Modified mRNA as a Treatment for Myocardial Infarction
Abstract
1. Introduction
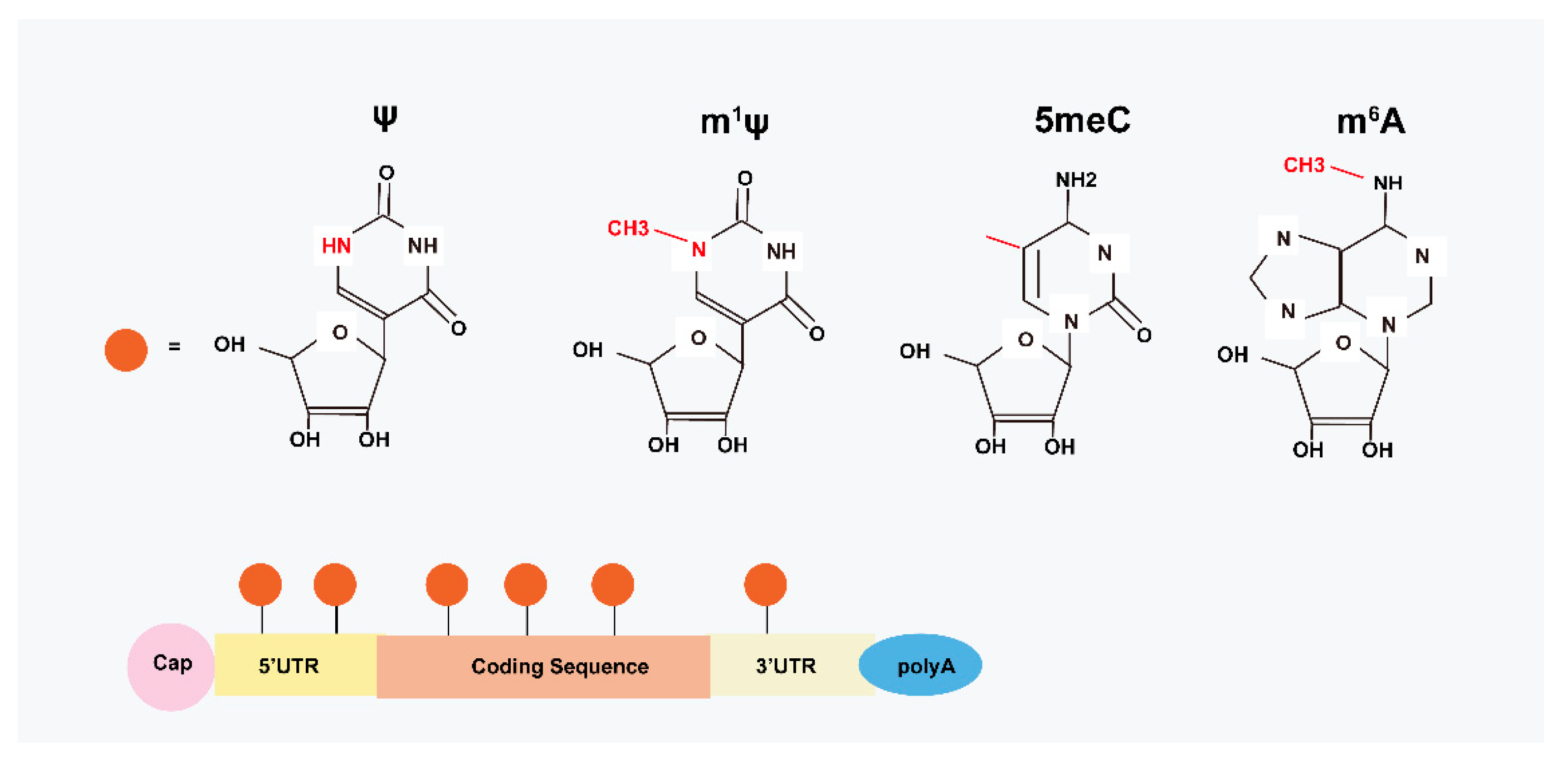
2. Structural Basis of modRNA
3. Delivery Vectors and Methods for modRNA in MI
3.1. The Viral Delivery Vectors
3.2. The Non-Viral Vectors
3.3. Delivery Method for modRNA
4. Modified mRNA-Based Therapy in MI Treatment
5. Limitations and Future Directions of modRNA-Based Therapy in MI Treatment
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update from the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Tsao, C.W.; Aday, A.W.; Almarzooq, Z.I.; Alonso, A.; Beaton, A.Z.; Bittencourt, M.S.; Boehme, A.K.; Buxton, A.E.; Carson, A.P.; Commodore-Mensah, Y.; et al. Heart Disease and Stroke Statistics—2022 Update: A Report from the American Heart Association. Circulation 2022, 145, e153–e639. [Google Scholar] [CrossRef]
- Notari, M.; Ventura-Rubio, A.; Bedford-Guaus, S.J.; Jorba, I.; Mulero, L.; Navajas, D.; Martí, M.; Raya, Á. The local microenvironment limits the regenerative potential of the mouse neonatal heart. Sci. Adv. 2018, 4, eaao5553. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef] [PubMed]
- Garnett, E.R.; Raines, R.T. Emerging biological functions of ribonuclease 1 and angiogenin. Crit. Rev. Biochem. Mol. Biol. 2021, 57, 244–260. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Medina, S.G.; Kushawah, G.; DeVore, M.L.; Castellano, L.A.; Hand, J.M.; Wright, M.; Bazzini, A.A. Translation affects mRNA stability in a codon-dependent manner in human cells. Elife 2019, 8, e45396. [Google Scholar] [CrossRef]
- Jain, A.; Mittal, S.; Tripathi, L.P.; Nussinov, R.; Ahmad, S. Host-pathogen protein-nucleic acid interactions: A comprehensive review. Comput. Struct. Biotechnol. J. 2022, 20, 4415–4436. [Google Scholar] [CrossRef]
- Li, W.; Wang, H.; Zheng, S.J. Roles of RNA Sensors in Host Innate Response to Influenza Virus and Coronavirus Infections. Int. J. Mol. Sci. 2022, 23, 8285. [Google Scholar] [CrossRef]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef]
- Ilan, L.; Osman, F.; Namer, L.S.; Eliahu, E.; Cohen-Chalamish, S.; Ben-Asouli, Y.; Banai, Y.; Kaempfer, R. PKR activation and eIF2α phosphorylation mediate human globin mRNA splicing at spliceosome assembly. Cell Res. 2017, 27, 688–704. [Google Scholar] [CrossRef]
- Donnelly, N.; Gorman, A.; Gupta, S.; Samali, A. The eIF2α kinases: Their structures and functions. Cell. Mol. Life Sci. 2013, 70, 3493–3511. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Y.L. Modified mRNA-Based Vaccines Against Coronavirus Disease 2019. Cell Transplant. 2022, 31, 9636897221090259. [Google Scholar] [CrossRef] [PubMed]
- Nwokeoji, A.O.; Chou, T.; Nwokeoji, E.A. Low Resource Integrated Platform for Production and Analysis of Capped mRNA. ACS Synth. Biol. 2022, 12, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.-H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef]
- Asrani, K.H.; Farelli, J.D.; Stahley, M.R.; Miller, R.L.; Cheng, C.J.; Subramanian, R.R.; Brown, J.M. Optimization of mRNA untranslated regions for improved expression of therapeutic mRNA. RNA Biol. 2018, 15, 756–762. [Google Scholar] [CrossRef] [PubMed]
- Sultana, N.; Hadas, Y.; Sharkar, M.T.K.; Kaur, K.; Magadum, A.; Kurian, A.A.; Hossain, N.; Alburquerque, B.; Ahmed, S.; Chepurko, E.; et al. Optimization of 5′ Untranslated Region of Modified mRNA for Use in Cardiac or Hepatic Ischemic Injury. Mol. Ther.-Methods Clin. Dev. 2020, 17, 622–633. [Google Scholar] [CrossRef] [PubMed]
- Sample, P.J.; Wang, B.; Reid, D.W.; Presnyak, V.; McFadyen, I.J.; Morris, D.R.; Seelig, G. Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nat. Biotechnol. 2019, 37, 803–809. [Google Scholar] [CrossRef]
- Suknuntha, K.; Tao, L.; Brok-Volchanskaya, V.; D’Souza, S.S.; Kumar, A.; Slukvin, I. Optimization of Synthetic mRNA for Highly Efficient Translation and its Application in the Generation of Endothelial and Hematopoietic Cells from Human and Primate Pluripotent Stem Cells. Stem Cell Rev. Rep. 2018, 14, 525–534. [Google Scholar] [CrossRef]
- von Niessen, A.G.O.; Poleganov, M.A.; Rechner, C.; Plaschke, A.; Kranz, L.M.; Fesser, S.; Diken, M.; Löwer, M.; Vallazza, B.; Beissert, T.; et al. Improving mRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3′ UTRs Identified by Cellular Library Screening. Mol. Ther. 2018, 27, 824–836. [Google Scholar] [CrossRef]
- Lima, S.A.; Chipman, L.B.; Nicholson, A.L.; Chen, Y.-H.; Yee, B.A.; Yeo, E.; Coller, J.; Pasquinelli, A.E. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 2017, 24, 1057–1063. [Google Scholar] [CrossRef]
- Jahnz-Wechmann, Z.; Framski, G.R.; Januszczyk, P.A.; Boryski, J. Base-Modified Nucleosides: Etheno Derivatives. Front. Chem. 2016, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.R.; Muramatsu, H.; Nallagatla, S.R.; Bevilacqua, P.C.; Sansing, L.H.; Weissman, D.; Karikó, K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010, 38, 5884–5892. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Tacuba, L.; Rojas, M.; Arias, C.F.; López, S. Rotavirus Controls Activation of the 2′-5′-Oligoadenylate Synthetase/RNase L Pathway Using at Least Two Distinct Mechanisms. J. Virol. 2015, 89, 12145–12153. [Google Scholar] [CrossRef]
- Uchida, S.; Kataoka, K.; Itaka, K. Screening of mRNA Chemical Modification to Maximize Protein Expression with Reduced Immunogenicity. Pharmaceutics 2015, 7, 137–151. [Google Scholar] [CrossRef]
- Sultana, N.; Sharkar, M.T.K.; Hadas, Y.; Chepurko, E.; Zangi, L. In Vitro Synthesis of Modified RNA for Cardiac Gene Therapy. Methods Mol. Biol. 2020, 2158, 281–294. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Li, B.; Luo, X.; Dong, Y. Effects of Chemically Modified Messenger RNA on Protein Expression. Bioconjugate Chem. 2016, 27, 849–853. [Google Scholar] [CrossRef]
- Fleming, A.M.; Xiao, S.; Burrows, C.J. Pseudouridine and N1-Methylpseudouridine Display pH-Independent Reaction Rates with Bisulfite Yielding Ribose Adducts. Org. Lett. 2022, 24, 6182–6185. [Google Scholar] [CrossRef]
- Loomis, K.H.; Lindsay, K.E.; Zurla, C.; Bhosle, S.M.; Vanover, D.A.; Blanchard, E.L.; Kirschman, J.L.; Bellamkonda, R.V.; Santangelo, P.J. In Vitro Transcribed mRNA Vaccines with Programmable Stimulation of Innate Immunity. Bioconjugate Chem. 2018, 29, 3072–3083. [Google Scholar] [CrossRef]
- Svitkin, Y.V.; Cheng, Y.M.; Chakraborty, T.; Presnyak, V.; John, M.; Sonenberg, N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017, 45, 6023–6036. [Google Scholar] [CrossRef] [PubMed]
- Parr, C.J.C.; Wada, S.; Kotake, K.; Kameda, S.; Matsuura, S.; Sakashita, S.; Park, S.; Sugiyama, H.; Kuang, Y.; Saito, H. N 1-Methylpseudouridine substitution enhances the performance of synthetic mRNA switches in cells. Nucleic Acids Res. 2020, 48, e35. [Google Scholar] [CrossRef] [PubMed]
- Mauger, D.M.; Cabral, B.J.; Presnyak, V.; Su, S.V.; Reid, D.W.; Goodman, B.; Link, K.; Khatwani, N.; Reynders, J.; Moore, M.J.; et al. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. USA 2019, 116, 24075–24083. [Google Scholar] [CrossRef] [PubMed]
- Furtado, D.; Cortez-Jugo, C.; Hung, Y.H.; Bush, A.I.; Caruso, F. mRNA Treatment Rescues Niemann–Pick Disease Type C1 in Patient Fibroblasts. Mol. Pharm. 2022, 19, 3987–3999. [Google Scholar] [CrossRef]
- Sultana, N.; Magadum, A.; Hadas, Y.; Kondrat, J.; Singh, N.; Youssef, E.; Calderon, D.; Chepurko, E.; Dubois, N.; Hajjar, R.J.; et al. Optimizing Cardiac Delivery of Modified mRNA. Mol. Ther. 2017, 25, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liao, J.; Mei, Z.; Liu, X.; Ge, J. Insight into Crosstalk between Ferroptosis and Necroptosis: Novel Therapeutics in Ischemic Stroke. Oxidative Med. Cell. Longev. 2021, 2021, 9991001. [Google Scholar] [CrossRef] [PubMed]
- Andries, O.; Mc Cafferty, S.; De Smedt, S.C.; Weiss, R.; Sanders, N.N.; Kitada, T. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 2015, 217, 337–344. [Google Scholar] [CrossRef]
- Perrino, C.; Barabási, A.-L.; Condorelli, G.; Davidson, S.M.; De Windt, L.; Dimmeler, S.; Engel, F.B.; Hausenloy, D.J.; Hill, J.A.; Van Laake, L.W.; et al. Epigenomic and transcriptomic approaches in the post-genomic era: Path to novel targets for diagnosis and therapy of the ischaemic heart? Position Paper of the European Society of Cardiology Working Group on Cellular Biology of the Heart. Cardiovasc. Res. 2017, 113, 725–736. [Google Scholar] [CrossRef]
- Magadum, A.; Singh, N.; Kurian, A.A.; Sharkar, M.T.K.; Chepurko, E.; Zangi, L. Ablation of a Single N-Glycosylation Site in Human FSTL 1 Induces Cardiomyocyte Proliferation and Cardiac Regeneration. Mol. Ther. Nucleic Acids 2018, 13, 133–143. [Google Scholar] [CrossRef]
- Yiangou, L.; Blanch-Asensio, A.; de Korte, T.; Miller, D.C.; van Meer, B.J.; Mol, M.P.H.; Brink, L.V.D.; Brandão, K.O.; Mummery, C.L.; Davis, R.P. Optogenetic Reporters Delivered as mRNA Facilitate Repeatable Action Potential and Calcium Handling Assessment in Human iPSC-Derived Cardiomyocytes. Stem Cells 2022, 40, 655–668. [Google Scholar] [CrossRef]
- Meng, J.; Moore, M.; Counsell, J.; Muntoni, F.; Popplewell, L.; Morgan, J. Optimized lentiviral vector to restore full-length dystrophin via a cell-mediated approach in a mouse model of Duchenne muscular dystrophy. Mol. Ther. Methods Clin. Dev. 2022, 25, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kelly, S.C.; Ivey, J.R.; Thorne, P.K.; Yamada, K.P.; Aikawa, T.; Mazurek, R.; Turk, J.R.; Silva, K.A.S.; Amin, A.R.; et al. Distribution of cardiomyocyte-selective adeno-associated virus serotype 9 vectors in swine following intracoronary and intravenous infusion. Physiol. Genom. 2022, 54, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Magadum, A.; Singh, N.; Kurian, A.A.; Munir, I.; Mehmood, T.; Brown, K.; Sharkar, M.T.K.; Chepurko, E.; Sassi, Y.; Oh, J.G.; et al. Pkm2 Regulates Cardiomyocyte Cell Cycle and Promotes Cardiac Regeneration. Circulation 2020, 141, 1249–1265. [Google Scholar] [CrossRef] [PubMed]
- Papayannakos, C.; Daniel, R. Understanding lentiviral vector chromatin targeting: Working to reduce insertional mutagenic potential for gene therapy. Gene Ther. 2012, 20, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Kieserman, J.M.; Myers, V.D.; Dubey, P.; Cheung, J.Y.; Feldman, A.M. Current Landscape of Heart Failure Gene Therapy. J. Am. Hear. Assoc. 2019, 8, e012239. [Google Scholar] [CrossRef]
- Scott, T.; Nel, L. Rabies Prophylactic and Treatment Options: An In Vitro Study of siRNA- and Aptamer-Based Therapeutics. Viruses 2021, 13, 881. [Google Scholar] [CrossRef]
- McBride, J.R.; Pennycook, T.J.; Pennycook, S.J.; Rosenthal, S.J. The Possibility and Implications of Dynamic Nanoparticle Surfaces. ACS Nano 2013, 7, 8358–8365. [Google Scholar] [CrossRef]
- Dadfar, S.M.; Roemhild, K.; Drude, N.I.; von Stillfried, S.; Knüchel, R.; Kiessling, F.; Lammers, T. Iron oxide nanoparticles: Diagnostic, therapeutic and theranostic applications. Adv. Drug Deliv. Rev. 2019, 138, 302–325. [Google Scholar] [CrossRef]
- Singh, R.D.; Hillestad, M.L.; Livia, C.; Li, M.; Alekseev, A.E.; Witt, T.A.; Stalboerger, P.G.; Yamada, S.; Terzic, A.; Behfar, A. M3RNA Drives Targeted Gene Delivery in Acute Myocardial Infarction. Tissue Eng. Part A 2019, 25, 145–158. [Google Scholar] [CrossRef]
- Zaitseva, T.S.; Alcazar, M.C.; Zamani, M.; Hou, L.; Sawamura, S.; Yakubov, E.; Hopkins, M.; Woo, Y.J.; Paukshto, M.V.; Huang, N.F. Aligned Nanofibrillar Scaffolds for Controlled Delivery of Modified mRNA. Tissue Eng. Part A 2019, 25, 121–130. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef] [PubMed]
- Evers, M.J.; Du, W.; Yang, Q.; Kooijmans, S.A.; Vink, A.; van Steenbergen, M.; Vader, P.; de Jager, S.C.; Fuchs, S.A.; Mastrobattista, E.; et al. Delivery of modified mRNA to damaged myocardium by systemic administration of lipid nanoparticles. J. Control. Release 2022, 343, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, I.C.; Eltoukhy, A.A.; Fish, K.M.; Nonnenmacher, M.; Ishikawa, K.; Chen, J.; Hajjar, R.J.; Anderson, D.G.; Costa, K.D. Myocardial Delivery of Lipidoid Nanoparticle Carrying modRNA Induces Rapid and Transient Expression. Mol. Ther. 2016, 24, 66–75. [Google Scholar] [CrossRef]
- Turnbull, I.C.; Eltoukhy, A.A.; Anderson, D.G.; Costa, K.D. Lipidoid mRNA Nanoparticles for Myocardial Delivery in Rodents. Methods Mol. Biol. 2016, 1521, 153–166. [Google Scholar] [CrossRef]
- Carlsson, L.; Clarke, J.C.; Yen, C.; Gregoire, F.; Albery, T.; Billger, M.; Egnell, A.-C.; Gan, L.-M.; Jennbacken, K.; Johansson, E.; et al. Biocompatible, Purified VEGF-A mRNA Improves Cardiac Function after Intracardiac Injection 1 Week Post-myocardial Infarction in Swine. Mol. Ther. Methods Clin. Dev. 2018, 9, 330–346. [Google Scholar] [CrossRef]
- Kaczmarek, J.C.; Patel, A.K.; Kauffman, K.J.; Fenton, O.S.; Webber, M.; Heartlein, M.W.; DeRosa, F.; Anderson, D.G. Polymer-Lipid Nanoparticles for Systemic Delivery of mRNA to the Lungs. Angew. Chem. Int. Ed. 2016, 55, 13808–13812. [Google Scholar] [CrossRef] [PubMed]
- Dib, N.; Khawaja, H.; Varner, S.; McCarthy, M.; Campbell, A. Cell Therapy for Cardiovascular Disease: A Comparison of Methods of Delivery. J. Cardiovasc. Transl. Res. 2010, 4, 177–181. [Google Scholar] [CrossRef]
- Mokhtari, B.; Aboutaleb, N.; Nazarinia, D.; Nikougoftar, M.; Tousi, S.M.T.R.; Molazem, M.; Azadi, M.-R. Comparison of the effects of intramyocardial and intravenous injections of human mesenchymal stem cells on cardiac regeneration after heart failure. Iran. J. Basic Med. Sci. 2020, 23, 879–885. [Google Scholar] [CrossRef]
- Firoozi, S.; Pahlavan, S.; Ghanian, M.-H.; Rabbani, S.; Barekat, M.; Nazari, A.; Pakzad, M.; Shekari, F.; Hassani, S.-N.; Moslem, F.; et al. Mesenchymal stem cell-derived extracellular vesicles alone or in conjunction with a SDKP-conjugated self-assembling peptide improve a rat model of myocardial infarction. Biochem. Biophys. Res. Commun. 2020, 524, 903–909. [Google Scholar] [CrossRef]
- Furtado, M.B.; Nim, H.T.; Boyd, S.; Rosenthal, N. View from the heart: Cardiac fibroblasts in development, scarring and regeneration. Development 2016, 143, 387–397. [Google Scholar] [CrossRef]
- Luger, D.; Lipinski, M.J.; Westman, P.C.; Glover, D.K.; Dimastromatteo, J.; Frias, J.C.; Albelda, M.T.; Sikora, S.; Kharazi, A.; Vertelov, G.; et al. Intravenously Delivered Mesenchymal Stem Cells: Systemic Anti-Inflammatory Effects Improve Left Ventricular Dysfunction in Acute Myocardial Infarction and Ischemic Cardiomyopathy. Circ. Res. 2017, 120, 1598–1613. [Google Scholar] [CrossRef]
- Prabhu, S.D.; Frangogiannis, N.G. The Biological Basis for Cardiac Repair After Myocardial Infarction. Circ. Res. 2016, 119, 91–112. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.C.; Kofidis, T. Adult stem cells for cardiac tissue engineering. J. Mol. Cell. Cardiol. 2011, 50, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Porrello, E.R.; Olson, E.N. A neonatal blueprint for cardiac regeneration. Stem Cell Res. 2014, 13, 556–570. [Google Scholar] [CrossRef]
- Trounson, A.; McDonald, C. Stem Cell Therapies in Clinical Trials: Progress and Challenges. Cell Stem Cell 2015, 17, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Petrus-Reurer, S.; Romano, M.; Howlett, S.; Jones, J.L.; Lombardi, G.; Saeb-Parsy, K. Immunological considerations and challenges for regenerative cellular therapies. Commun. Biol. 2021, 4, 798. [Google Scholar] [CrossRef] [PubMed]
- Magadum, A.; Kaur, K.; Zangi, L. mRNA-Based Protein Replacement Therapy for the Heart. Mol. Ther. 2019, 27, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Yla-Herttuala, E.; Laidinen, S.; Laakso, H.; Liimatainen, T. Quantification of myocardial infarct area based on TRAFFn relaxation time maps-comparison with cardiovascular magnetic resonance late gadolinium enhancement, T1ρ and T2 in vivo. J. Cardiovasc. Magn. Reson. 2018, 20, 34. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, L.; Wang, S.; Cheng, H.; Xu, L.; Pei, G.; Wang, Y.; Fu, C.; Jiang, Y.; He, C.; et al. Signaling pathways and targeted therapy for myocardial infarction. Signal Transduct. Target. Ther. 2022, 7, 78. [Google Scholar] [CrossRef]
- Wei, K.; Serpooshan, V.; Hurtado, C.; Diez-Cuñado, M.; Zhao, M.; Maruyama, S.; Zhu, W.; Fajardo, G.; Noseda, M.; Nakamura, K.; et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature 2015, 525, 479–485. [Google Scholar] [CrossRef]
- Gallego-Colon, E.; Villalba, M.; Tonkin, J.; Cruz, F.; Bernal, J.A.; Jimenez-Borregureo, L.J.; Schneider, M.D.; Lara-Pezzi, E.; Rosenthal, N. Intravenous delivery of adeno-associated virus 9-encoded IGF-1Ea propeptide improves post-infarct cardiac remodelling. npj Regen. Med. 2016, 1, 16001. [Google Scholar] [CrossRef]
- Zangi, L.; Oliveira, M.S.; Ye, L.Y.; Ma, Q.; Sultana, N.; Hadas, Y.; Chepurko, E.; Später, D.; Zhou, B.; Chew, W.L.; et al. Insulin-Like Growth Factor 1 Receptor-Dependent Pathway Drives Epicardial Adipose Tissue Formation After Myocardial Injury. Circulation 2017, 135, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Hadas, Y.; Vincek, A.S.; Youssef, E.; Żak, M.M.; Chepurko, E.; Sultana, N.; Sharkar, M.T.K.; Guo, N.; Komargodski, R.; Kurian, A.A.; et al. Altering Sphingolipid Metabolism Attenuates Cell Death and Inflammatory Response After Myocardial Infarction. Circulation 2020, 141, 916–930. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ma, Q.; King, J.S.; Sun, Y.; Xu, B.; Zhang, X.; Zohrabian, S.; Guo, H.; Cai, W.; Li, G.; et al. aYAP modRNA reduces cardiac inflammation and hypertrophy in a murine ischemia-reperfusion model. Life Sci. Alliance 2019, 3, e201900424. [Google Scholar] [CrossRef] [PubMed]
- Zangi, L.; Lui, K.O.; Von Gise, A.; Ma, Q.; Ebina, W.; Ptaszek, L.M.; Später, D.; Xu, H.; Tabebordbar, M.; Gorbatov, R.; et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013, 31, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Lui, K.O.; Zangi, L.; Silva, E.A.; Bu, L.; Sahara, M.; Li, R.A.; Mooney, D.; Chien, K.R. Driving vascular endothelial cell fate of human multipotent Isl1+ heart progenitors with VEGF modified mRNA. Cell Res. 2013, 23, 1172–1186. [Google Scholar] [CrossRef] [PubMed]
- Ai, X.; Yan, B.; Witman, N.; Gong, Y.; Yang, L.; Tan, Y.; Chen, Y.; Liu, M.; Lu, T.; Luo, R.; et al. Transient secretion of VEGF protein from transplanted hiPSC-CMs enhances engraftment and improves rat heart function post MI. Mol. Ther. 2022, 31, 211–229. [Google Scholar] [CrossRef]
- Kaur, K.; Hadas, Y.; Kurian, A.A.; Żak, M.M.; Yoo, J.; Mahmood, A.; Girard, H.; Komargodski, R.; Io, T.; Santini, M.P.; et al. Direct reprogramming induces vascular regeneration post muscle ischemic injury. Mol. Ther. 2021, 29, 3042–3058. [Google Scholar] [CrossRef]
- Yu, Z.; Witman, N.; Wang, W.; Li, D.; Yan, B.; Deng, M.; Wang, X.; Wang, H.; Zhou, G.; Liu, W.; et al. Cell-mediated delivery of VEGF modified mRNA enhances blood vessel regeneration and ameliorates murine critical limb ischemia. J. Control. Release 2019, 310, 103–114. [Google Scholar] [CrossRef]
- Magadum, A.; Singh, N.; Kurian, A.A.; Sharkar, M.T.K.; Sultana, N.; Chepurko, E.; Kaur, K.; Żak, M.M.; Hadas, Y.; Lebeche, D.; et al. Therapeutic Delivery of Pip4k2c-Modified mRNA Attenuates Cardiac Hypertrophy and Fibrosis in the Failing Heart. Adv. Sci. 2021, 8, 2004661. [Google Scholar] [CrossRef]
- Yamakawa, H.; Ieda, M. Cardiac regeneration by direct reprogramming in this decade and beyond. Inflamm. Regen. 2021, 41, 20. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.E.; Cober, N.D.; Dai, Z.; Stewart, D.J.; Zhao, Y.-Y. Endothelial cells in the pathogenesis of pulmonary arterial hypertension. Eur. Respir. J. 2021, 58, 2003957. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-L.; Leblond, A.-L.; Turner, E.C.; Kumar, A.H.; Martin, K.; Whelan, D.; O’Sullivan, D.M.; Caplice, N.M. Synthetic Chemically Modified mRNA-Based Delivery of Cytoprotective Factor Promotes Early Cardiomyocyte Survival Post-Acute Myocardial Infarction. Mol. Pharm. 2015, 12, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Kondrat, J.; Sultana, N.; Zangi, L. Synthesis of Modified mRNA for Myocardial Delivery. Methods Mol. Biol. 2016, 1521, 127–138. [Google Scholar] [CrossRef]
- Hadas, Y.; Sultana, N.; Youssef, E.; Sharkar, M.T.K.; Kaur, K.; Chepurko, E.; Zangi, L. Optimizing Modified mRNA In Vitro Synthesis Protocol for Heart Gene Therapy. Mol. Ther. Methods Clin. Dev. 2019, 14, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Kaur, K.; Zangi, L. Modified mRNA as a Therapeutic Tool for the Heart. Cardiovasc. Drugs Ther. 2020, 34, 871–880. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Schneller, J.L.; Frassetto, A.; Liang, S.; Zhu, X.; Park, J.-S.; Theisen, M.; Hong, S.-J.; Zhou, J.; Rajendran, R.; et al. Systemic Messenger RNA Therapy as a Treatment for Methylmalonic Acidemia. Cell Rep. 2017, 21, 3548–3558. [Google Scholar] [CrossRef]
- Lo, N.; Xu, X.; Soares, F.; He, H.H. The Basis and Promise of Programmable RNA Editing and Modification. Front. Genet. 2022, 13, 834413. [Google Scholar] [CrossRef]
- Gan, L.-M.; Lagerström-Fermér, M.; Carlsson, L.G.; Arfvidsson, C.; Egnell, A.-C.; Rudvik, A.; Kjaer, M.; Collén, A.; Thompson, J.D.; Joyal, J.; et al. Intradermal delivery of modified mRNA encoding VEGF-A in patients with type 2 diabetes. Nat. Commun. 2019, 10, 871. [Google Scholar] [CrossRef]
- Anttila, V.; Saraste, A.; Knuuti, J.; Jaakkola, P.; Hedman, M.; Svedlund, S.; Lagerström-Fermér, M.; Kjaer, M.; Jeppsson, A.; Gan, L.-M. Synthetic mRNA Encoding VEGF-A in Patients Undergoing Coronary Artery Bypass Grafting: Design of a Phase 2a Clinical Trial. Mol. Ther. Methods Clin. Dev. 2020, 18, 464–472. [Google Scholar] [CrossRef]

| Functions | Genes | Delivery Materials | Delivery Methods | Cells | Animals | Effects |
|---|---|---|---|---|---|---|
| Promoting proliferation | FSTL1 | Sucrose-citrate buffer | IM | CMs | Mice | Increased cardiac function, decreased scar size, and increased capillary density [39] |
| PKM2 | Sucrose-citrate buffer | IM | CMs | Mice | Enhanced cardiac function and improved long-term survival [43] | |
| Promoting differentiation | IGF1 | RNAiMAX | IM | EPCs | Mice | Governed epicardial adipose tissue formation in the context of myocardial injury by redirecting the fate of Wt1+ lineage cells [72] |
| Promoting angiogenesis | VEGFA | RNAiMAX | IM | EPCs | Mice | Improved heart function and enhanced long-term survival [75] |
| VEGF | Sucrose-citrate buffer | IM | iPSC-CMs | Rats | Improved heart function and enhanced long-term survival of recipients [77] | |
| VEGFA | Citrate-saline | IM | CMs | Pigs, mice | Improved systolic ventricular function and limited myocardial damage, left ventricular ejection fraction, border zone arteriolar and capillary density increased, and myocardial fibrosis decreased [55] | |
| VEGFA | RNAiMAX | IM | Isl1+ progenitors | Mice | Driven endothelial specification, engraftment, and survival following transplantation [76] | |
| 7G | Sucrose-citrate buffer | IM | MSCs | Mice | Improved cardiac function, scar size, long-term survival, and capillary density [78] | |
| Inhibiting fibrosis | PIP4K2C | Sucrose-citrate buffer | IM | CMs, fibroblasts | Mice | Attenuating cardiac hypertrophy and fibrosis, enhanced long-term survival |
| Promoting survival | AC | RNAiMAX | IM | CMs | Mice | Improved heart function, longer survival, and reduced scar size [73] |
| aYAP | Saline | IM | CMs | Mice | Improved heart function and suppressed cardiac hypertrophy [74] | |
| IGF-1 | Polyethylenimine-based nanoparticle | IM | CMs | Mice | Promoted CM survival and abrogated cell apoptosis under hypoxia-induced apoptosis conditions [83] | |
| Promoting delivery efficiency | - | LNP | i.v. via the tail vein | Fibroblasts | Mice | Most targeted cells were cardiac fibroblasts but also some CMs and macrophages [52] |
| eGFP | FLNP | IM or intracoronary administration | - | Rats, pigs | Increasing modRNA expression in heart [53] | |
| GEVIs, GECIs | Lipofectamine stem transfection reagent | In vitro | hiPSC-CMs | - | Delivered strong and stable signals in hiPSC-CMs [40] | |
| Varies | RNAiMAX | IM | - | Mice | Effective synthesis of modRNA for in vivo use [84] | |
| Luciferase | Sucrose-citrate buffer | IM | - | Mice | Optimized modRNA amount, time, and delivery [35] | |
| Luciferase | Sucrose-citrate buffer | IM | - | Mice | Increased translation by replacing 5′ UTR [16] | |
| eGFP | RNAiMAX | In vitro | - | - | Improved in vitro transcription [26] | |
| EGFP, mCherry, Fluc | Alginate, nanomater | IM | - | Pigs | Optimized M3RNA delivery into myocardium [49] | |
| HGF | Nanofibrillar scaffolds | IM | Fibroblasts | Mice | Improved translation efficiency [50] | |
| GFP, luciferase | Sucrose-citrate buffer | In vitro | CMs | - | Improved modRNA yield and translation efficiency, reduced its immunogenicity [85] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wu, M.; Guo, H. Modified mRNA as a Treatment for Myocardial Infarction. Int. J. Mol. Sci. 2023, 24, 4737. https://doi.org/10.3390/ijms24054737
Wang Y, Wu M, Guo H. Modified mRNA as a Treatment for Myocardial Infarction. International Journal of Molecular Sciences. 2023; 24(5):4737. https://doi.org/10.3390/ijms24054737
Chicago/Turabian StyleWang, Yu, Meiping Wu, and Haidong Guo. 2023. "Modified mRNA as a Treatment for Myocardial Infarction" International Journal of Molecular Sciences 24, no. 5: 4737. https://doi.org/10.3390/ijms24054737
APA StyleWang, Y., Wu, M., & Guo, H. (2023). Modified mRNA as a Treatment for Myocardial Infarction. International Journal of Molecular Sciences, 24(5), 4737. https://doi.org/10.3390/ijms24054737





