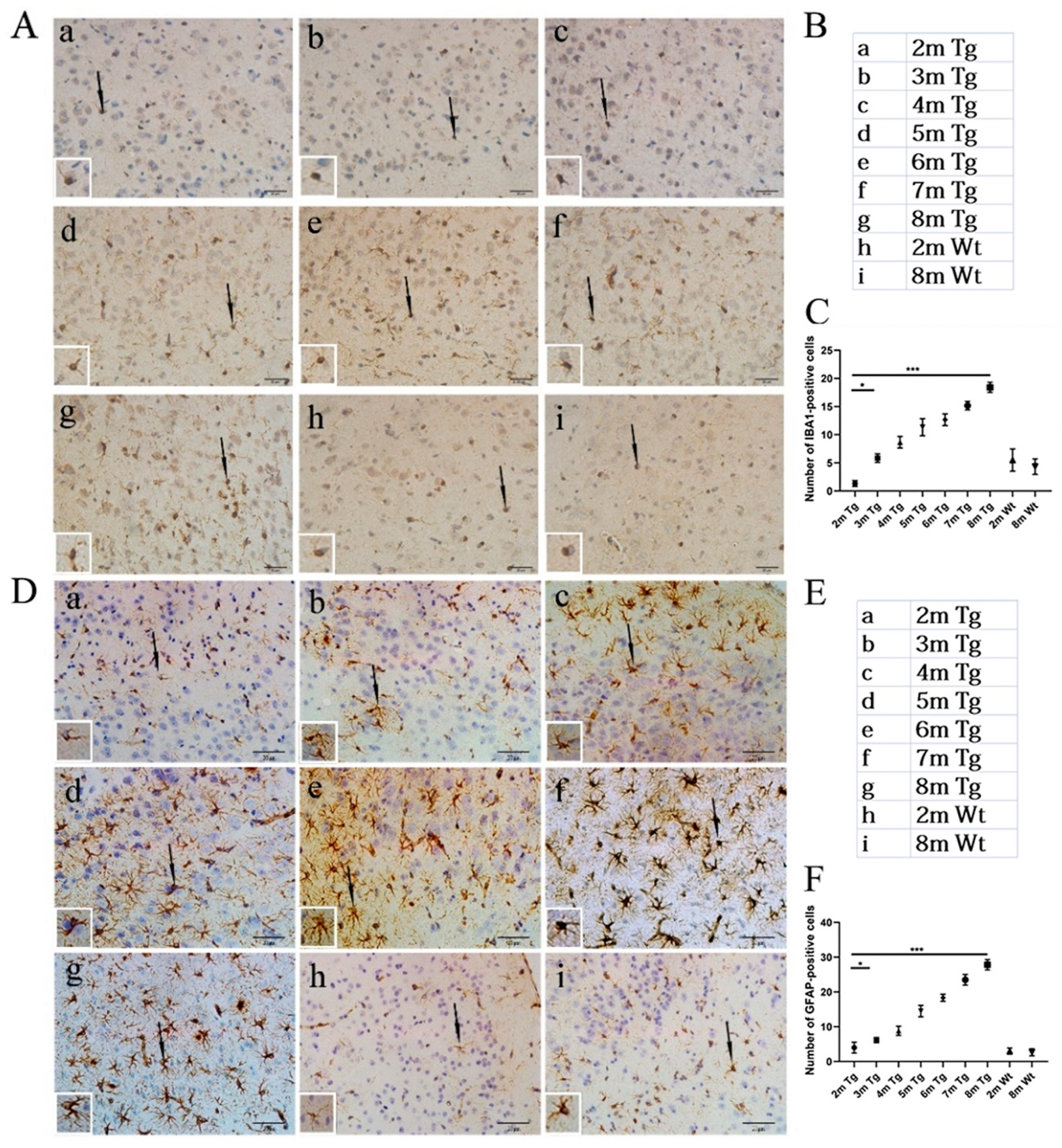
2.2. Age-Related Changes in Microglia and Astrocytes in the Entorhinal Cortexes of the APP/PS1 and Wt Mice
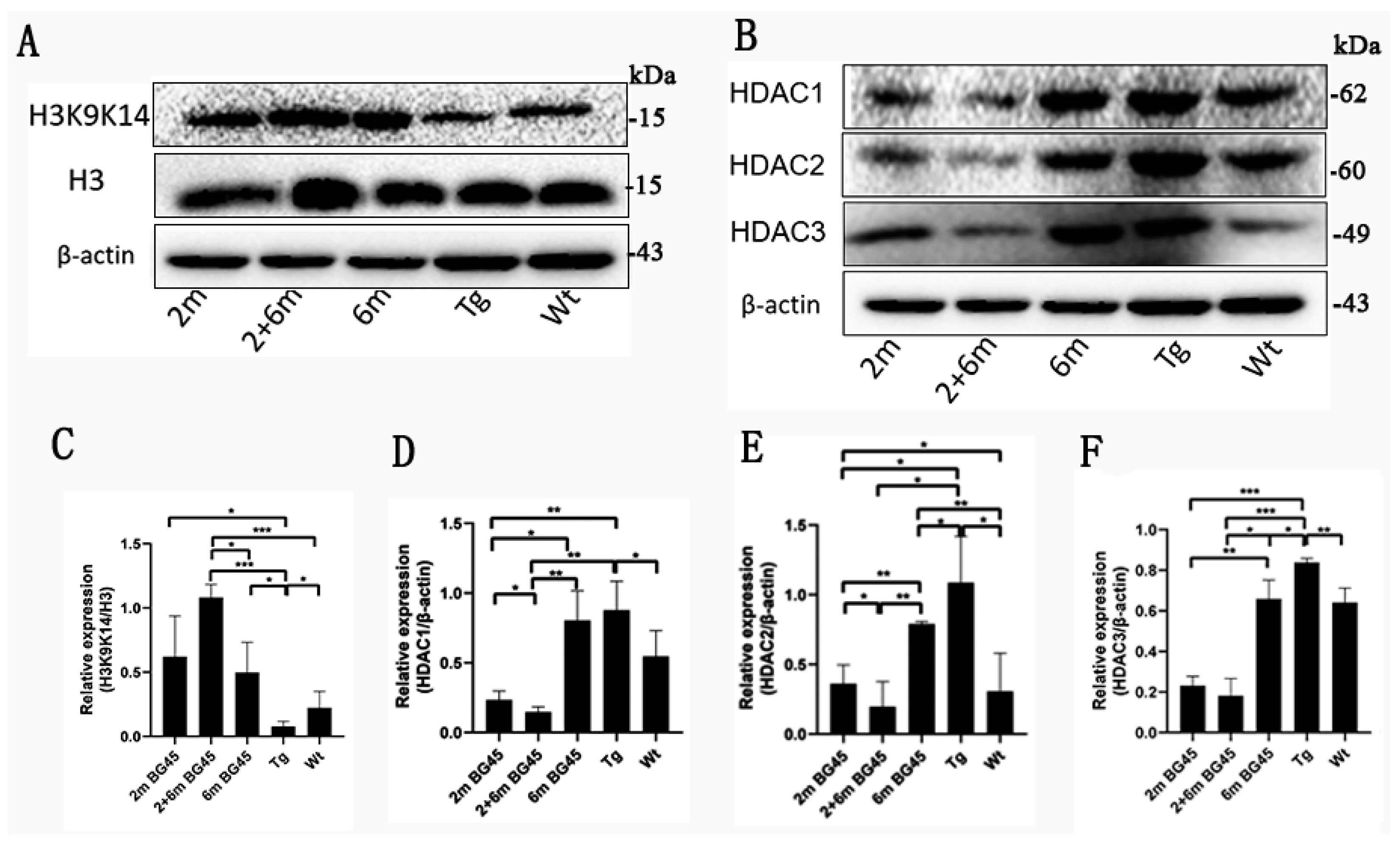
2.3. BG45 Promoted H3K9K14/H3 Acetylation and Inhibited the Expression of HDAC1, HDAC2, and HDAC3 in the Entorhinal Cortexes of the APP/PS1 Mice
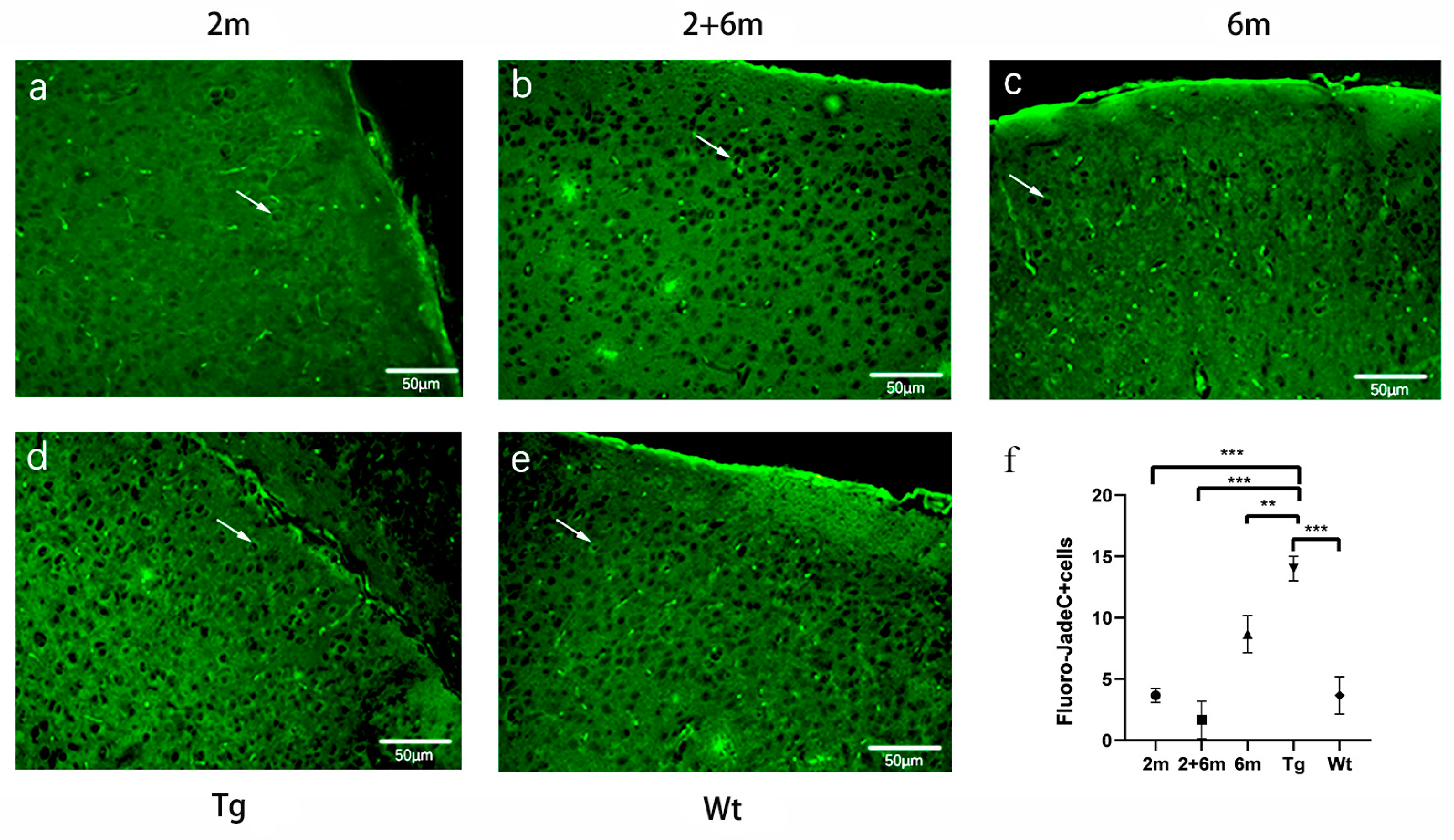
2.4. BG45 Alleviated Neuronal Degeneration in the Entorhinal Cortexes of the APP/PS1 Mice
2.5. BG45 Reduced Aβ Deposition and Downregulated p-tau Expression in the Entorhinal Cortexes of the APP/PS1 Mice
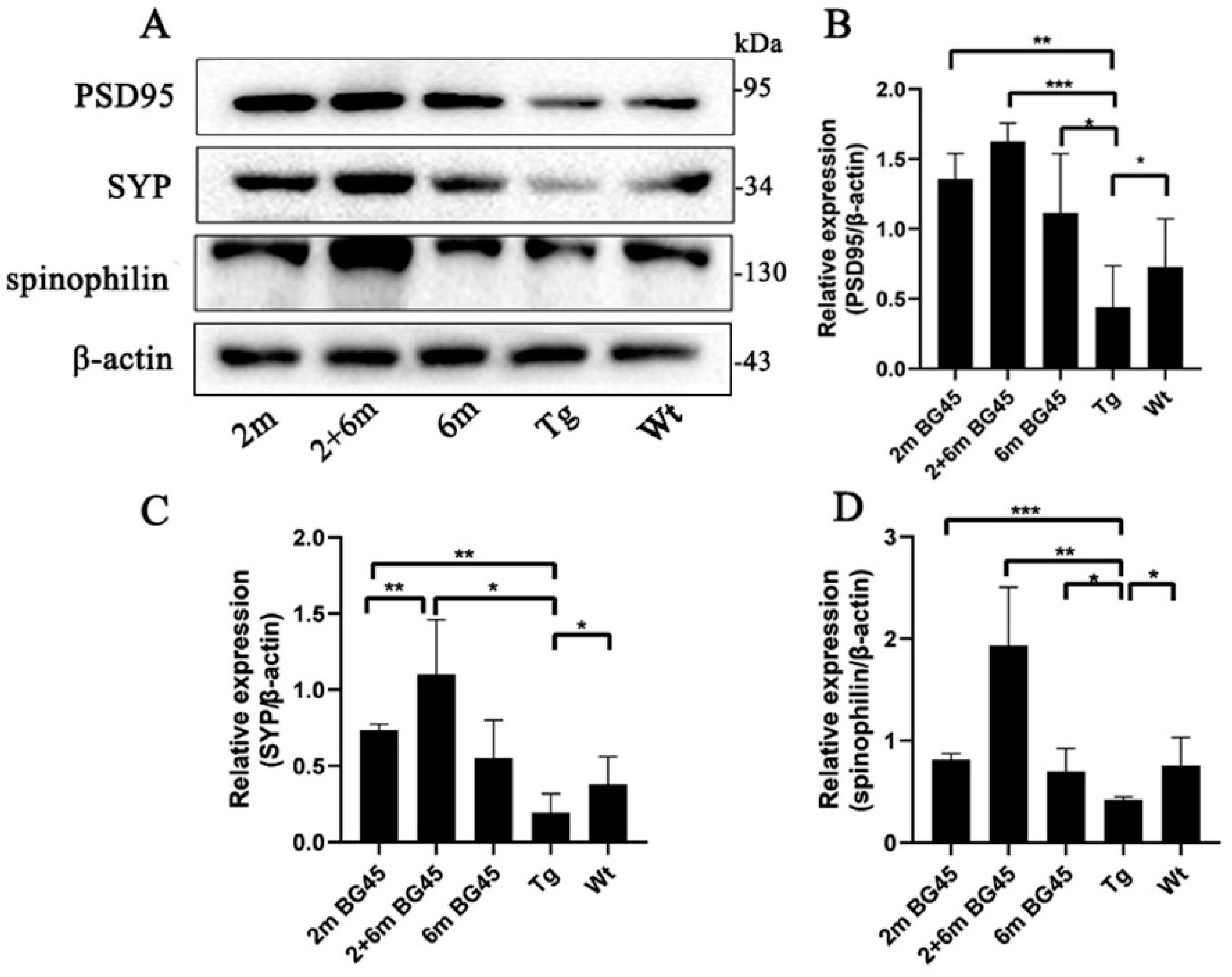
2.6. BG45 Increased Synaptic Protein Expression in the Entorhinal Corteesx of the APP/PS1 Mice
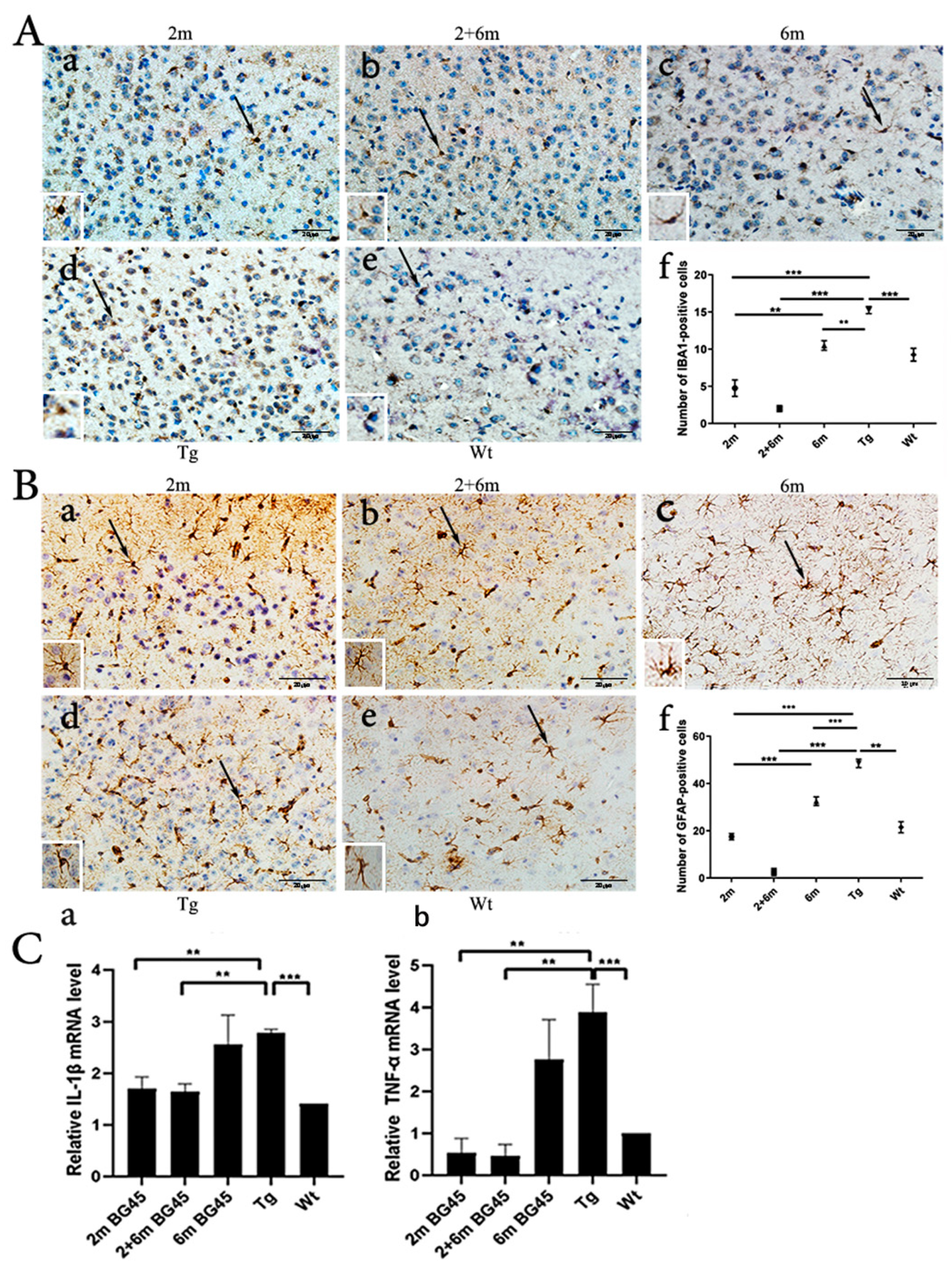
2.7. BG45 Decreased the Number of IBA1-Positive Microglia and GFAP-Positive Astrocytes and Downregulated the Levels of IL-1β and TNF-α Gene Expression in the Entorhinal Cortexes of the APP/PS1 Mice
2.8. BG45 Changed the Expression of Key Factors in the CREB/BDNF/NF-kB Pathway in the Entorhinal Cortexes of the APP/PS1 Mice
3. Discussion
4. Materials and Methods
4.1. Animals and Drug Administration
4.2. Immunohistochemistry Staining
4.3. Western Blotting
4.4. Fluoro-Jade C Staining
4.5. Real-Time Quantitative Polymerase Chain Reaction
- TNF-α
- 5-GACGTGGAACTGGCAGAAGAG-3
- 5-TTGGTGGTTTGTGAGTGTGAG-3
- IL-1β
- 5-GCCCATCCTCTGTGACTCAT-3
- 5-AGGCCACAGGTATTTTGTCG-3
- GAPDH
- 5-GAGCCCTTCCACAATGCCAAAGTT-3
- 5-TGTGATGGGTGTGAACCACGAGAA-3
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barnes, J.; Dickerson, B.C.; Frost, C.; Jiskoot, L.C.; Wolk, D.; van der Flier, W.M. Alzheimer’s disease first symptoms are age dependent: Evidence from the NACC dataset. Alzheimers Dement. 2015, 11, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer’s association in USA 2020, Alzheimer’s disease facts and figures. Alzheimers Dement. 2020, 16, 391–460. [CrossRef] [PubMed]
- Oboudiyat, C.; Glazer, H.; Seifan, A.; Greer, C.; Isaacson, R.S. Alzheimer’s Disease. Semin. Neurol. 2013, 33, 313–329. [Google Scholar] [CrossRef] [PubMed]
- Janelsins, M.C.; Mastrangelo, M.A.; Oddo, S.; LaFerla, F.M.; Federoff, H.J.; Bowers, W.J. Early correlation of microglial activation with enhanced tumor necrosis factor-alpha and monocyte chemoattractant protein-1 expression specifically within the entorhinal cortex of triple transgenic Alzheimer’s disease mice. J. Neuroinflamm 2005, 2, 23. [Google Scholar] [CrossRef]
- Belfiore, R.; Rodin, A.; Ferreira, E.; Velazquez, R.; Branca, C.; Caccamo, A.; Oddo, S. Temporal and regional progression of Alzheimer’s disease-like pathology in 3xTg-AD mice. Aging Cell 2019, 18, e12873. [Google Scholar] [CrossRef]
- Duffy, A.M.; Morales-Corraliza, J.; Bermudez-Hernandez, K.M.; Schaner, M.J.; Magagna-Poveda, A.; Mathews, P.M.; Scharfman, H.E. Entorhinal cortical defects in Tg2576 mice are present as early as 2–4 months of age. Neurobiol. Aging 2015, 36, 134–148. [Google Scholar] [CrossRef]
- Garcia-Alloza, M.; Robbins, E.M.; Zhang-Nunes, S.X.; Purcell, S.M.; Betensky, R.A.; Raju, S.; Prada, C.; Greenberg, S.M.; Bacskai, B.J.; Frosch, M.P. Characterization of amyloid deposition in the APPswe/PS1dE9 mouse model of Alzheimer disease. Neurobiol. Dis. 2006, 24, 516–524. [Google Scholar] [CrossRef]
- Alzheimers, A. 2012 Alzheimer’s disease facts and figures. Alzheimers Dement. 2012, 8, 131–168. [Google Scholar]
- Nikolac Perkovic, M.; Videtic Paska, A.; Konjevod, M.; Kouter, K.; Svob Strac, D.; Nedic Erjavec, G.; Pivac, N. Epigenetics of Alzheimer’s Disease. Biomolecules 2021, 11, 195. [Google Scholar] [CrossRef]
- Fukuda, H.; Sano, N.; Muto, S.; Horikoshi, M. Simple histone acetylation plays a complex role in the regulation of gene expression. Brief. Funct. Genom. Proteom. 2006, 5, 190–208. [Google Scholar] [CrossRef]
- Wu, X.; Chen, P.S.; Dallas, S.; Wilson, B.; Block, M.L.; Wang, C.C.; Kinyamu, H.; Lu, N.; Gao, X.; Leng, Y.; et al. Histone deacetylase inhibitors up-regulate astrocyte GDNF and BDNF gene transcription and protect dopaminergic neurons. Int. J. Neuropsychopharmacol. 2008, 11, 1123–1134. [Google Scholar] [CrossRef] [PubMed]
- Peserico, A.; Simone, C. Physical and Functional HAT/HDAC Interplay Regulates Protein Acetylation Balance. J. Biomed. Biotechnol. 2011, 2011, 371832. [Google Scholar] [CrossRef] [PubMed]
- Wey, H.Y.; Gilbert, T.M.; Zürcher, N.R.; She, A.; Bhanot, A.; Taillon, B.D.; Schroeder, F.A.; Wang, C.; Haggarty, S.J.; Hooker, J.M. Insights into neuroepigenetics through human histone deacetylase PET imaging. Sci. Transl. Med. 2016, 8, 351ra106. [Google Scholar] [CrossRef]
- Anderson, K.W.; Chen, J.; Wang, M.; Mast, N.; Pikuleva, I.A.; Turko, I.V. Quantification of Histone Deacetylase Isoforms in Human Frontal Cortex, Human Retina, and Mouse Brain. PLoS ONE 2015, 10, e0126592. [Google Scholar] [CrossRef]
- Sharma, S.; Sarathlal, K.C.; Taliyan, R. Epigenetics in Neurodegenerative Diseases: The Role of Histone Deacetylases. CNS Neurol. Disord. Drug Targets 2019, 18, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Gupt, R.; Ambast, R.K.; Kumar, P. Histone deacetylase in neuropathology. Adv. Clin. Chem. 2021, 104, 151–231. [Google Scholar]
- de Ruijter, A.J.; van Gennip, A.H.; Caron, H.N.; Kemp, S.; van Kuilenburg, A.B. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003, 370, 737–749. [Google Scholar] [CrossRef]
- Kumar, V.; Kundu, S.; Singh, A.; Singh, S. Understanding the Role of Histone Deacetylase and their Inhibitors in Neurodegenerative Disorders: Current Targets and Future Perspective. Curr. Neuropharmacol. 2022, 20, 158–178. [Google Scholar] [CrossRef]
- Yamakawa, H.; Cheng, J.; Penney, J.; Gao, F.; Rueda, R.; Wang, J.; Yamakawa, S.; Kritskiy, O.; Gjoneska, E.; Tsai, L.H. The Transcription Factor Sp3 Cooperates with HDAC2 to Regulate Synaptic Function and Plasticity in Neurons. Cell Rep. 2017, 20, 1319–1334. [Google Scholar] [CrossRef]
- Guan, J.S.; Haggarty, S.J.; Giacometti, E.; Dannenberg, J.H.; Joseph, N.; Gao, J.; Nieland, T.J.; Zhou, Y.; Wang, X.; Mazitschek, R.; et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 2009, 459, 55–60. [Google Scholar] [CrossRef]
- Janczura, K.J.; Volmar, C.H.; Sartor, G.C.; Rao, S.J.; Ricciardi, N.R.; Lambert, G.; Brothers, S.P.; Wahlestedt, C. Inhibition of HDAC3 reverses Alzheimer’s disease-related pathologies in vitro and in the 3xTg-AD mouse model. Proc. Natl. Acad. Sci. USA 2018, 115, E11148–E11157. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, S.; Yu, L.; Jin, J.; Ye, X.; Liu, Y.; Xu, Y. HDAC3 negatively regulates spatial memory in a mouse model of Alzheimer’s disease. Aging Cell 2017, 16, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Bose, P.; Dai, Y.; Grant, S. Histone deacetylase inhibitor (HDACI) mechanisms of action: Emerging insights. Pharmacol. Ther. 2014, 143, 323–336. [Google Scholar] [CrossRef]
- Gräff, J.; Rei, D.; Guan, J.S.; Wang, W.Y.; Seo, J.; Hennig, K.M.; Nieland, T.J.; Fass, D.M.; Kao, P.F.; Kahn, M.; et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature 2012, 483, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Takada, N.; Nakamura, Y.; Ikeda, K.; Takaoka, N.; Hisaoka-Nakashima, K.; Sanoh, S.; Kotake, Y.; Nakata, Y.; Morioka, N. Treatment with Histone Deacetylase Inhibitor Attenuates Peripheral Inflammation-Induced Cognitive Dysfunction and Microglial Activation: The Effect of SAHA as a Peripheral HDAC Inhibitor. Neurochem. Res. 2021, 46, 2285–2296. [Google Scholar] [CrossRef] [PubMed]
- Kaldun, J.C.; Sprecher, S.G. Initiated by CREB: Resolving Gene Regulatory Programs in Learning and Memory Switch in Cofactors and Transcription Regulators between Memory Consolidation and Maintenance Network. Bioessays 2019, 41, e1900045. [Google Scholar] [CrossRef] [PubMed]
- Benito, E.; Barco, A. CREB’s control of intrinsic and synaptic plasticity: Implications for CREB-dependent memory models. Trends Neurosci. 2010, 33, 230–240. [Google Scholar] [CrossRef]
- Esvald, E.E.; Tuvikene, J.; Sirp, A.; Patil, S.; Bramham, C.R.; Timmusk, T. CREB Family Transcription Factors Are Major Mediators of BDNF Transcriptional Autoregulation in Cortical Neurons. J. Neurosci. 2020, 40, 1405–1426. [Google Scholar] [CrossRef]
- Bi, Q.R.; Hou, J.J.; Qi, P.; Ma, C.H.; Shen, Y.; Feng, R.H.; Yan, B.P.; Wang, J.W.; Shi, X.J.; Zheng, Y.Y.; et al. Venenum Bufonis induces rat neuroinflammation by activiating NF-κB pathway and attenuation of BDNF. J. Ethnopharmacol. 2016, 186, 103–110. [Google Scholar] [CrossRef]
- Lima Giacobbo, B.; Doorduin, J.; Klein, H.C.; Dierckx, R.A.J.O.; Bromberg, E.; de Vries, E.F.J. Brain-Derived Neurotrophic Factor in Brain Disorders: Focus on Neuroinflammation. Mol. Neurobiol. 2019, 56, 3295–3312. [Google Scholar] [CrossRef]
- Minami, J.; Suzuki, R.; Mazitschek, R.; Gorgun, G.; Ghosh, B.; Cirstea, D.; Hu, Y.; Mimura, N.; Ohguchi, H.; Cottini, F.; et al. Histone deacetylase 3 as a novel therapeutic target in multiple myeloma. Leukemia 2014, 28, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Takehara-Nishiuchi, K. Entorhinal cortex and consolidated memory. Neurosci. Res. 2014, 84, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Pennanen, C.; Kivipelto, M.; Tuomainen, S.; Hartikainen, P.; Hänninen, T.; Laakso, M.P.; Hallikainen, M.; Vanhanen, M.; Nissinen, A.; Helkala, E.L.; et al. Hippocampus and entorhinal cortex in mild cognitive impairment and early AD. Neurobiol. Aging 2004, 25, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Chen, L.; Liu, J.; Chen, J.; Wang, C.; Guo, Y.; Yu, X.; Zhang, C.; Chu, H.; Ma, H. A Class I HDAC Inhibitor Rescues Synaptic Damage and Neuron Loss in APP-Transfected Cells and APP/PS1 Mice through the GRIP1/AMPA Pathway. Molecules 2022, 27, 4160. [Google Scholar] [CrossRef]
- Han, Y.; Chen, L.; Guo, Y.; Wang, C.; Zhang, C.; Kong, L.; Ma, H. Class I HDAC Inhibitor Improves Synaptic Proteins and Repairs Cytoskeleton Through Regulating Synapse-Related Genes In vitro and In vivo. Front. Aging Neurosci. 2021, 12, 619866. [Google Scholar] [CrossRef]
- Kosel, F.; Pelley, J.M.S.; Franklin, T.B. Behavioural and psychological symptoms of dementia in mouse models of Alzheimer’s disease-related pathology. Neurosci. Biobehav. Rev. 2020, 112, 634–647. [Google Scholar] [CrossRef]
- Hansen, D.V.; Hanson, J.E.; Sheng, M. Microglia in Alzheimer’s disease. J. Cell Biol. 2018, 217, 459–472. [Google Scholar] [CrossRef]
- Delgado-Morales, R.; Esteller, M. Opening up the DNA methylome of dementia. Mol. Psychiatry 2017, 22, 485–496. [Google Scholar] [CrossRef]
- Lahiri, D.K.; Maloney, B.; Zawia, N.H. The LEARn model: An epigenetic explanation for idiopathic neurobiological diseases. Mol. Psychiatry 2009, 14, 992–1003. [Google Scholar] [CrossRef]
- Martínez-Iglesias, O.; Naidoo, V.; Carrera, I.; Cacabelos, R. Epigenetic Studies in the Male APP/BIN1/COPS5 Triple-Transgenic Mouse Model of Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 2446. [Google Scholar] [CrossRef]
- Narayan, P.J.; Lill, C.; Faull, R.; Curtis, M.A.; Dragunow, M. Increased acetyl and total histone levels in post-mortem Alzheimer’s disease brain. Neurobiol. Dis. 2015, 74, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Klein, H.U.; McCabe, C.; Gjoneska, E.; Sullivan, S.E.; Kaskow, B.J.; Tang, A.; Smith, R.V.; Xu, J.; Pfenning, A.R.; Bernstein, B.E.; et al. Epigenome-wide study uncovers large-scale changes in histone acetylation driven by tau pathology in aging and Alzheimer’s human brains. Nat. Neurosci. 2019, 22, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Marzi, S.J.; Leung, S.K.; Ribarska, T.; Hannon, E.; Smith, A.R.; Pishva, E.; Poschmann, J.; Moore, K.; Troakes, C.; Al-Sarraj, S.; et al. A histone acetylome-wide association study of Alzheimer’s disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat. Neurosci. 2018, 21, 1618–1627. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, C.; Wang, J.; Huang, Y.; Shen, D.; Hu, Y.; Chu, H.; Yu, X.; Zhang, L.; Ma, H. A Class I HDAC Inhibitor BG45 Alleviates Cognitive Impairment through the CaMKII/ITPKA/Ca Signaling Pathway. Pharmaceuticals 2022, 15, 1481. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yun, F.; Sui, J.; Liang, W.; Shen, D.; Zhang, Q. HAT- and HDAC-Targeted Protein Acetylation in the Occurrence and Treatment of Epilepsy. Biomedicines 2022, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Citraro, R.; Leo, A.; De Caro, C.; Nesci, V.; Gallo Cantafio, M.E.; Amodio, N.; Mattace Raso, G.; Lama, A.; Russo, R.; Calignano, A.; et al. Effects of Histone Deacetylase Inhibitors on the Development of Epilepsy and Psychiatric Comorbidity in WAG/Rij Rats. Mol. Neurobiol. 2020, 57, 408–421. [Google Scholar] [CrossRef] [PubMed]
- Amidfar, M.; de Oliveira, J.; Kucharska, E.; Budni, J.; Kim, Y.K. The role of CREB and BDNF in neurobiology and treatment of Alzheimer’s disease. Life Sci. 2020, 257, 118020. [Google Scholar] [CrossRef]
- Zhang, H.; Han, Y.; Zhang, L.; Jia, X.; Niu, Q. The GSK-3β/β-Catenin Signaling-Mediated Brain-Derived Neurotrophic Factor Pathway Is Involved in Aluminum-Induced Impairment of Hippocampal LTP In Vivo. Biol. Trace Elem. Res. 2021, 199, 4635–4645. [Google Scholar] [CrossRef]
- Yang, W.; Liu, Y.; Xu, Q.Q.; Xian, Y.F.; Lin, Z.X. Sulforaphene Ameliorates Neuroinflammation and Hyperphosphorylated Tau Protein via Regulating the PI3K/Akt/GSK-3 beta Pathway in Experimental Models of Alzheimer’s Disease. Oxidative Med. Cell. Longev. 2020, 2020, 4754195. [Google Scholar] [CrossRef]
- Elliott, E.; Atlas, R.; Lange, A.; Ginzburg, I. Brain-derived neurotrophic factor induces a rapid dephosphorylation of tau protein through a PI-3Kinase signalling mechanism. Eur. J. Neurosci. 2005, 22, 1081–1089. [Google Scholar] [CrossRef]
- Yoshii, A.; Constantine-Paton, M. Postsynaptic BDNF-TrkB Signaling in Synapse Maturation, Plasticity, and Disease. Dev. Neurobiol. 2010, 70, 304–322. [Google Scholar] [CrossRef] [PubMed]
- Andero, R.; Choi, D.; Ressler, K. BDNF-TrkB receptor regulation of distributed adult neural plasticity, memory formation, and psychiatric disorders. Prog. Mol. Biol. Transl. Sci. 2014, 122, 169–192. [Google Scholar] [PubMed]
- Chen, J.J.; Wang, T.; An, C.D.; Jiang, C.Y.; Zhao, J.; Li, S. Brain-derived neurotrophic factor: A mediator of inflammation-associated neurogenesis in Alzheimer’s disease. Rev. Neurosci. 2016, 27, 793–811. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Xiang, Y.; Qu, X.; Liu, H.; Liu, C.; Li, G.; Han, L.; Qin, X. Apelin-13 Suppresses Neuroinflammation Against Cognitive Deficit in a Streptozotocin-Induced Rat Model of Alzheimer’s Disease Through Activation of BDNF-TrkB Signaling Pathway. Front. Pharmacol. 2019, 10, 395. [Google Scholar] [CrossRef] [PubMed]
- Phan, M.L.; Gergues, M.M.; Mahidadia, S.; Jimenez-Castillo, J.; Vicario, D.S.; Bieszczad, K.M. HDAC3 Inhibitor RGFP966 Modulates Neuronal Memory for Vocal Communication Signals in a Songbird Model. Front. Syst. Neurosci. 2017, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Hervera, A.; Zhou, L.; Palmisano, I.; McLachlan, E.; Kong, G.; Hutson, T.H.; Danzi, M.C.; Lemmon, V.P.; Bixby, J.L.; Matamoros-Angles, A.; et al. PP4-dependent HDAC3 dephosphorylation discriminates between axonal regeneration and regenerative failure. EMBO J. 2019, 38, e101032. [Google Scholar] [CrossRef]
- Soria Lopez, J.A.; Gonzalez, H.M.; Leger, G.C. Alzheimer’s disease. Handb. Clin. Neurol. 2019, 167, 231–255. [Google Scholar]
- Reiss, A.B.; Arain, H.A.; Stecker, M.M.; Siegart, N.M.; Kasselman, L.J. Amyloid toxicity in Alzheimer’s disease. Rev. Neurosci. 2018, 29, 613–627. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, C.; Wang, C.; Huang, Y.; Liu, J.; Chu, H.; Ren, X.; Kong, L.; Ma, H. Thioredoxin-1 Is a Target to Attenuate Alzheimer-Like Pathology in Diabetic Encephalopathy by Alleviating Endoplasmic Reticulum Stress and Oxidative Stress. Front. Physiol. 2021, 12, 651105. [Google Scholar] [CrossRef]
- Huang, Q.; Zhang, C.; Dong, S.; Han, J.; Qu, S.; Xie, T.; Zhao, H.; Shi, Y. Asafoetida exerts neuroprotective effect on oxidative stress induced apoptosis through PI3K/Akt/GSK3 beta/Nrf2/HO-1 pathway. Chin. Med. 2022, 17, 83. [Google Scholar] [CrossRef]
- Ikenari, T.; Kurata, H.; Satoh, T.; Hata, Y.; Mori, T. Evaluation of Fluoro-Jade C Staining: Specificity and Application to Damaged Immature Neuronal Cells in the Normal and Injured Mouse Brain. Neuroscience 2020, 425, 146–156. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Shen, D.; Hu, Y.; Chen, J.; Liu, J.; Huang, Y.; Yu, X.; Chu, H.; Zhang, C.; Yin, L.; et al. Selective Targeting of Class I HDAC Reduces Microglial Inflammation in the Entorhinal Cortex of Young APP/PS1 Mice. Int. J. Mol. Sci. 2023, 24, 4805. https://doi.org/10.3390/ijms24054805
Wang C, Shen D, Hu Y, Chen J, Liu J, Huang Y, Yu X, Chu H, Zhang C, Yin L, et al. Selective Targeting of Class I HDAC Reduces Microglial Inflammation in the Entorhinal Cortex of Young APP/PS1 Mice. International Journal of Molecular Sciences. 2023; 24(5):4805. https://doi.org/10.3390/ijms24054805
Chicago/Turabian StyleWang, Chunyang, Di Shen, Yingqiu Hu, Jie Chen, Jingyun Liu, Yufei Huang, Xuebin Yu, Haiying Chu, Chenghong Zhang, Liangwei Yin, and et al. 2023. "Selective Targeting of Class I HDAC Reduces Microglial Inflammation in the Entorhinal Cortex of Young APP/PS1 Mice" International Journal of Molecular Sciences 24, no. 5: 4805. https://doi.org/10.3390/ijms24054805
APA StyleWang, C., Shen, D., Hu, Y., Chen, J., Liu, J., Huang, Y., Yu, X., Chu, H., Zhang, C., Yin, L., Liu, Y., & Ma, H. (2023). Selective Targeting of Class I HDAC Reduces Microglial Inflammation in the Entorhinal Cortex of Young APP/PS1 Mice. International Journal of Molecular Sciences, 24(5), 4805. https://doi.org/10.3390/ijms24054805




