Organizational Principles of the Centrifugal Projections to the Olfactory Bulb
Abstract
1. Introduction
2. Results
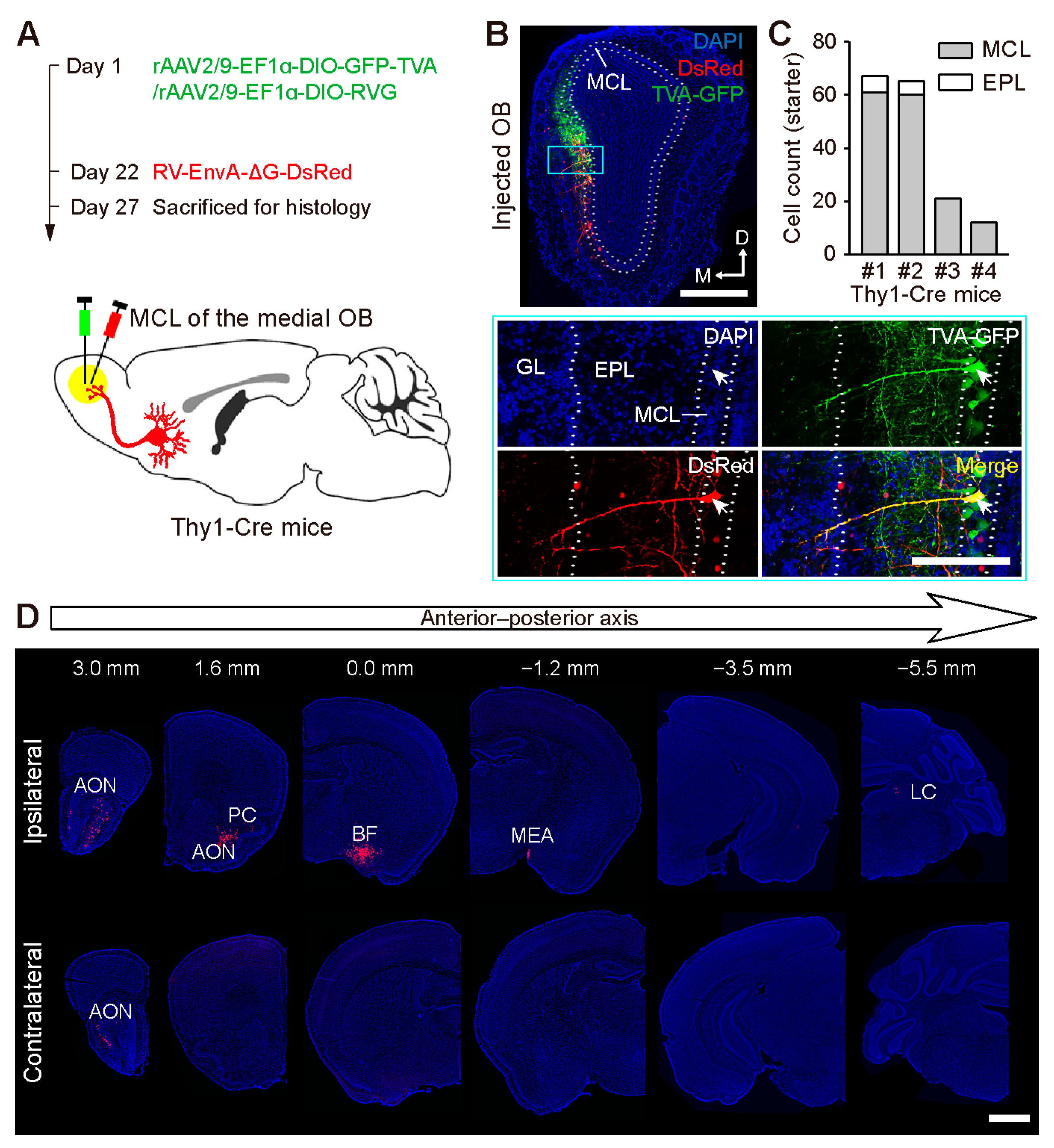
2.1. Brain-Wide Distribution of the Centrifugal Input to the OB
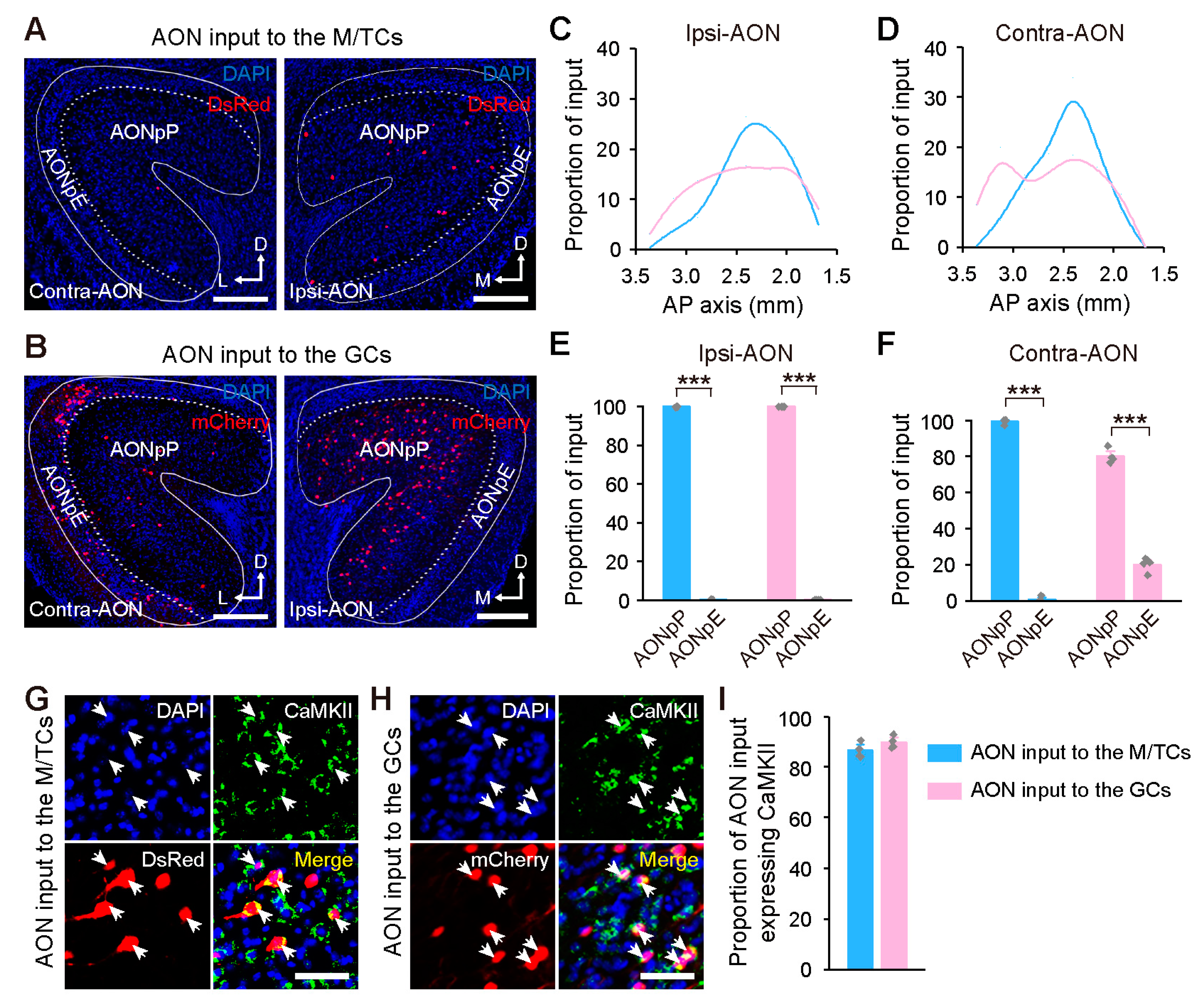
2.2. Centrifugal Input from the Primary Olfactory Areas to the OB
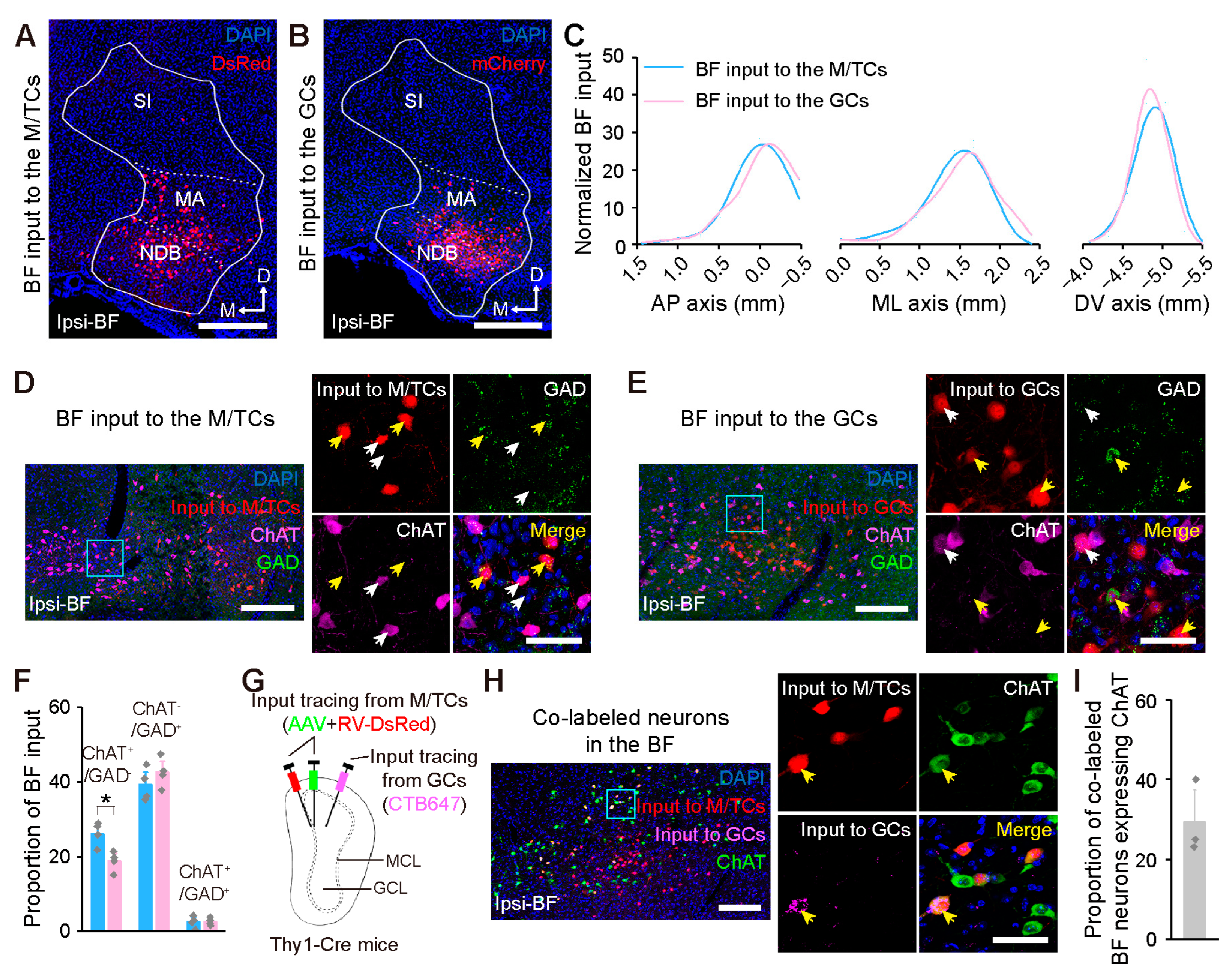
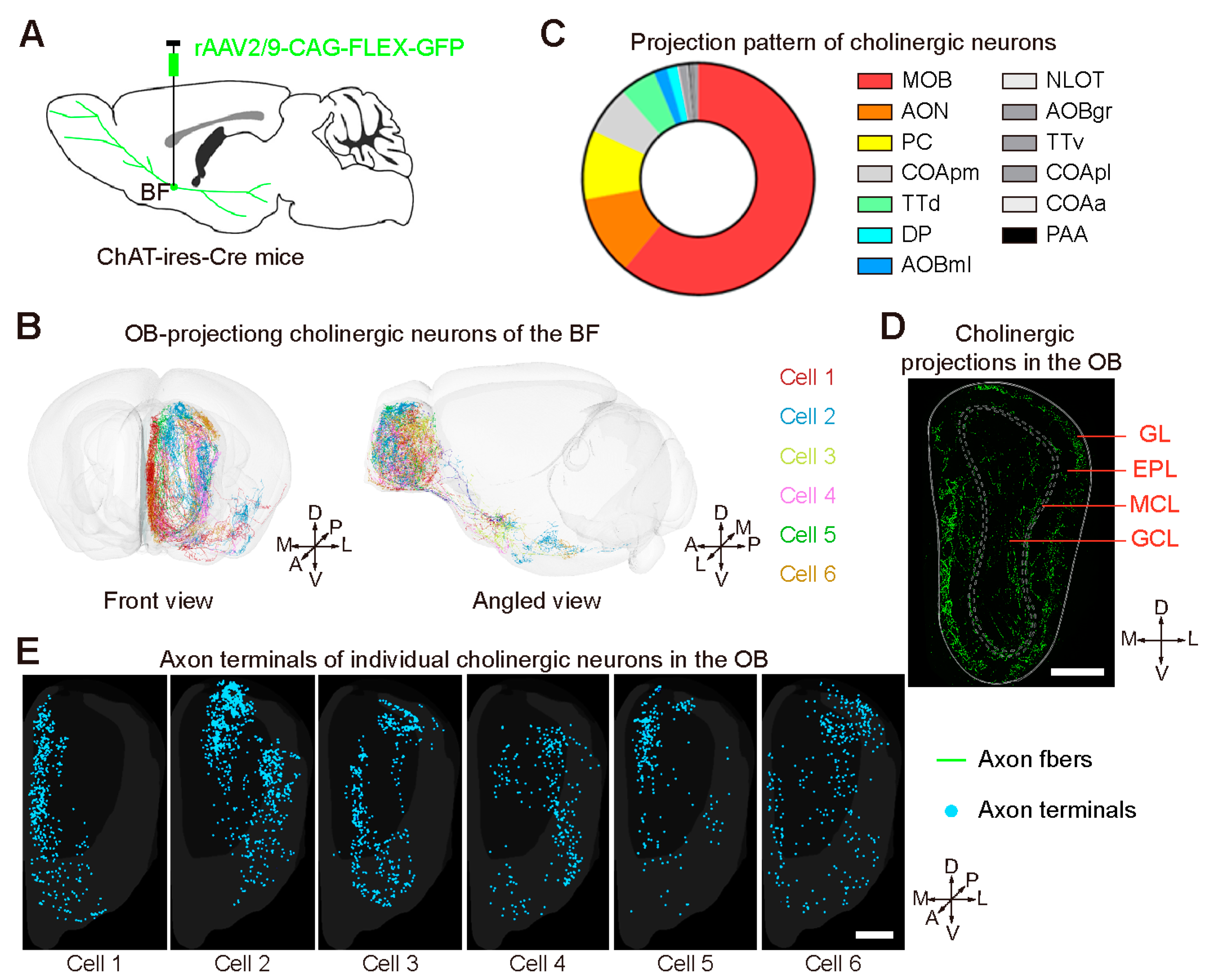
2.3. Centrifugal Input from the Neuromodulatory Areas to the OB
3. Discussion
4. Materials and Methods
4.1. Animals and Stereotaxic Surgeries
4.2. Retrograde Tracing Strategies
4.3. Histology and Imaging
4.4. Whole-Brain Imaging
4.5. Tracing Data Analysis
4.6. Reconstruction and Analysis of the Neurons
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Agranular insular cortex |
| AMY | Amygdalar area |
| AOBgr | Granular cell layer of the accessory olfactory bulb |
| AOBml | Mitral cell layer of the accessory olfactory bulb |
| AON | Anterior olfactory nucleus |
| AONpE | Anterior olfactory nucleus pars externa |
| AONpP | Anterior olfactory nucleus pars principalis |
| AP | Anterior–posterior |
| APC | Anterior piriform cortex |
| BF | Basal forebrain |
| COA | Cortical amygdalar nucleus |
| COAa | Anterior part of the cortical amygdalar nucleus |
| COApl | Posterolateral part of the cortical amygdalar nucleus |
| COApm | Posteromedial part of the cortical amygdalar nucleus |
| DR | Dorsal raphe nucleus |
| DV | Dorsal–ventral |
| EP | Endopiriform nucleus |
| EPL | External plexiform layer |
| fMOST | Fluorescence micro-optical sectioning tomography |
| GCL | Granular cell layer |
| GCs | Granular neurons |
| GL | Glomerular layer |
| HIP | Hippocampal region |
| HY | Hypothalamus |
| LC | Locus coeruleus |
| LEC | Lateral entorhinal cortex |
| MA | Magnocellular preoptic nucleus |
| MEA | Medial amygdalar nucleus |
| MCL | Mitral cell layer |
| ML | Medial–lateral |
| MOB | Main olfactory bulb |
| MS | Medial septum |
| M/TCs | Mitral/tufted cells |
| NDB | Diagonal band nucleus |
| NLOT | Nucleus of the lateral olfactory tract |
| OB | Olfactory bulb |
| OT | Olfactory tubercle |
| PBS | Phosphate-buffered saline |
| PC | Piriform cortex |
| PERI | Perirhinal cortex |
| PFA | Paraformaldehyde |
| PPC | Posterior piriform cortex |
| SI | Substantia innominata |
| TT | Taenia tecta |
| TTd | Dorsal part of the taenia tecta |
| TTv | Ventral part of the taenia tecta |
| VTA | Ventral tegmental area |
References
- Quintela, M.R.; Bauer, J.; Wallhorn, L.; Le, K.; Brunert, D.; Rothermel, M. Dynamic impairment of olfactory behavior and signaling mediated by an olfactory corticofugal system. J. Neurosci. 2020, 40, 7269–7285. [Google Scholar] [CrossRef]
- Ogg, M.C.; Ross, J.M.; Bendahmane, M.; Fletcher, M.L. Olfactory bulb acetylcholine release dishabituates odor responses and reinstates odor investigation. Nat. Commun. 2018, 9, 1868. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.D.; Li, W. Top-down influences on visual processing. Nat. Rev. Neurosci. 2013, 14, 350–363. [Google Scholar] [CrossRef] [PubMed]
- Petreanu, L.; Gutnisky, D.A.; Huber, D.; Xu, N.L.; O’Connor, D.H.; Tian, L.; Looger, L.; Svoboda, K. Activity in motor-sensory projections reveals distributed coding in somatosensation. Nature 2012, 489, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.D.; Sigman, M. Brain states: Top-down influences in sensory processing. Neuron 2007, 54, 677–696. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Sakano, H. How is the olfactory map formed and interpreted in the mammalian brain? Annu. Rev. Neurosci. 2011, 34, 467–499. [Google Scholar] [CrossRef]
- Cleland, T.A.; Linster, C. Central olfactory structures. Handb. Clin. Neurol. 2019, 164, 79–96. [Google Scholar]
- Wen, P.; Rao, X.; Xu, L.Y.; Zhang, Z.; Jia, F.; He, X.; Xu, F. Cortical organization of centrifugal afferents to the olfactory bulb: Mono- and trans-synaptic tracing with recombinant neurotropic viral tracers. Neurosci. Bull. 2019, 35, 709–723. [Google Scholar] [CrossRef]
- Chen, Z.; Padmanabhan, K. Top-down control of inhibitory granule cells in the main olfactory bulb reshapes neural dynamics giving rise to a diversity of computations. Front. Comput. Neurosci. 2020, 14, 59. [Google Scholar] [CrossRef]
- Otazu, G.H.; Chae, H.; Davis, M.B.; Albeanu, D.F. Cortical feedback decorrelates olfactory bulb output in awake mice. Neuron 2015, 86, 1461–1477. [Google Scholar] [CrossRef]
- Matsutani, S.; Yamamoto, N. Centrifugal innervation of the mammalian olfactory bulb. Anat. Sci. Int. 2008, 83, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.J.; Macrides, F. The organization of centrifugal projections from the anterior olfactory nucleus, ventral hippocampal rudiment, and piriform cortex to the main olfactory bulb in the hamster: An autoradiog raphic study. J. Comp. Neurol. 1981, 203, 475–493. [Google Scholar] [CrossRef]
- Davis, B.J.; Macrides, F.; Youngs, W.M.; Schneider, S.P.; Rosene, D.L. Efferents and centrifugal afferents of the main and accessory olfactory bulbs in the hamster. Brain Res. Bull. 1978, 3, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Marla, B.; Luskin, P.J.L. The topographic organization of associational fibers of the olfactory system in the rat, including centrifugal fibers to the olfactory bulb. J. Comp. Neurol. 1983, 216, 264–291. [Google Scholar]
- Schoenfeld, T.A.; Macrides, F. Topographic organization of connections between the main olfactory bulb and pars externa of the anterior olfactory nucleus in the hamster. J. Comp. Neurol. 1984, 227, 121–135. [Google Scholar] [CrossRef]
- Miyamichi, K.; Shlomai-Fuchs, Y.; Shu, M.; Weissbourd, B.C.; Luo, L.; Mizrahi, A. Dissecting local circuits: Parvalbumin interneurons underlie broad feedback control of olfactory bulb output. Neuron 2013, 80, 1232–1245. [Google Scholar] [CrossRef] [PubMed]
- Boyd, A.M.; Sturgill, J.F.; Poo, C.; Isaacson, J.S. Cortical feedback control of olfactory bulb circuits. Neuron 2012, 76, 1161–1174. [Google Scholar] [CrossRef] [PubMed]
- Balu, R.; Pressler, R.T.; Strowbridge, B.W. Multiple modes of synaptic excitation of olfactory bulb granule cells. J. Neurosci. 2007, 27, 5621–5632. [Google Scholar] [CrossRef]
- Grobman, M.; Dalal, T.; Lavian, H.; Shmuel, R.; Belelovsky, K.; Xu, F.; Korngreen, A.; Haddad, R. A mirror-symmetric excitatory link coordinates odor maps across olfactory bulbs and enables odor perceptual unity. Neuron 2018, 99, 800–813. [Google Scholar] [CrossRef]
- Illig, K.R. Corticofugal projections from the anterior olfactory nucleus target olfactory bulb principal cells. Nat. Preced. 2011. [Google Scholar] [CrossRef]
- Markopoulos, F.; Rokni, D.; Gire, D.H.; Murthy, V.N. Functional properties of cortical feedback projections to the olfactory bulb. Neuron 2012, 76, 1175–1188. [Google Scholar] [CrossRef]
- Vinograd, A.; Tasaka, G.I.; Kreines, L.; Weiss, Y.; Mizrahi, A. The pre-synaptic landscape of mitral/tufted cells of the main olfactory bulb. Front. Neuroanat. 2019, 13, 58. [Google Scholar] [CrossRef]
- Brunert, D.; Rothermel, M. Extrinsic neuromodulation in the rodent olfactory bulb. Cell Tissue Res. 2021, 383, 507–524. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, K.; Osakada, F.; Tarabrina, A.; Kizer, E.; Callaway, E.M.; Gage, F.H.; Sejnowski, T.J. Centrifugal inputs to the main olfactory bulb revealed through whole brain circuit-mapping. Front. Neuroanat. 2018, 12, 115. [Google Scholar] [CrossRef] [PubMed]
- Zandt, E.E.; Cansler, H.L.; Denson, H.B.; Wesson, D.W. Centrifugal innervation of the olfactory bulb: A reappraisal. eNeuro 2019, 6, ENEURO.0390-18.2019. [Google Scholar]
- Padmanabhan, K.; Osakada, F.; Tarabrina, A.; Kizer, E.; Callaway, E.M.; Gage, F.H.; Sejnowski, T.J. Diverse representations of olfactory information in centrifugal feedback projections. J. Neurosci. 2016, 36, 7535–7545. [Google Scholar] [CrossRef]
- Mohedano-Moriano, A.; de la Rosa-Prieto, C.; Saiz-Sanchez, D.; Ubeda-Banon, I.; Pro-Sistiaga, P.; de Moya-Pinilla, M.; Martinez-Marcos, A. Centrifugal telencephalic afferent connections to the main and accessory olfactory bulbs. Front. Neuroanat. 2012, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Zeppilli, S.; Ackels, T.; Attey, R.; Klimpert, N.; Ritola, K.D.; Boeing, S.; Crombach, A.; Schaefer, A.T.; Fleischmann, A. Molecular characterization of projection neuron subtypes in the mouse olfactory bulb. Elife 2021, 10, e65445. [Google Scholar] [CrossRef]
- Brunjes, P.C.; Illig, K.R.; Meyer, E.A. A field guide to the anterior olfactory nucleus (cortex). Brain Res. Rev. 2005, 50, 305–335. [Google Scholar] [CrossRef]
- Bekkers, J.M.; Suzuki, N. Neurons and circuits for odor processing in the piriform cortex. Trends Neurosci. 2013, 36, 429–438. [Google Scholar] [CrossRef]
- Wilson, D.A.; Sullivan, R.M. Cortical processing of odor objects. Neuron 2011, 72, 506–519. [Google Scholar] [CrossRef]
- Zheng, Y.; Tao, S.; Liu, Y.; Liu, J.; Sun, L.; Zheng, Y.; Tian, Y.; Su, P.; Zhu, X.; Xu, F. Basal forebrain-dorsal hippocampus cholinergic circuit regulates olfactory associative learning. Int. J. Mol. Sci. 2022, 23, 8472. [Google Scholar] [CrossRef]
- Suyama, H.; Egger, V.; Lukas, M. Top-down acetylcholine signaling via olfactory bulb vasopressin cells contributes to social discrimination in rats. Commun. Biol. 2021, 4, 603. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Z.; Chen, J.; Manyande, A.; Haddad, R.; Liu, Q.; Xu, F. Cell-type-specific whole-brain direct inputs to the anterior and posterior piriform cortex. Front. Neural Circuit 2020, 14, 4. [Google Scholar] [CrossRef]
- Ahrlund-Richter, S.; Xuan, Y.; van Lunteren, J.A.; Kim, H.; Ortiz, C.; Pollak Dorocic, I.; Meletis, K.; Carlen, M. A whole-brain atlas of monosynaptic input targeting four different cell types in the medial prefrontal cortex of the mouse. Nat. Neurosci. 2019, 22, 657–668. [Google Scholar] [CrossRef]
- Roland, B.; Deneux, T.; Franks, K.M.; Bathellier, B.; Fleischmann, A. Odor identity coding by distributed ensembles of neurons in the mouse olfactory cortex. Elife 2017, 6, e26337. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, W.; Wang, W.; Fan, H.; Hou, R.; Chen, Y.; Chen, Z.; Ge, C.; Duan, S.; Compte, A.; et al. Active information maintenance in working memory. Elife 2019, 8, e43191. [Google Scholar] [CrossRef]
- Aqrabawi, A.J.; Kim, J.C. Olfactory memory representations are stored in the anterior olfactory nucleus. Nat. Commun. 2020, 11, 1246. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Barkai, E.; Schiller, J. Plasticity of olfactory bulb inputs mediated by dendritic NMDA-spikes in rodent piriform cortex. Elife 2021, 10, e70383. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Tan, J.; Qin, C.; Lu, Y.; Ding, C.; Luo, M. Precise circuitry links bilaterally symmetric olfactory maps. Neuron 2008, 58, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Dalal, T.; Gupta, N.; Haddad, R. Bilateral and unilateral odor processing and odor perception. Commun. Biol. 2020, 3, 150. [Google Scholar] [CrossRef]
- Miyamichi, K.; Amat, F.; Moussavi, F.; Wang, C.; Wickersham, I.; Wall, N.R.; Taniguchi, H.; Tasic, B.; Huang, Z.J.; He, Z.; et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature 2011, 472, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Mooney, R.; Katz, L.C. Synaptic integration of olfactory information in mouse anterior olfactory nucleus. J. Neurosci. 2006, 26, 12023–12032. [Google Scholar] [CrossRef] [PubMed]
- Kermen, F.; Lal, P.; Faturos, N.G.; Yaksi, E. Interhemispheric connections between olfactory bulbs improve odor detection. PLoS Biol. 2020, 18, e3000701. [Google Scholar] [CrossRef]
- Illig, K.R.; Eudy, J.D. Contralateral projections of the rat anterior olfactory nucleus. J. Comp. Neurol. 2009, 512, 115–123. [Google Scholar] [CrossRef]
- Ma, M.; Luo, M. Optogenetic activation of basal forebrain cholinergic neurons modulates neuronal excitability and sensory responses in the main olfactory bulb. J. Neurosci. 2012, 32, 10105–10116. [Google Scholar] [CrossRef]
- Case, D.T.; Burton, S.D.; Gedeon, J.Y.; Williams, S.G.; Urban, N.N.; Seal, R.P. Layer- and cell type-selective co-transmission by a basal forebrain cholinergic projection to the olfactory bulb. Nat. Commun. 2017, 8, 652. [Google Scholar] [CrossRef]
- Chapuis, J.; Wilson, D.A. Cholinergic modulation of olfactory pattern separation. Neurosci. Lett. 2013, 545, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, M.L.; Wilson, D.A. Experience modifies olfactory acuity: Acetylcholine-dependent learning decreases behavioral generalization between similar odorants. J. Neurosci. 2002, 22, RC201. [Google Scholar] [CrossRef] [PubMed]
- Rothermel, M.; Carey, R.M.; Puche, A.; Shipley, M.T.; Wachowiak, M. Cholinergic inputs from basal forebrain add an excitatory bias to odor coding in the olfactory bulb. J. Neurosci. 2014, 34, 4654–4664. [Google Scholar] [CrossRef]
- Picciotto, M.R.; Higley, M.J.; Mineur, Y.S. Acetylcholine as a neuromodulator: Cholinergic signaling shapes nervous system function and behavior. Neuron 2012, 76, 116–129. [Google Scholar] [CrossRef]
- Kasa, P.; Hlavati, I.; Dobo, E.; Wolff, A.; Joo, F.; Wolff, J.R. Synaptic and non-synaptic cholinergic innervation of the various types of neurons in the main olfactory bulb of adult rat: Immunocytochemistry of choline acetyltransferase. Neuroscience 1995, 66, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, M.; Kiyokage, E.; Sohn, J.; Hioki, H.; Harada, T.; Toida, K. Structural basis for cholinergic regulation of neural circuits in the mouse olfactory bulb. J. Comp. Neurol. 2017, 525, 574–591. [Google Scholar] [CrossRef]
- Lchikawa, T.; Hirata, Y. Organization of choline acetyltransferase-containing structures in the forebrain of the rat. J. Neurosci. 1986, 6, 281–292. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, R.D.; Vijayaraghavan, S. Paying attention to smell: Cholinergic signaling in the olfactory bulb. Front. Synaptic Neurosci. 2014, 6, 21. [Google Scholar] [PubMed]
- Luo, L.; Callaway, E.M.; Svoboda, K. Genetic dissection of neural circuits- a decade of progress. Neuron 2018, 98, 256–281. [Google Scholar] [CrossRef]
- Callaway, E.M.; Luo, L. Monosynaptic circuit tracing with glycoprotein-deleted rabies viruses. J. Neurosci. 2015, 35, 8979–8985. [Google Scholar] [CrossRef]
- Zhu, X.; Lin, K.; Liu, Q.; Yue, X.; Mi, H.; Huang, X.; He, X.; Wu, R.; Zheng, D.; Wei, D.; et al. Rabies virus pseudotyped with CVS-N2C glycoprotein as a powerful tool for retrograde neuronal network tracing. Neurosci. Bull. 2020, 36, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Reardon, T.R.; Murray, A.J.; Turi, G.F.; Wirblich, C.; Croce, K.R.; Schnell, M.J.; Jessell, T.M.; Losonczy, A. Rabies virus CVS-N2c(deltaG) strain enhances retrograde synaptic transfer and neuronal viability. Neuron 2016, 89, 711–724. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, H.; Wen, P.; Zhu, X.; Wang, L.; Liu, Q.; Wang, J.; He, X.; Wang, H.; Xu, F. Whole-brain mapping of the inputs and outputs of the medial part of the olfactory tubercle. Front. Neural Circuit 2017, 11, 52. [Google Scholar] [CrossRef]
- Li, X.; Yu, B.; Sun, Q.; Zhang, Y.; Ren, M.; Zhang, X.; Li, A.; Yuan, J.; Madisen, L.; Luo, Q.; et al. Generation of a whole-brain atlas for the cholinergic system and mesoscopic projectome analysis of basal forebrain cholinergic neurons. Proc. Natl. Acad. Sci. USA 2018, 115, 415–420. [Google Scholar] [CrossRef]
- Wee, R.W.S.; MacAskill, A.F. Biased connectivity of brain-wide inputs to ventral subiculum output neurons. Cell Rep. 2020, 30, 3644–3654.e6. [Google Scholar] [CrossRef] [PubMed]



| Virus Name | Resource | Titers |
|---|---|---|
| rAAV2/9-EF1α-DIO-GFP-TVA | Fuqiang Xu group [34,60] | 6.50 × 1012 vg/mL |
| rAAV2/9-EF1α-DIO-RVG | Fuqiang Xu group [34,60] | 5.00 × 1012 vg/mL |
| RV-EnvA-ΔG-DsRed | Fuqiang Xu group [34,60] | 4.70 × 108 PFU/mL |
| RV-ΔG-mCherry | Fuqiang Xu group [8,58] | 6.00 × 107 PFU/ML |
| Antibodies, Recombinant Proteins, and Chemicals | Resource | Identification |
|---|---|---|
| Anti-CaMKIIα goat polyclonal antibody | Abcam | Cat# ab87597 |
| Anti-ChAT goat polyclonal antibody | Millipore | Cat# ab144p |
| Anti-GAD65/67 rabbit monoclonal antibody | Abcam | Cat# ab183999 |
| Donkey anti-goat Alexa Fluor 488 | Jackson ImmunoResearch | Cat# 705-545-147 |
| Donkey anti-goat Alexa Fluor 647 | Jackson ImmunoResearch | Cat# 705-605-147 |
| Donkey anti-rabbit fluorescein FITC | Jackson ImmunoResearch | Cat# 711-095-152 |
| 4’,6-Diamidino-2-phenylindole dihydrochloride (DAPI) | Beyotime | Cat# C1002 |
| Paraformaldehyde (PFA) | Sigma | Cat# 158127 |
| Cholera toxin subunit B (recombinant), Alexa Fluor 647 conjugate (CTB647) | Invitrogen | Cat# C34778 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Li, X.; Chen, F.; Liu, Q.; Xu, F. Organizational Principles of the Centrifugal Projections to the Olfactory Bulb. Int. J. Mol. Sci. 2023, 24, 4579. https://doi.org/10.3390/ijms24054579
Wang L, Li X, Chen F, Liu Q, Xu F. Organizational Principles of the Centrifugal Projections to the Olfactory Bulb. International Journal of Molecular Sciences. 2023; 24(5):4579. https://doi.org/10.3390/ijms24054579
Chicago/Turabian StyleWang, Li, Xiangning Li, Fengming Chen, Qing Liu, and Fuqiang Xu. 2023. "Organizational Principles of the Centrifugal Projections to the Olfactory Bulb" International Journal of Molecular Sciences 24, no. 5: 4579. https://doi.org/10.3390/ijms24054579
APA StyleWang, L., Li, X., Chen, F., Liu, Q., & Xu, F. (2023). Organizational Principles of the Centrifugal Projections to the Olfactory Bulb. International Journal of Molecular Sciences, 24(5), 4579. https://doi.org/10.3390/ijms24054579






