Genome-Wide Investigation of Apyrase (APY) Genes in Peanut (Arachis hypogaea L.) and Functional Characterization of a Pod-Abundant Expression Promoter AhAPY2-1p
Abstract
1. Introduction
2. Results
2.1. Identification, Genomic Locations, and Physicochemical Properties of AhAPYs
2.2. Gene Structure, Conserved Motifs, and Protein 3D Structure Simulation
2.3. Phylogenetic and Gene Duplication Analysis
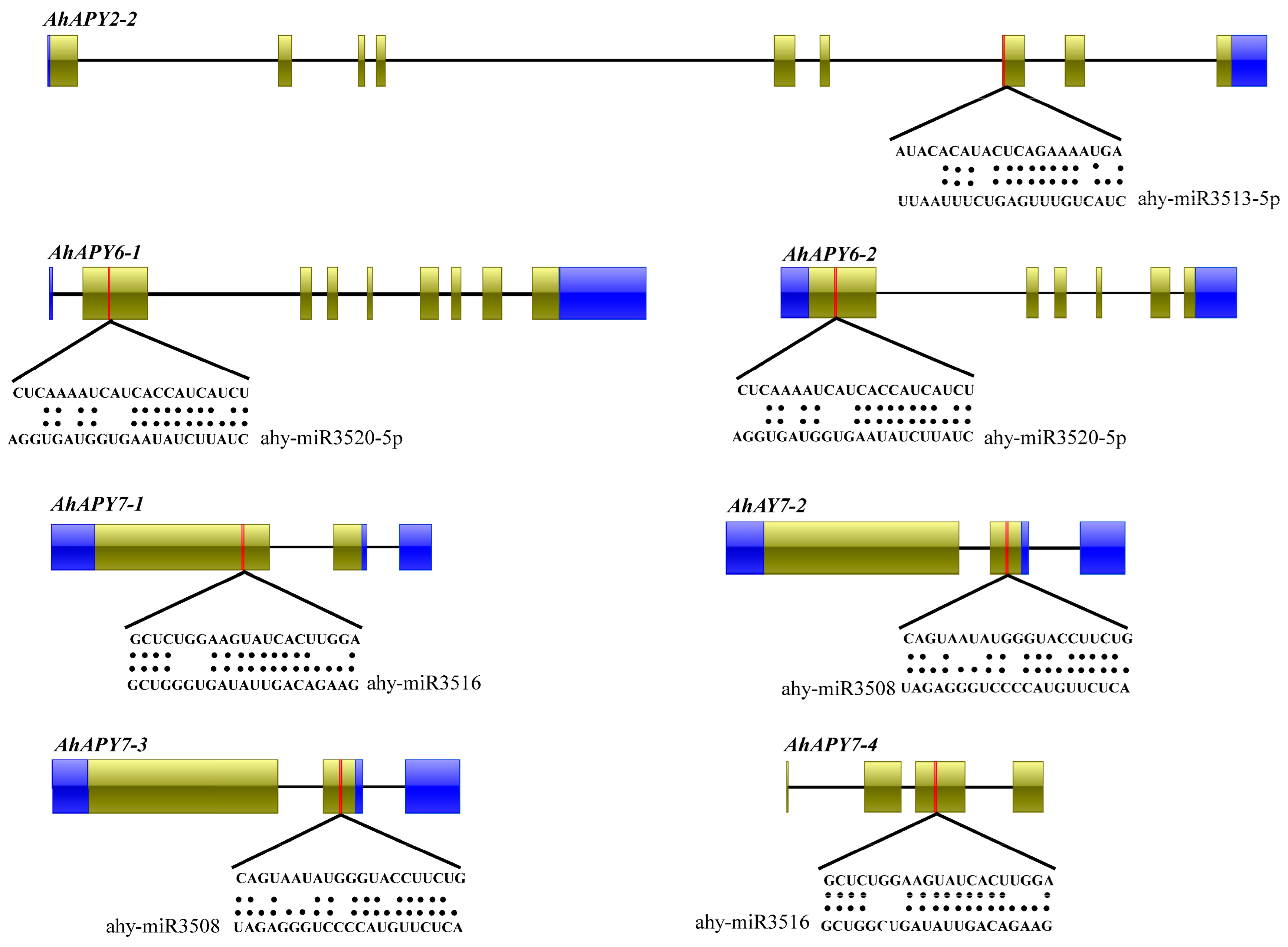
2.4. Prediction of miRNAs Targeting AhAPYs and Analysis of Putative Protein–Protein Interactions
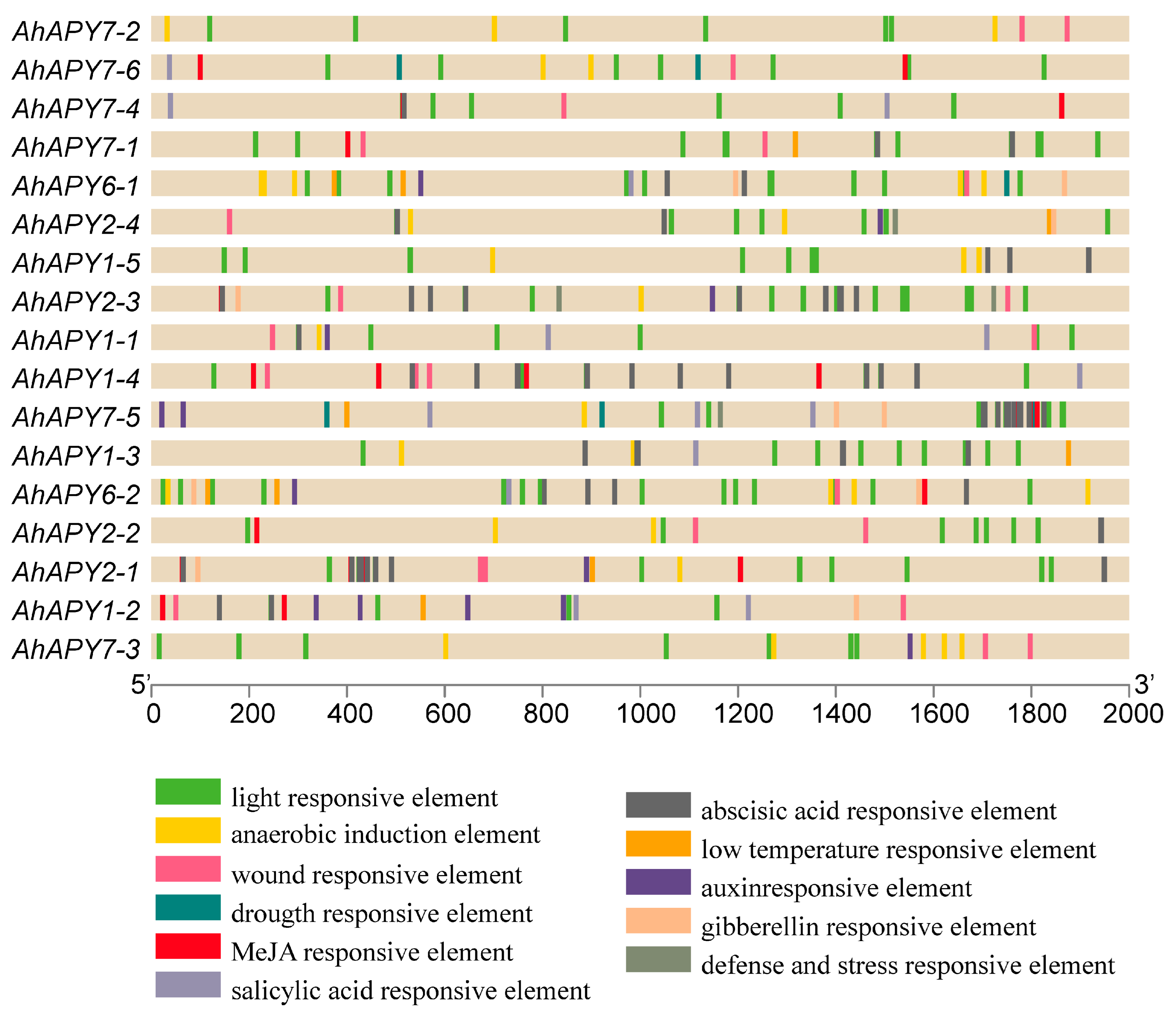
2.5. Analysis of Cis-Regulatory Elements
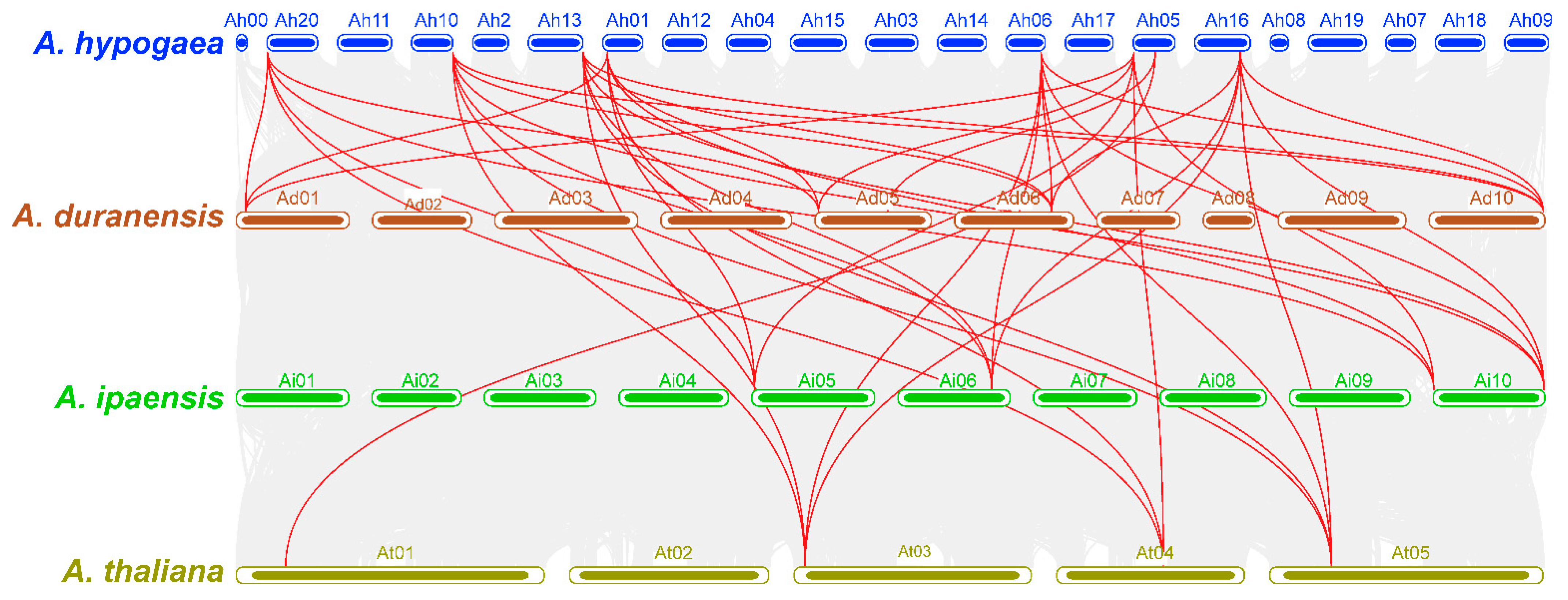
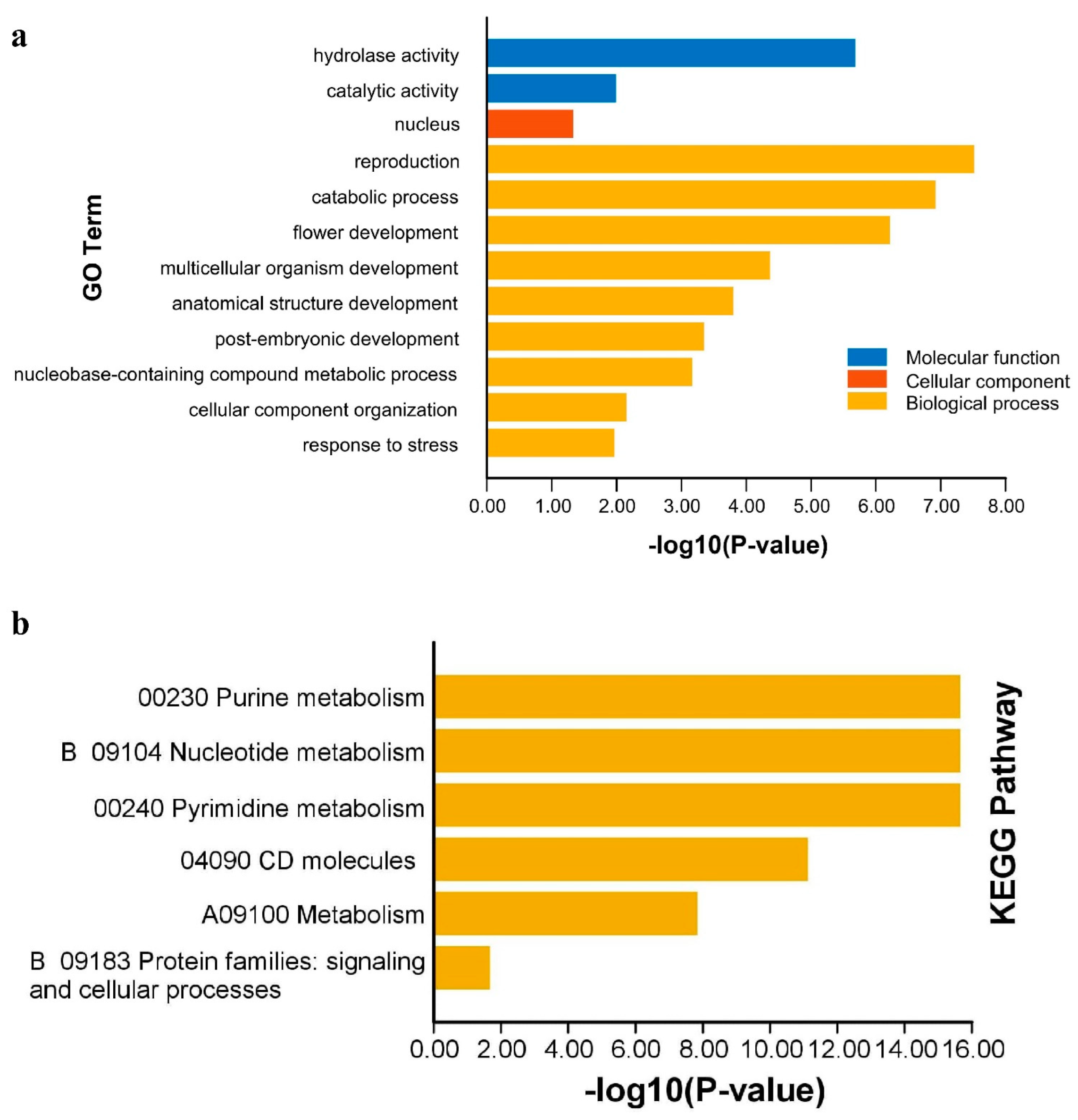
2.6. Synteny and Functional Annotation Analysis
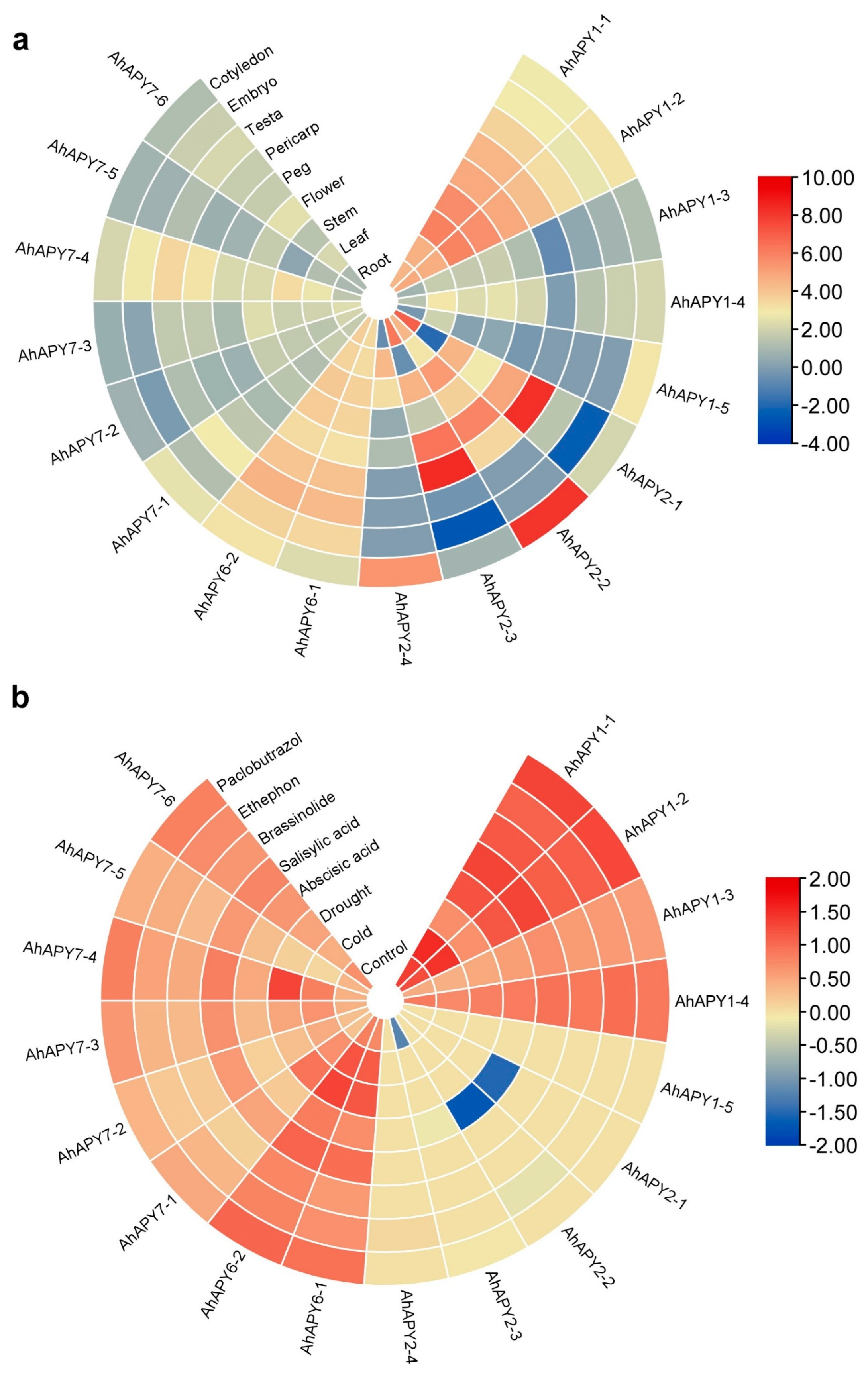
2.7. Expression Analysis
2.8. Selection of Gene for Promoter Cloning and In Silico Analysis of Promoter
2.9. Cloning and Genetic Transformation
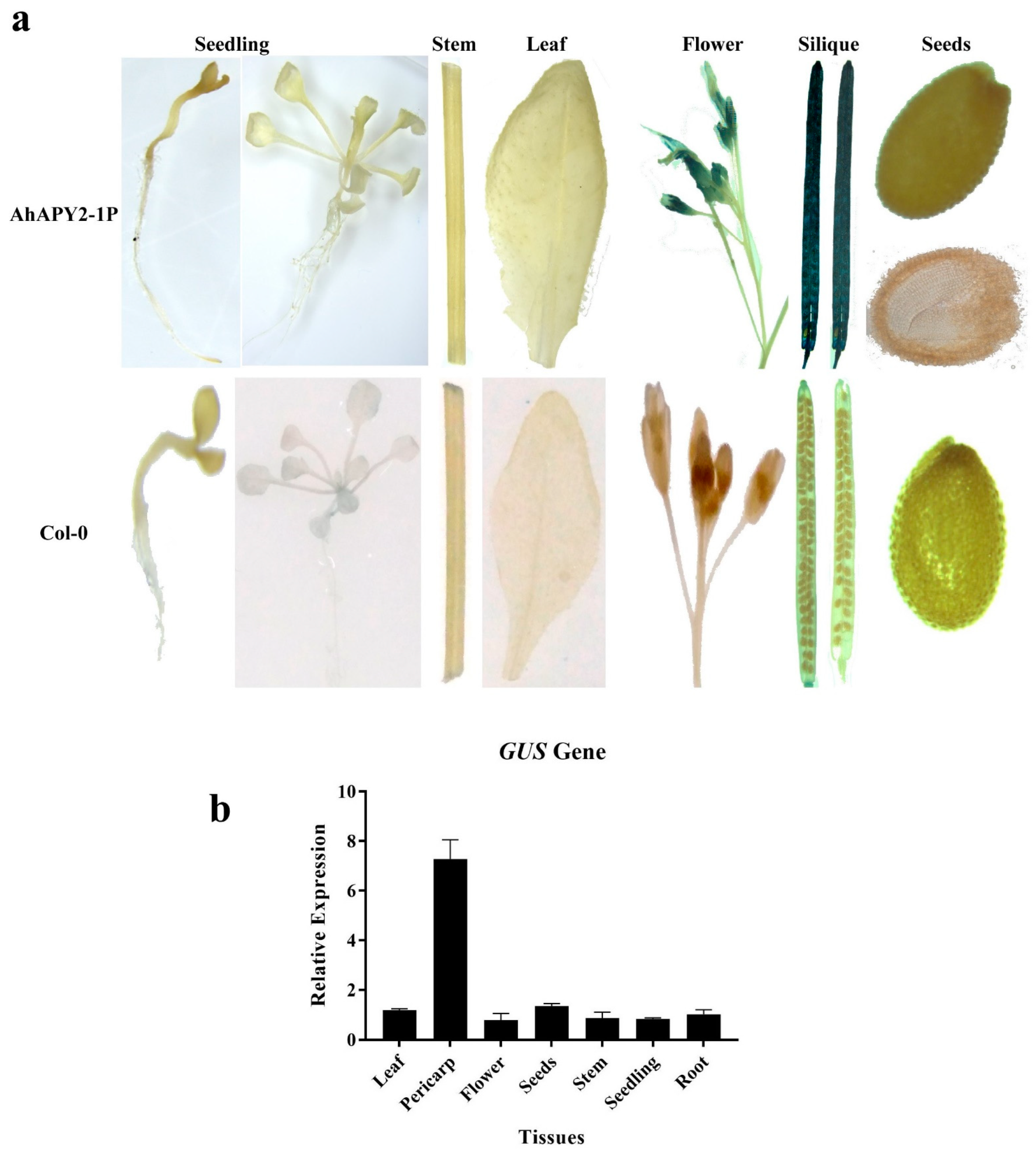
2.10. Functional Study of Promoter Activity in Transgenic Plants
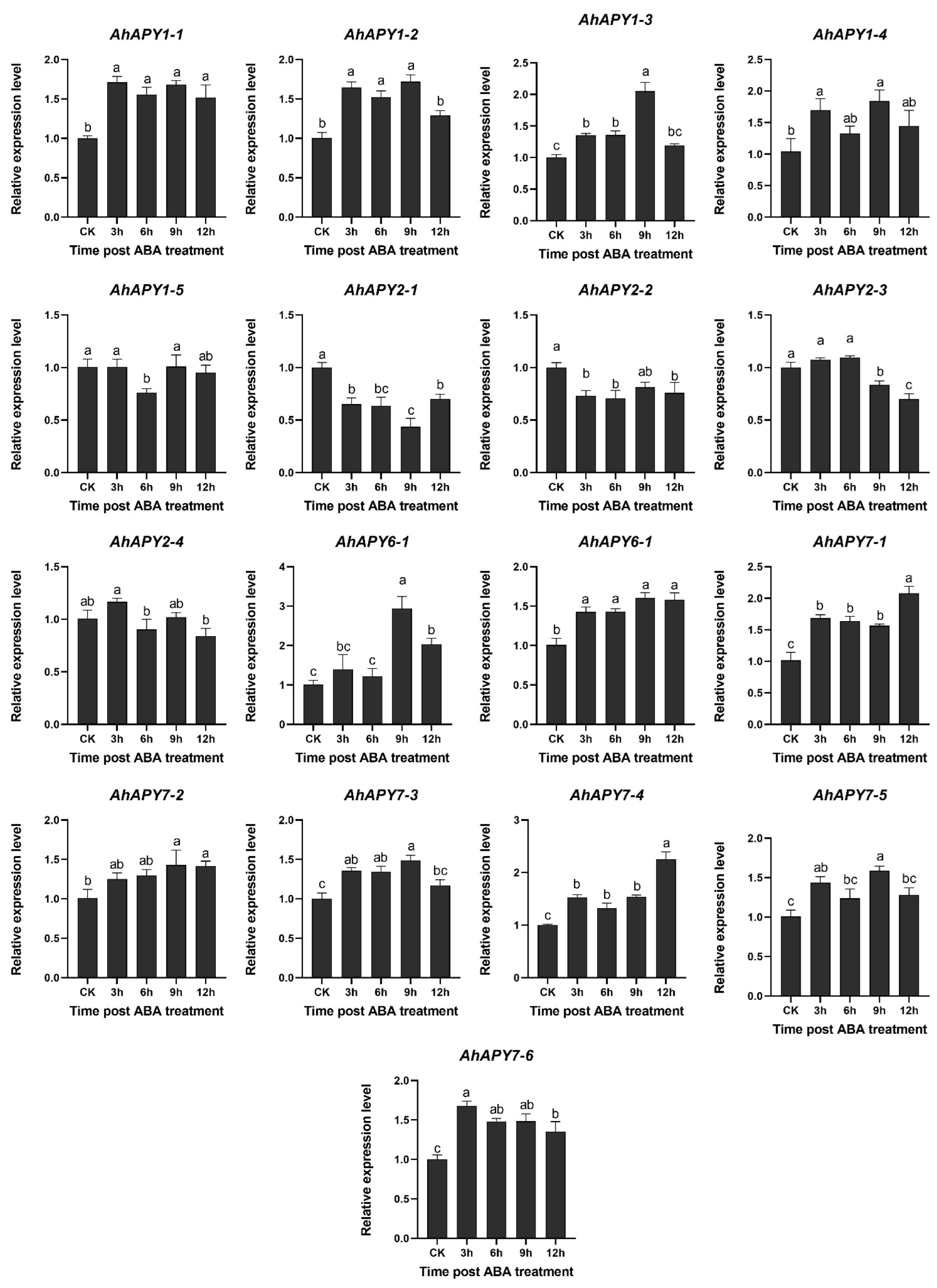
2.11. Response of AhAPY2-1 Promoter to Hormones
3. Discussions
4. Materials and Methods
4.1. Identification of APYs/NTPs from Peanut Genome
4.2. Genomic Positions, Structure, and Physicochemical Properties of APYs
4.3. Phylogeny and Duplication Analysis
4.4. Protein–Protein Interaction Analysis and Putative miRNAs Targeting AhAPYs
4.5. Promoter Cis-Elements Analysis
4.6. Comparative Genome Synteny and Functional Annotation Analysis
4.7. Expression Analysis of AhAPYs
4.8. Plant Materials and Cloning of AhAPY2-1 Promoter
4.9. Study of Promoter Activity in Transgenic Plants
4.10. Statistical Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Varshney, R.K.; Pandey, M.K.; Puppala, N. The Peanut Genome: An Introduction. In The Peanut Genome; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–6. [Google Scholar]
- FAOSTAT. Food and Agriculture Organization of the United Nations (FAO). 2020. Available online: https://fenix.fao.org/faostat/internal/en/#data/QCL/visualize (accessed on 11 August 2022).
- Kumar, R.; Pandey, M.K.; Roychoudhry, S.; Nayyar, H.; Kepinski, S.; Varshney, R.K. Peg biology: Deciphering the molecular regulations involved during peanut peg development. Front. Plant Sci. 2019, 10, 1289. [Google Scholar] [CrossRef]
- Haro, R.J.; Mantese, A.; Otegui, M.E. Peg viability and pod set in peanut: Response to impaired pegging and water deficit. Flora 2011, 206, 865–871. [Google Scholar] [CrossRef]
- Soni, P.; Gangurde, S.S.; Ortega-Beltran, A.; Kumar, R.; Parmar, S.; Sudini, H.K.; Lei, Y.; Ni, X.; Huai, D.; Fountain, J.C. Functional biology and molecular mechanisms of host-pathogen interactions for aflatoxin contamination in groundnut (Arachis hypogaea L.) and maize (Zea mays L.). Front. Microbiol. 2020, 11, 227. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.K.; Kumar, R.; Pandey, A.K.; Soni, P.; Gangurde, S.S.; Sudini, H.K.; Fountain, J.C.; Liao, B.; Desmae, H.; Okori, P. Mitigating aflatoxin contamination in groundnut through a combination of genetic resistance and post-harvest management practices. Toxins 2019, 11, 315. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Chung, C.Y.-L.; Li, M.-W.; Wong, F.-L.; Wang, X.; Liu, A.; Wang, Z.; Leung, A.K.-Y.; Wong, T.-H.; Tong, S.-W. A reference-grade wild soybean genome. Nat. Commun. 2019, 10, 1216. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Tang, H.; Zhang, Q.; Hua, X.; Ma, X.; Zhu, F.; Jones, T.; Zhu, X.; Bowers, J. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 2018, 50, 1565–1573. [Google Scholar] [CrossRef]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.-C.; Zhang, L.; Zhang, X.; Tang, R. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.; Ren, L.; Farmer, A.D.; Pandey, M.K. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef]
- Song, H.; Wang, P.; Lin, J.-Y.; Zhao, C.; Bi, Y.; Wang, X. Genome-wide identification and characterization of WRKY gene family in peanut. Front. Plant Sci. 2016, 7, 534. [Google Scholar] [CrossRef]
- Gao, C.; Sun, J.; Wang, C.; Dong, Y.; Xiao, S.; Wang, X.; Jiao, Z.J.P.O. Genome-wide analysis of basic/helix-loop-helix gene family in peanut and assessment of its roles in pod development. PLoS ONE 2017, 12, e0181843. [Google Scholar] [CrossRef]
- Wang, Z.; Yan, L.; Wan, L.; Huai, D.; Kang, Y.; Shi, L.; Jiang, H.; Lei, Y.; Liao, B. Genome-wide systematic characterization of bZIP transcription factors and their expression profiles during seed development and in response to salt stress in peanut. BMC Genom. 2019, 20, 51. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Li, K.; Ning, L.; He, J.; Ma, X.; Li, Z.; Zhang, X.; Yin, D. Genome-wide analysis of the growth-regulating factor family in peanut (Arachis hypogaea L.). Int. J. Mol. Sci. 2019, 20, 4120. [Google Scholar] [CrossRef]
- Tang, Y.; Du, G.; Xiang, J.; Hu, C.; Li, X.; Wang, W.; Zhu, H.; Qiao, L.; Zhao, C.; Wang, J. Genome-wide identification of auxin response factor (ARF) gene family and the miR160-ARF18-mediated response to salt stress in peanut (Arachis hypogaea L.). Genomics 2022, 114, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Sharif, Y.; Chen, K.; Wang, L.; Fu, H.; Zhuang, Y.; Chitikineni, A.; Chen, H.; Zhang, C.; Varshney, R.K. Genome-wide characterization of ascorbate peroxidase (APX) gene family in peanut (Arachis hypogea L.) revealed their crucial role in growth and multiple stress tolerance. Front. Plant Sci. 2022, 13, 962182. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Qin, S.; Guo, Y.; Chen, Y.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Genome-wide analysis of the AP2/ERF gene family in physic nut and overexpression of the JcERF011 gene in rice increased its sensitivity to salinity stress. PLoS ONE 2016, 11, e0150879. [Google Scholar] [CrossRef]
- Cao, Y.; Tanaka, K.; Nguyen, C.T.; Stacey, G. Extracellular ATP is a central signaling molecule in plant stress responses. Curr. Opin. Plant Biol. 2014, 20, 82–87. [Google Scholar] [CrossRef]
- Veerappa, R.; Slocum, R.D.; Siegenthaler, A.; Wang, J.; Clark, G.; Roux, S. Ectopic expression of a pea apyrase enhances root system architecture and drought survival in Arabidopsis and soybean. Plant Cell Environ. 2019, 42, 337–353. [Google Scholar] [CrossRef]
- Masoud, H.M.; Helmy, M.S.; Darwish, D.A.; Abdel-Monsef, M.M.; Ibrahim, M.A. Apyrase with anti-platelet aggregation activity from the nymph of the camel tick Hyalomma dromedarii. Exp. Appl. Acarol. 2020, 80, 349–361. [Google Scholar] [CrossRef]
- Proietti, M.; Perruzza, L.; Scribano, D.; Pellegrini, G.; D’Antuono, R.; Strati, F.; Raffaelli, M.; Gonzalez, S.F.; Thelen, M.; Hardt, W.-D. ATP released by intestinal bacteria limits the generation of protective IgA against enteropathogens. Nat. Commun. 2019, 10, 250. [Google Scholar] [CrossRef] [PubMed]
- Da’dara, A.A.; Bhardwaj, R.; Ali, Y.B.; Skelly, P.J. Schistosome tegumental ecto-apyrase (SmATPDase1) degrades exogenous pro-inflammatory and pro-thrombotic nucleotides. PeerJ 2014, 2, e316. [Google Scholar] [CrossRef] [PubMed]
- Chiu, T.-Y.; Lao, J.; Manalansan, B.; Loqué, D.; Roux, S.J.; Heazlewood, J.L. Biochemical characterization of Arabidopsis APYRASE family reveals their roles in regulating endomembrane NDP/NMP homoeostasis. Biochem. J. 2015, 472, 43–54. [Google Scholar] [CrossRef]
- Dunkley, T.P.; Watson, R.; Griffin, J.L.; Dupree, P.; Lilley, K.S. Localization of organelle proteins by isotope tagging (LOPIT). Mol. Cell. Proteom. 2004, 3, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Leal, D.B.; Streher, C.A.; Neu, T.N.; Bittencourt, F.P.; Leal, C.A.; da Silva, J.E.; Morsch, V.M.; Schetinger, M.R. Characterization of NTPDase (NTPDase1; ecto-apyrase; ecto-diphosphohydrolase; CD39; EC 3.6. 1.5) activity in human lymphocytes. Biochim. Et Biophys. Acta 2005, 1721, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Yang, J. Functional Analyses of Arabidopsis Apyrases 3 through 7; The University of Texas: Austin, TX, USA, 2011. [Google Scholar]
- Raza, A.; Tabassum, J.; Fakhar, A.Z.; Sharif, R.; Chen, H.; Zhang, C.; Ju, L.; Fotopoulos, V.; Siddique, K.H.; Singh, R.K. Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit. Rev. Biotechnol. 2022, 1–28, online ahead of print. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, C.; Zhang, X.; Deng, S.; Zhao, R.; Shen, X.; Chen, S. Extracellular ATP signaling and homeostasis in plant cells. Plant Signal. Behav. 2012, 7, 566–569. [Google Scholar] [CrossRef]
- Clark, G.; Fraley, D.; Steinebrunner, I.; Cervantes, A.; Onyirimba, J.; Liu, A.; Torres, J.; Tang, W.; Kim, J.; Roux, S.J. Extracellular nucleotides and apyrases regulate stomatal aperture in Arabidopsis. Plant Physiol. 2011, 156, 1740–1753. [Google Scholar] [CrossRef]
- Yang, X.; Wang, B.; Farris, B.; Clark, G.; Roux, S.J. Modulation of root skewing in Arabidopsis by apyrases and extracellular ATP. Plant Cell Physiol. 2015, 56, 2197–2206. [Google Scholar] [CrossRef]
- Lim, M.H.; Wu, J.; Yao, J.; Gallardo, I.F.; Dugger, J.W.; Webb, L.J.; Huang, J.; Salmi, M.L.; Song, J.; Clark, G. Apyrase suppression raises extracellular ATP levels and induces gene expression and cell wall changes characteristic of stress responses. Plant Physiol. 2014, 164, 2054–2067. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wu, J.; Romanovicz, D.; Clark, G.; Roux, S.J.J.P.p.; biochemistry. Co-regulation of exine wall patterning, pollen fertility and anther dehiscence by Arabidopsis apyrases 6 and 7. Plant Physiol. Biochem. 2013, 69, 62–73. [Google Scholar] [CrossRef]
- Shiraishi, T. Suppression of defense response related to plant cell wall. Jpn. Agric. Res. Q. 2013, 47, 21–27. [Google Scholar] [CrossRef]
- Deng, S.; Sun, J.; Zhao, R.; Ding, M.; Zhang, Y.; Sun, Y.; Wang, W.; Tan, Y.; Liu, D.; Ma, X. Populus euphratica APYRASE2 enhances cold tolerance by modulating vesicular trafficking and extracellular ATP in Arabidopsis plants. Plant Physiol. 2015, 169, 530–548. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, X.; Deng, S.; Zhang, C.; Wang, M.; Ding, M.; Zhao, R.; Shen, X.; Zhou, X.; Lu, C.J.P.o. Extracellular ATP signaling is mediated by H2O2 and cytosolic Ca2+ in the salt response of Populus euphratica cells. PLoS ONE 2012, 7, e53136. [Google Scholar] [CrossRef] [PubMed]
- Alam, I.; Lee, D.-G.; Kim, K.-H.; Park, C.-H.; Sharmin, S.A.; Lee, H.; Oh, K.-W.; Yun, B.-W.; Lee, B.-H. Proteome analysis of soybean roots under waterlogging stress at an early vegetative stage. J. Biosci. 2010, 35, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, M.K.; Gangurde, S.S.; Pandey, M.K.; Niral, V.; Sudha, R.; Jerard, B.A.; Kadke, G.N.; Sabana, A.A.; Muralikrishna, K.S.; Samsudeen, K. Insights on Genetic Diversity, Population Structure, and Linkage Disequilibrium in Globally Diverse Coconut Accessions Using Genotyping-by-Sequencing. OMICS A J. Integr. Biol. 2021, 25, 796–809. [Google Scholar] [CrossRef]
- Jadhav, M.P.; Gangurde, S.S.; Hake, A.A.; Yadawad, A.; Mahadevaiah, S.S.; Pattanashetti, S.K.; Gowda, M.; Shirasawa, K.; Varshney, R.K.; Pandey, M.K. Genotyping-by-Sequencing Based Genetic Mapping Identified Major and Consistent Genomic Regions for Productivity and Quality Traits in Peanut. Front. Plant Sci. 2021, 12, 668020. [Google Scholar] [CrossRef]
- Hong, Y.; Pandey, M.K.; Lu, Q.; Liu, H.; Gangurde, S.S.; Li, S.; Liu, H.; Li, H.; Liang, X.; Varshney, R.K. Genetic diversity and distinctness based on morphological and SSR markers in peanut. Agron. J. 2021, 113, 4648–4660. [Google Scholar] [CrossRef]
- Gangurde, S.S.; Nayak, S.N.; Joshi, P.; Purohit, S.; Sudini, H.K.; Chitikineni, A.; Hong, Y.; Guo, B.; Chen, X.; Pandey, M.K. Comparative transcriptome analysis identified candidate genes for late leaf spot resistance and cause of defoliation in groundnut. Int. J. Mol. Sci. 2021, 22, 4491. [Google Scholar] [CrossRef]
- Sharma, M.; Gangurde, S.S.; Salgotra, R.K.; Kumar, B.; Singh, A.K.; Pandey, M.K. Genetic mapping for grain quality and yield-attributed traits in Basmati rice using SSR-based genetic map. J. Biosci. 2021, 46, 50. [Google Scholar] [CrossRef]
- Pandey, M.K.; Gangurde, S.S.; Sharma, V.; Pattanashetti, S.K.; Naidu, G.K.; Faye, I.; Hamidou, F.; Desmae, H.; Kane, N.A.; Yuan, M. Improved genetic map identified major QTLs for drought tolerance-and iron deficiency tolerance-related traits in groundnut. Genes 2020, 12, 37. [Google Scholar] [CrossRef]
- Khan, S.A.; Chen, H.; Deng, Y.; Chen, Y.; Zhang, C.; Cai, T.; Ali, N.; Mamadou, G.; Xie, D.; Guo, B. High-density SNP map facilitates fine mapping of QTLs and candidate genes discovery for Aspergillus flavus resistance in peanut (Arachis hypogaea). Theor. Appl. Genet. 2020, 133, 2239–2257. [Google Scholar] [CrossRef]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Ding, Y.; Ma, Y.; Liu, N.; Xu, J.; Hu, Q.; Li, Y.; Wu, Y.; Xie, S.; Zhu, L.; Min, L. microRNAs involved in auxin signalling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum). Plant J. 2017, 91, 977–994. [Google Scholar] [CrossRef]
- Sun, X.; Xu, L.; Wang, Y.; Yu, R.; Zhu, X.; Luo, X.; Gong, Y.; Wang, R.; Limera, C.; Zhang, K. Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genom. 2015, 16, 197. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef]
- Grace, M.L.; Chandrasekharan, M.B.; Hall, T.C.; Crowe, A.J. Sequence and spacing of TATA box elements are critical for accurate initiation from the β-phaseolin promoter. J. Biol. Chem. 2004, 279, 8102–8110. [Google Scholar] [CrossRef] [PubMed]
- Shirsat, A.; Wilford, N.; Croy, R.; Boulter, D.J.M.; MGG, G.G. Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. Mol. Gen. Genet. 1989, 215, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Tanaka, K.; Liang, Y.; Cao, Y.; Lee, S.Y.; Stacey, G. Extracellular ATP, a danger signal, is recognized by DORN1 in Arabidopsis. Biochem. J. 2014, 463, 429–437. [Google Scholar] [CrossRef]
- Roux, S.J.; Steinebrunner, I. Extracellular ATP: An unexpected role as a signaler in plants. Trends Plant Sci. 2007, 12, 522–527. [Google Scholar] [CrossRef]
- Tanaka, K.; Gilroy, S.; Jones, A.M.; Stacey, G. Extracellular ATP signaling in plants. Trends Cell Biol. 2010, 20, 601–608. [Google Scholar] [CrossRef]
- Clark, G.B.; Morgan, R.O.; Fernandez, M.-P.; Salmi, M.L.; Roux, S.J. Breakthroughs spotlighting roles for extracellular nucleotides and apyrases in stress responses and growth and development. Plant Sci. 2014, 225, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Song, C.J.; Steinebrunner, I.; Wang, X.; Stout, S.C.; Roux, S. Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in Arabidopsis. Plant Physiol. 2006, 140, 1222–1232. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V.; Shang, Z.; Shin, R.; Thompson, E.; Rubio, L.; Laohavisit, A.; Mortimer, J.C.; Chivasa, S.; Slabas, A.R.; Glover, B. Plant extracellular ATP signalling by plasma membrane NADPH oxidase and Ca2+ channels. Plant J. 2009, 58, 903–913. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Deng, S.; Zhang, Y.; Zhang, X.; Sun, J.; Wang, M.; Zhao, R.; Jing, Y.; Shen, X.; Chen, S. Cloning of apyrase gene PeAPY2 from Populus euphratica and the salt tolerance of the transformed cells. Genom. Appl. Biol. 2013, 32, 276–284. [Google Scholar]
- Liu, W.; Ni, J.; Shah, F.A.; Ye, K.; Hu, H.; Wang, Q.; Wang, D.; Yao, Y.; Huang, S.; Hou, J. Genome-wide identification, characterization and expression pattern analysis of APYRASE family members in response to abiotic and biotic stresses in wheat. PeerJ 2019, 7, e7622. [Google Scholar] [CrossRef] [PubMed]
- Gangurde, S.S.; Wang, H.; Yaduru, S.; Pandey, M.K.; Fountain, J.C.; Chu, Y.; Isleib, T.; Holbrook, C.C.; Xavier, A.; Culbreath, A.K. Nested-association mapping (NAM)-based genetic dissection uncovers candidate genes for seed and pod weights in peanut (Arachis hypogaea). Plant Biotechnol. J. 2020, 18, 1457–1471. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Janila, P.; Vishwakarma, M.K.; Khan, A.W.; Manohar, S.S.; Gangurde, S.S.; Variath, M.T.; Shasidhar, Y.; Pandey, M.K.; Varshney, R.K. Whole-genome resequencing-based QTL-seq identified candidate genes and molecular markers for fresh seed dormancy in groundnut. Plant Biotechnol. J. 2020, 18, 992–1003. [Google Scholar] [CrossRef]
- Tayade, A.D.; Motagi, B.N.; Jadhav, M.P.; Nadaf, A.S.; Koti, R.V.; Gangurde, S.S.; Sharma, V.; Varshney, R.K.; Pandey, M.K.; Bhat, R.S. Genetic mapping of tolerance to iron deficiency chlorosis in peanut (Arachis hypogaea L.). Euphytica 2022, 218, 46. [Google Scholar] [CrossRef]
- Zhang, K.; Ma, J.; Gangurde, S.S.; Hou, L.; Xia, H.; Li, N.; Pan, J.; Tian, R.; Huang, H.; Wang, X. Targeted metabolome analysis reveals accumulation of metabolites in testa of four peanut germplasms. Front. Plant Sci. 2022, 13, 992124. [Google Scholar] [CrossRef]
- Shasidhar, Y.; Variath, M.; Vishwakarma, M.; Manohar, S.; Gangurde, S.; Sriswathi, M.; Sudini, H.; Dobariya, K.; Bera, S.; Radhakrishnan, T. Improvement of three Indian popular groundnut varieties for foliar disease resistance and high oleic acid using SSR markers and SNP array in marker-assisted backcrossing. Crop J. 2020, 8, 1–15. [Google Scholar] [CrossRef]
- Bomireddy, D.; Gangurde, S.S.; Variath, M.T.; Janila, P.; Manohar, S.S.; Sharma, V.; Parmar, S.; Deshmukh, D.; Reddisekhar, M.; Reddy, D.M. Discovery of Major Quantitative Trait Loci and Candidate Genes for Fresh Seed Dormancy in Groundnut. Agronomy 2022, 12, 404. [Google Scholar] [CrossRef]
- Guo, Y.; Qiu, L.-J. Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 2013, 8, e76809. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Raza, A.; Gao, A.; Jia, Z.; Zhang, Y.; Hussain, M.A.; Mehmood, S.S.; Cheng, Y.; Lv, Y.; Zou, X. Genome-Wide Analysis and Expression Profile of Superoxide Dismutase (SOD) Gene Family in Rapeseed (Brassica napus L.) under Different Hormones and Abiotic Stress Conditions. Antioxidants 2021, 10, 1182. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, Q.; Chen, K.; Zhao, S.; Zhang, C.; Pan, R.; Cai, T.; Deng, Y.; Wang, X.; Chen, Y. Integrated microRNA and transcriptome profiling reveals a miRNA-mediated regulatory network of embryo abortion under calcium deficiency in peanut (Arachis hypogaea L.). BMC Genom. 2019, 20, 392. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Zhang, B. micro RNA evolution and expression analysis in polyploidized cotton genome. Plant Biotechnol. J. 2015, 13, 421–434. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Maruyama-Nakashita, A.; Nakamura, Y.; Watanabe-Takahashi, A.; Inoue, E.; Yamaya, T.; Takahashi, H. Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 2005, 42, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014, 202, 35–49. [Google Scholar] [CrossRef]
- Chen, Y.; Song, W.; Xie, X.; Wang, Z.; Guan, P.; Peng, H.; Jiao, Y.; Ni, Z.; Sun, Q.; Guo, W. A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the triticeae tribe as a pilot practice in the plant pangenomic era. Mol. Plant 2020, 13, 1694–1708. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, H.; Cai, T.; Deng, Y.; Zhuang, R.; Zhang, N.; Zeng, Y.; Zheng, Y.; Tang, R.; Pan, R. Overexpression of a novel peanut NBS-LRR gene A h RRS 5 enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Biotechnol. J. 2017, 15, 39–55. [Google Scholar] [CrossRef]
- Karthik, S.; Pavan, G.; Sathish, S.; Siva, R.; Kumar, P.S.; Manickavasagam, M. Genotype-independent and enhanced in planta Agrobacterium tumefaciens-mediated genetic transformation of peanut [Arachis hypogaea (L.)]. 3 Biotech 2018, 8, 202. [Google Scholar] [CrossRef]
- Wei, H.H.; Yu, S.T.; Wang, Z.W.; Yang, Z.; Song, G.S.; Wang, X.Z.; Sun, X.S.; Wang, C.T. In Planta Genetic Transformation to Produce CRISPRed High-Oleic Peanut. Prepr. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Raza, A.; Salehi, H.; Rahman, M.A.; Zahid, Z.; Haghjou, M.M.; Najafi-Kakavand, S.; Charagh, S.; Osman, H.S.; Albaqami, M.; Zhuang, Y.; et al. Plant hormones and neurotransmitter interactions mediate antioxidant defenses under induced oxidative stress in plants. Front. Plant Sci. 2022, 13, 961872. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, 39–49. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A.J.N.a.r. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 2006, 64, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Sharif, Y.; Chen, H.; Deng, Y.; Ali, N.; Khan, S.; Zhang, C.; Xie, W.; Chen, K.; Cai, T.; Yang, Q. Cloning and Functional Characterization of a Pericarp Abundant Expression Promoter (AhGLP17-1P) From Peanut (Arachis hypogaea L.). Front. Genet. 2022, 12, 821281. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
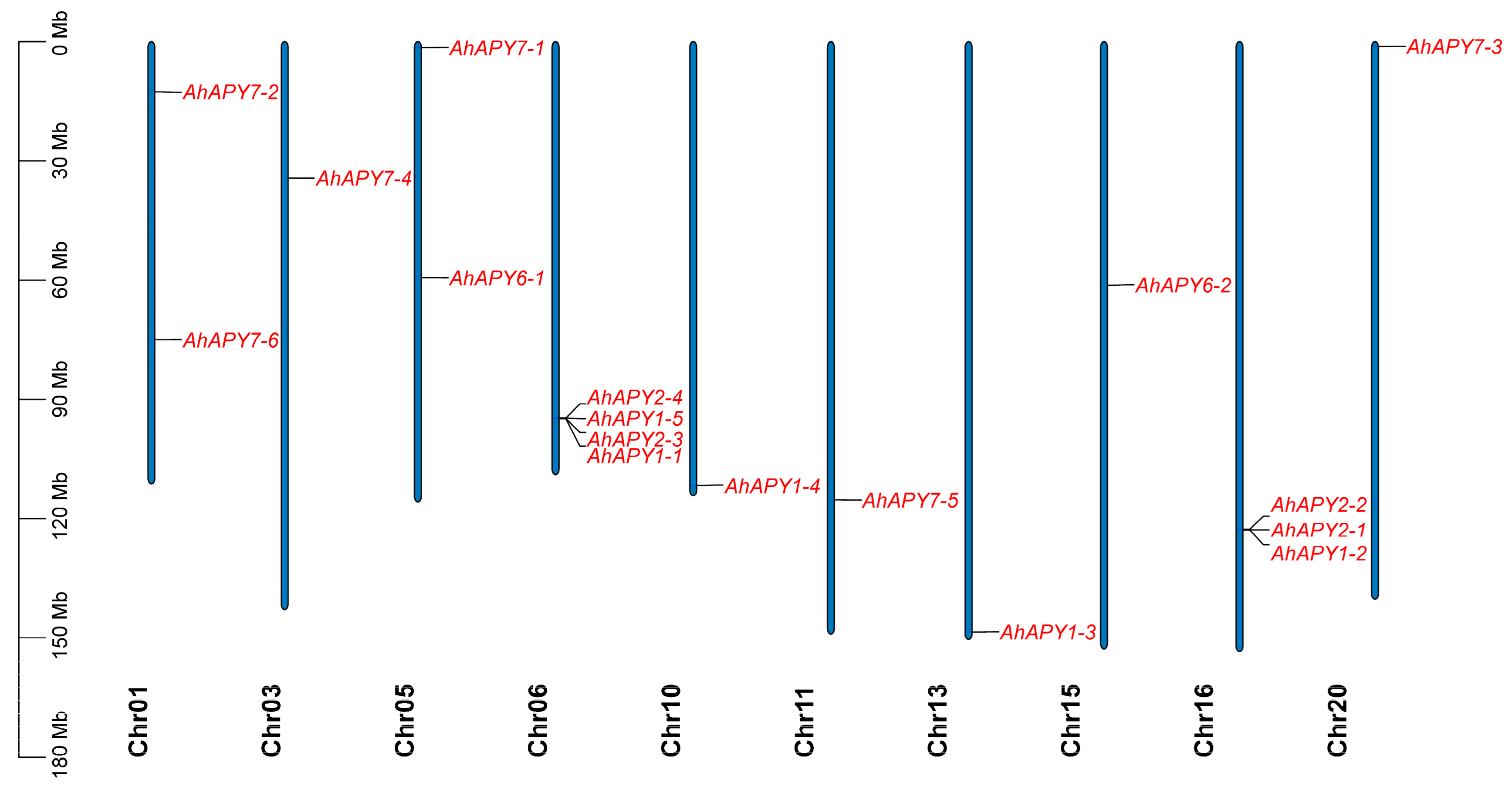
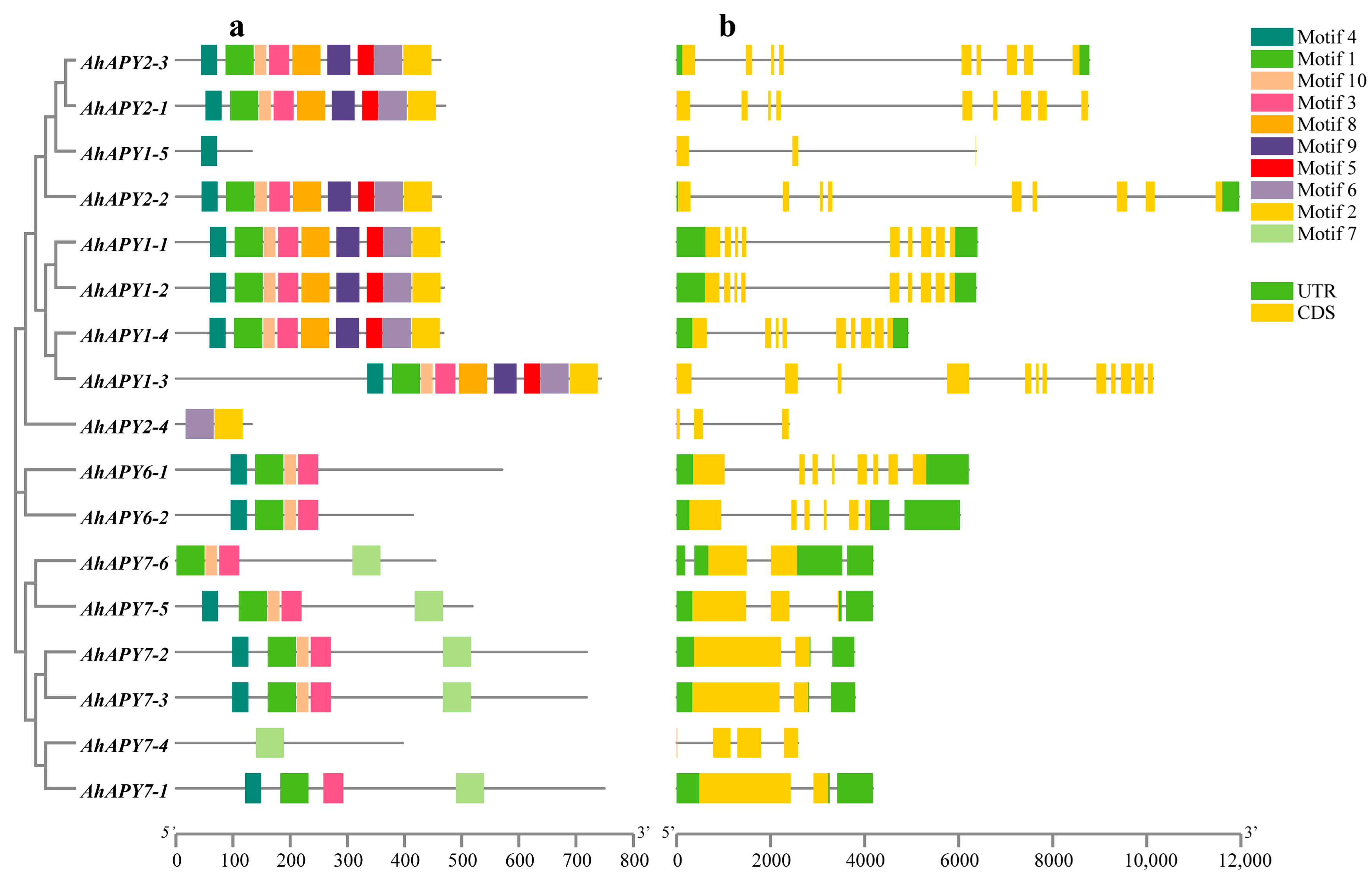
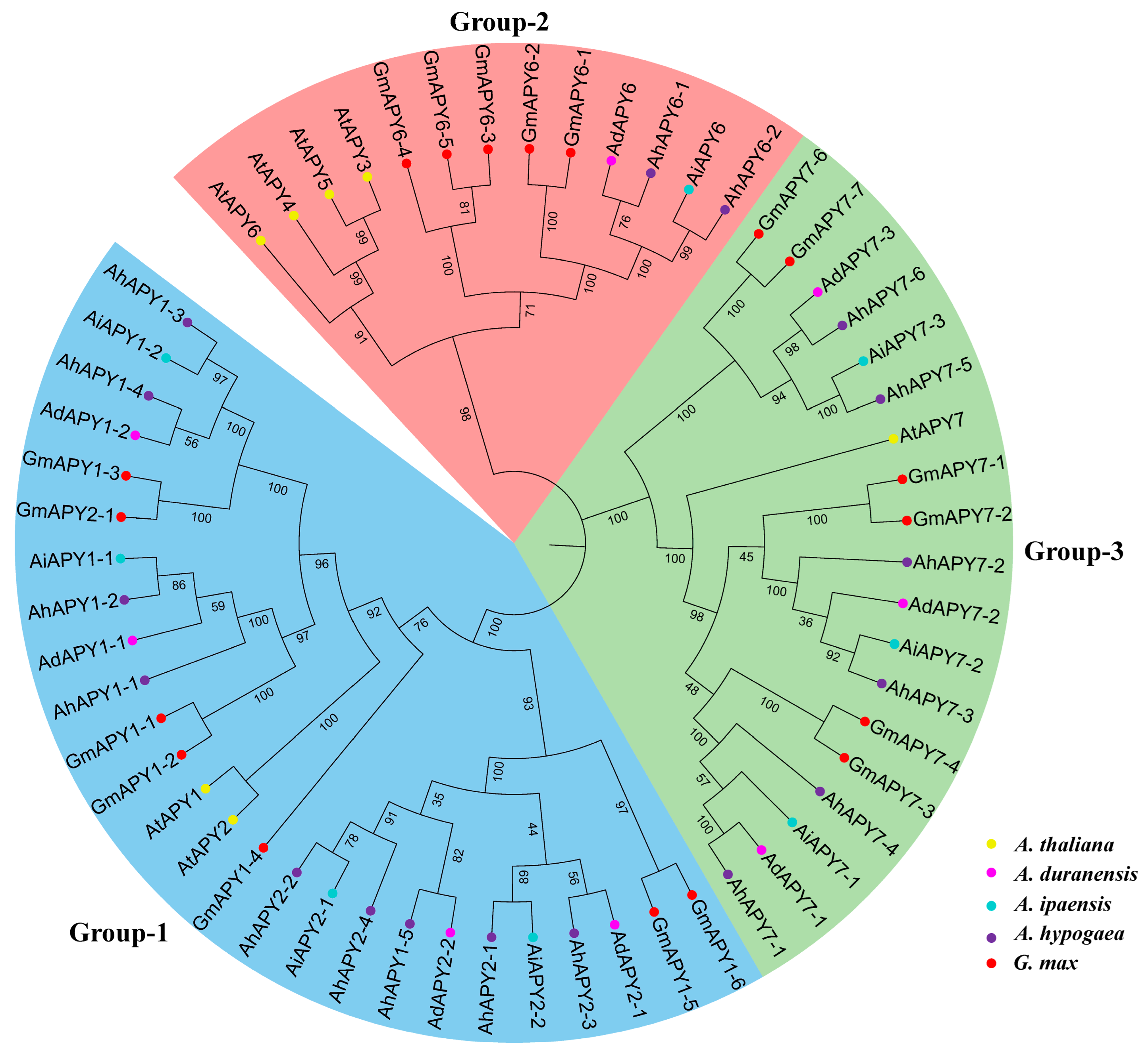
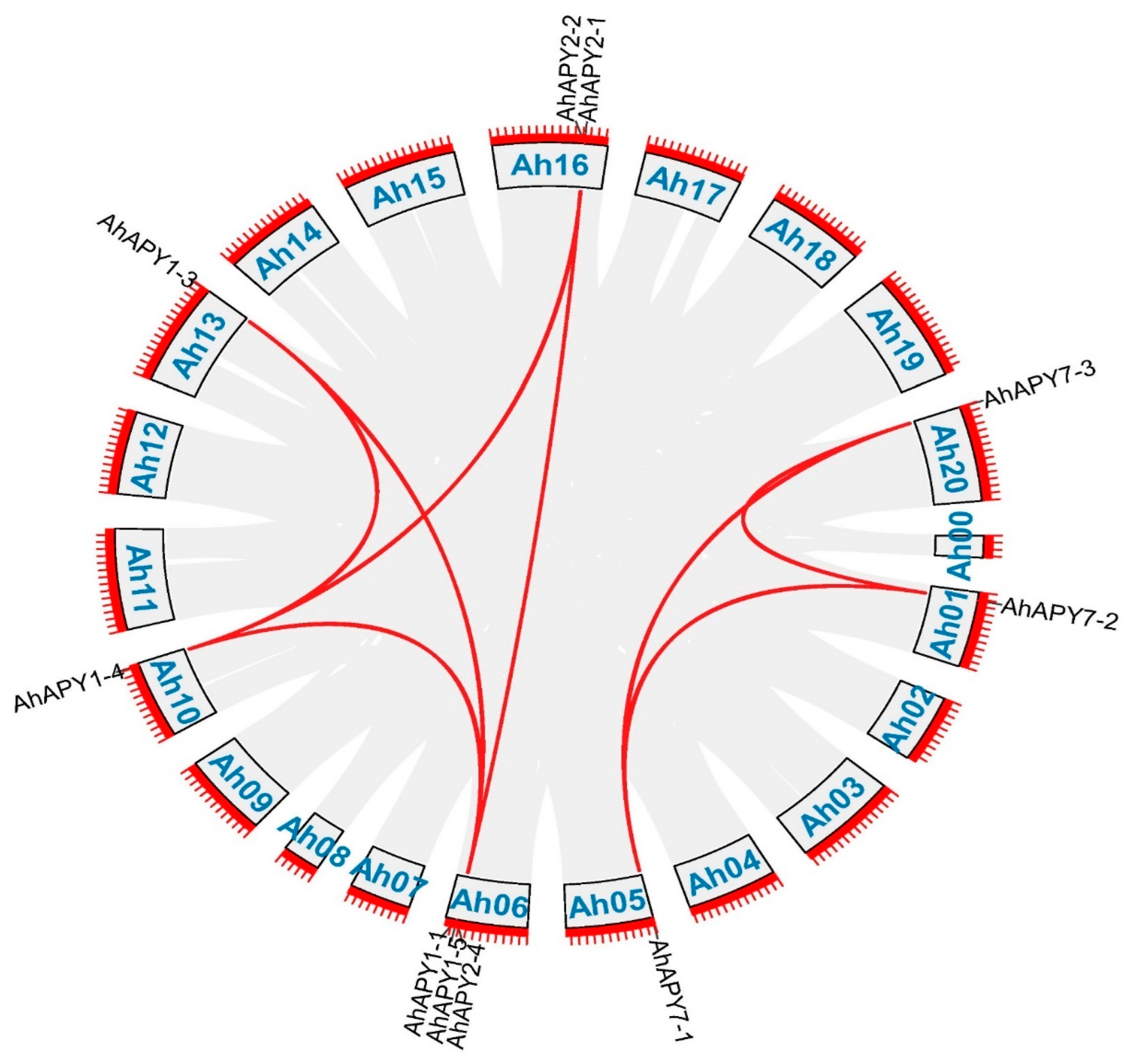


| ID | Renamed | Location | Protein Length (aa) | CDS Length (bp) | Gene Length (bp) | Exons | MW | pI | SC Localization |
|---|---|---|---|---|---|---|---|---|---|
| AH06G22700.1 | AhAPY1-1 | 6 (94877757…94883069) - | 469 | 1410 | 6399 | 9 | 51.4852 | 5.23 | Chloroplast |
| AH16G28170.1 | AhAPY1-2 | 16 (122861075…122866396) - | 469 | 1410 | 6372 | 9 | 50.957 | 5.77 | Chloroplast |
| AH13G57910.1 | AhAPY1-3 | 13 (148543072…148553205) + | 744 | 2235 | 10,134 | 12 | 80.6055 | 6.64 | Plasma Membrane |
| AH10G29400.1 | AhAPY1-4 | 10 (111719060…111723323) - | 468 | 1407 | 4925 | 9 | 51.0344 | 8.05 | Chloroplast |
| AH06G22670.1 | AhAPY1-5 | 6 (94715552…94721920) - | 133 | 402 | 6369 | 3 | 15.1463 | 7.89 | Extracellular/Mitochondrial/Nuclear |
| AH16G28160.1 | AhAPY2-1 | 16 (122829171…122837922) - | 471 | 1416 | 8752 | 9 | 52.2322 | 5.38 | Plasma Membrane |
| AH16G28150.1 | AhAPY2-2 | 16 (122697499…122709079) - | 464 | 1395 | 11,962 | 9 | 51.5263 | 5.18 | Plasma Membrane |
| AH06G22690.1 | AhAPY2-3 | 6 (94841125…94849566) - | 463 | 1392 | 8779 | 9 | 51.4852 | 5.23 | Plasma Membrane/Vacuole |
| AH06G22660.1 | AhAPY2-4 | 6 (94712005…94714391) - | 133 | 402 | 2387 | 3 | 14.9691 | 4.69 | Cytoplasmic/Extracellular |
| AH05G19000.1 | AhAPY6-1 | 5 (59386454…59391410) - | 571 | 1716 | 6212 | 8 | 63.0676 | 9.3 | Plasma Membrane |
| AH15G15910.1 | AhAPY6-2 | 15 (61240024…61243866) + | 415 | 1248 | 4526 | 6 | 45.4304 | 9.69 | Plasma Membrane |
| AH05G01440.1 | AhAPY7-1 | 5 (1495293…1498029) + | 750 | 2253 | 4222 | 2 | 82.3927 | 9.6 | Plasma Membrane |
| AH01G09830.1 | AhAPY7-2 | 1 (12576046…12578507) - | 719 | 2160 | 3829 | 2 | 80.8545 | 9.1 | Plasma Membrane |
| AH20G01300.1 | AhAPY7-3 | 20 (1188522…1190990) - | 719 | 2160 | 3801 | 2 | 80.8184 | 9.1 | Plasma Membrane |
| AH03G20330.1 | AhAPY7-4 | 3 (34322499…34325082) + | 397 | 1194 | 2584 | 4 | 43.2395 | 9.94 | Plasma Membrane |
| AH11G21920.1 | AhAPY7-5 | 11 (115305466…115308581) + | 519 | 1560 | 4173 | 3 | 57.6698 | 7.93 | Plasma Membrane |
| AH01G18780.1 | AhAPY7-6 | 1 (75010452…75012336) + | 454 | 1365 | 4229 | 2 | 50.1875 | 7.6 | Plasma Membrane |
| Seq_1 | Seq_2 | Ka | Ks | Ka_Ks | Selection Pressure | Time |
|---|---|---|---|---|---|---|
| AhAPY1-1 | AhAPY1-3 | 0.10308 | 0.76791 | 0.13424 | Purifying | 47.28514 |
| AhAPY1-4 | AhAPY1-3 | 0.00954 | 0.03517 | 0.27128 | Purifying | 2.165719 |
| AhAPY1-4 | AhAPY2-1 | 0.32445 | 1.1245 | 0.28852 | Purifying | 69.24287 |
| AhAPY1-5 | AhAPY1-4 | 0.3099 | 2.06906 | 0.14978 | Purifying | 127.405 |
| AhAPY2-4 | AhAPY2-2 | 0.04945 | 0.07096 | 0.69685 | Purifying | 4.36958 |
| AhAPY7-1 | AhAPY7-3 | 0.20883 | 0.71442 | 0.2923 | Purifying | 43.99162 |
| AhAPY7-2 | AhAPY7-1 | 0.20803 | 0.70387 | 0.29555 | Purifying | 43.34174 |
| AhAPY7-2 | AhAPY7-3 | 0.00362 | 0.02045 | 0.17706 | Purifying | 1.259094 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharif, Y.; Mamadou, G.; Yang, Q.; Cai, T.; Zhuang, Y.; Chen, K.; Deng, Y.; Khan, S.A.; Ali, N.; Zhang, C.; et al. Genome-Wide Investigation of Apyrase (APY) Genes in Peanut (Arachis hypogaea L.) and Functional Characterization of a Pod-Abundant Expression Promoter AhAPY2-1p. Int. J. Mol. Sci. 2023, 24, 4622. https://doi.org/10.3390/ijms24054622
Sharif Y, Mamadou G, Yang Q, Cai T, Zhuang Y, Chen K, Deng Y, Khan SA, Ali N, Zhang C, et al. Genome-Wide Investigation of Apyrase (APY) Genes in Peanut (Arachis hypogaea L.) and Functional Characterization of a Pod-Abundant Expression Promoter AhAPY2-1p. International Journal of Molecular Sciences. 2023; 24(5):4622. https://doi.org/10.3390/ijms24054622
Chicago/Turabian StyleSharif, Yasir, Gandeka Mamadou, Qiang Yang, Tiecheng Cai, Yuhui Zhuang, Kun Chen, Ye Deng, Shahid Ali Khan, Niaz Ali, Chong Zhang, and et al. 2023. "Genome-Wide Investigation of Apyrase (APY) Genes in Peanut (Arachis hypogaea L.) and Functional Characterization of a Pod-Abundant Expression Promoter AhAPY2-1p" International Journal of Molecular Sciences 24, no. 5: 4622. https://doi.org/10.3390/ijms24054622
APA StyleSharif, Y., Mamadou, G., Yang, Q., Cai, T., Zhuang, Y., Chen, K., Deng, Y., Khan, S. A., Ali, N., Zhang, C., Raza, A., Chen, H., Varshney, R. K., & Zhuang, W. (2023). Genome-Wide Investigation of Apyrase (APY) Genes in Peanut (Arachis hypogaea L.) and Functional Characterization of a Pod-Abundant Expression Promoter AhAPY2-1p. International Journal of Molecular Sciences, 24(5), 4622. https://doi.org/10.3390/ijms24054622









