PEGylated Liposomes Loaded with Carbamate Inhibitor ANP0903 Trigger Apoptosis by Enhancing ER Stress in HepG2 Cancer Cells
Abstract
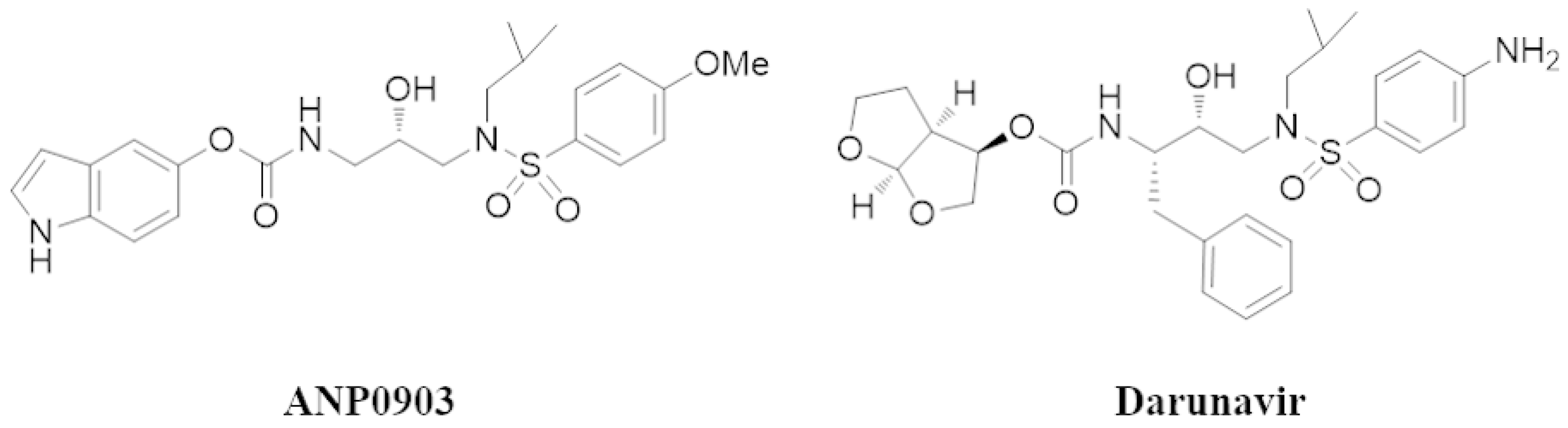
1. Introduction
2. Results
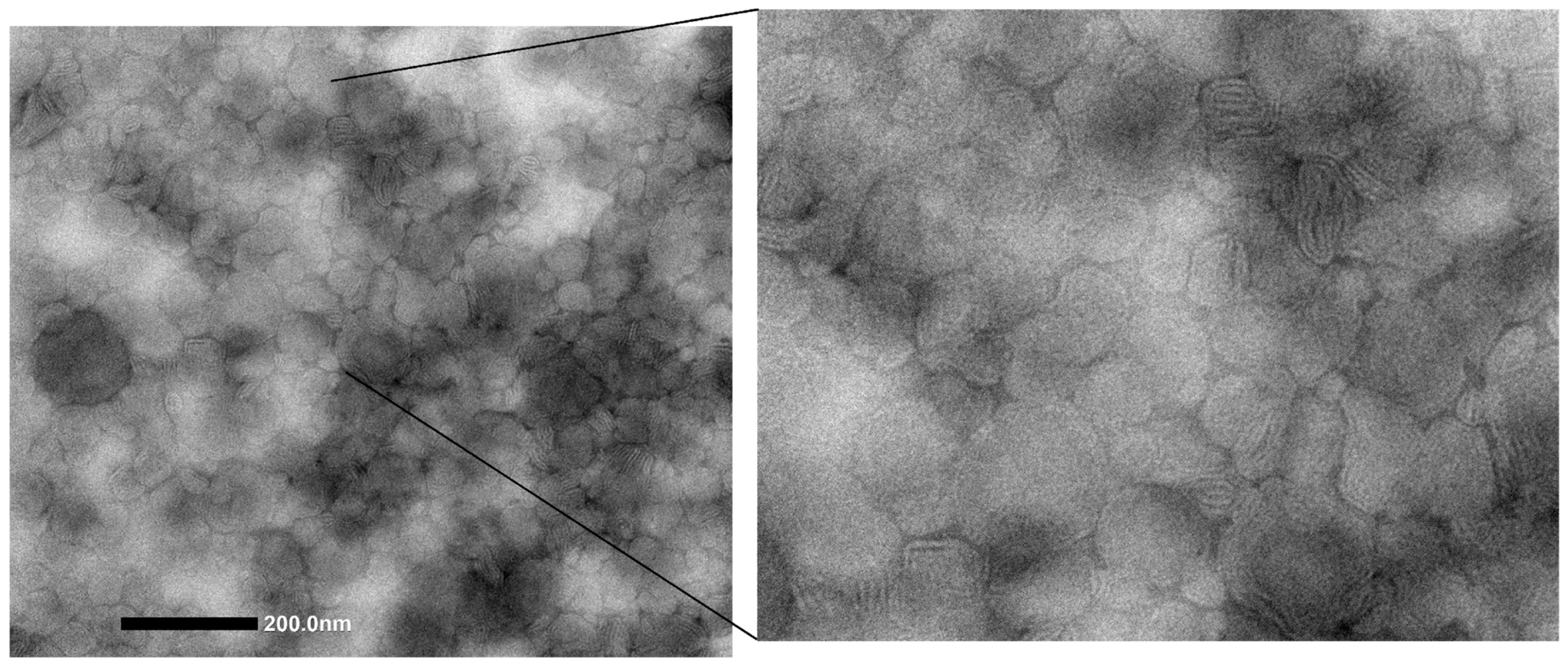
2.1. Vesicle Production and Characterization
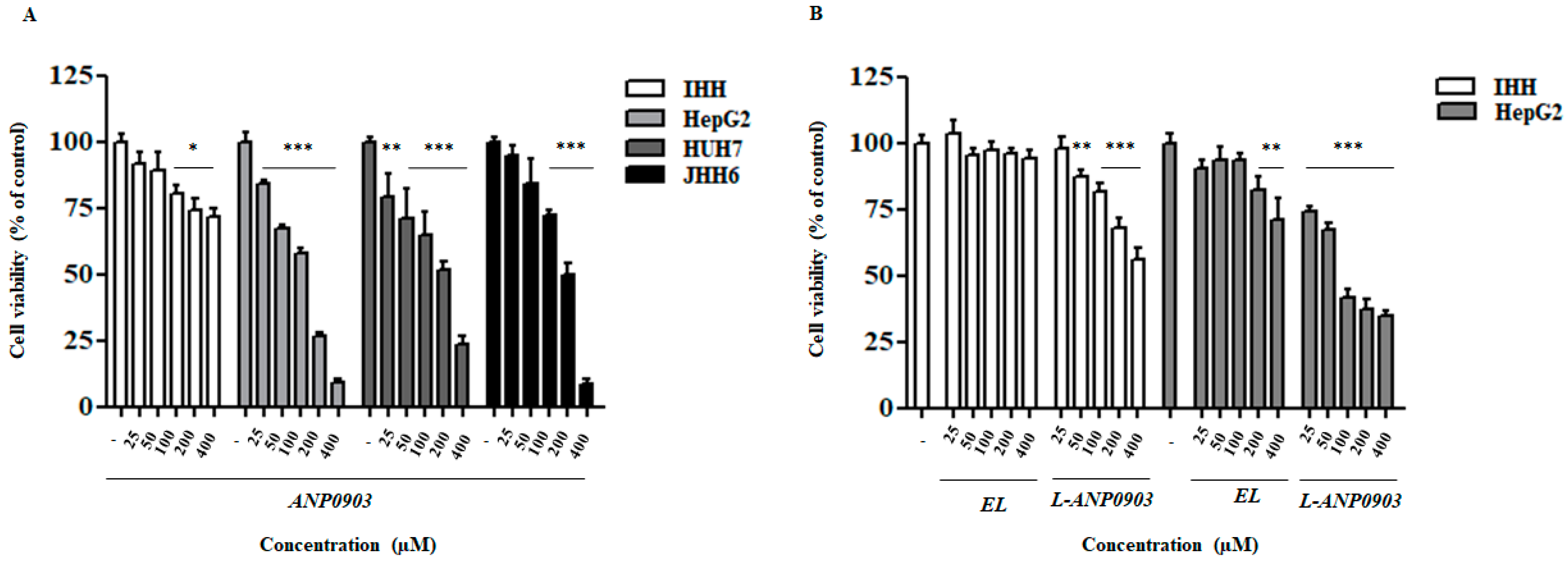
2.2. ANP0903 Affected Viability of Hepatoma Cell Lines in a Dose-Dependent Manner
2.3. ANP0903 Induced Morphological Alterations in HepG2-Treated Cells
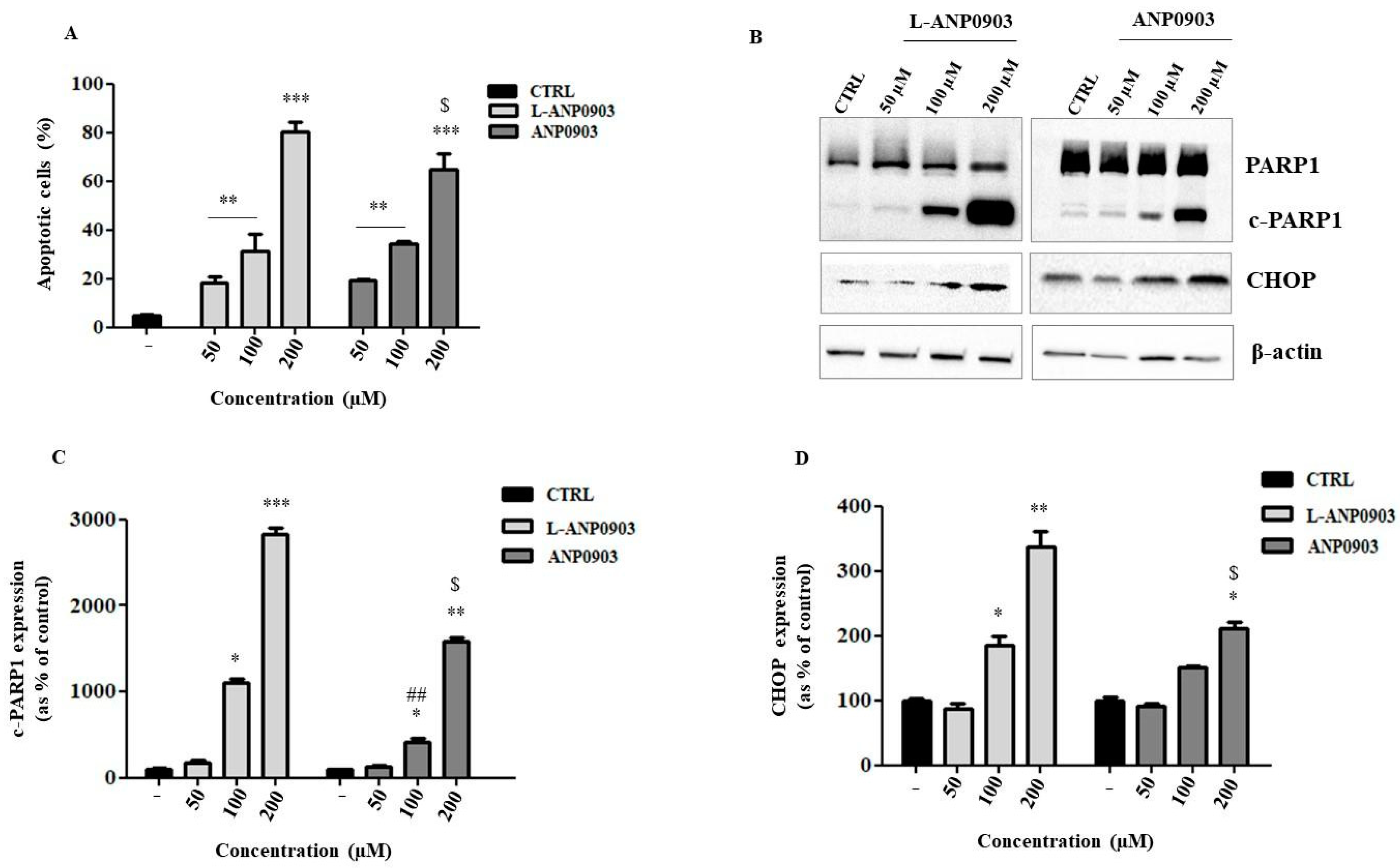
2.4. Treatment of ANP0903 Significantly Induced Apoptosis in Hepatoma Cells
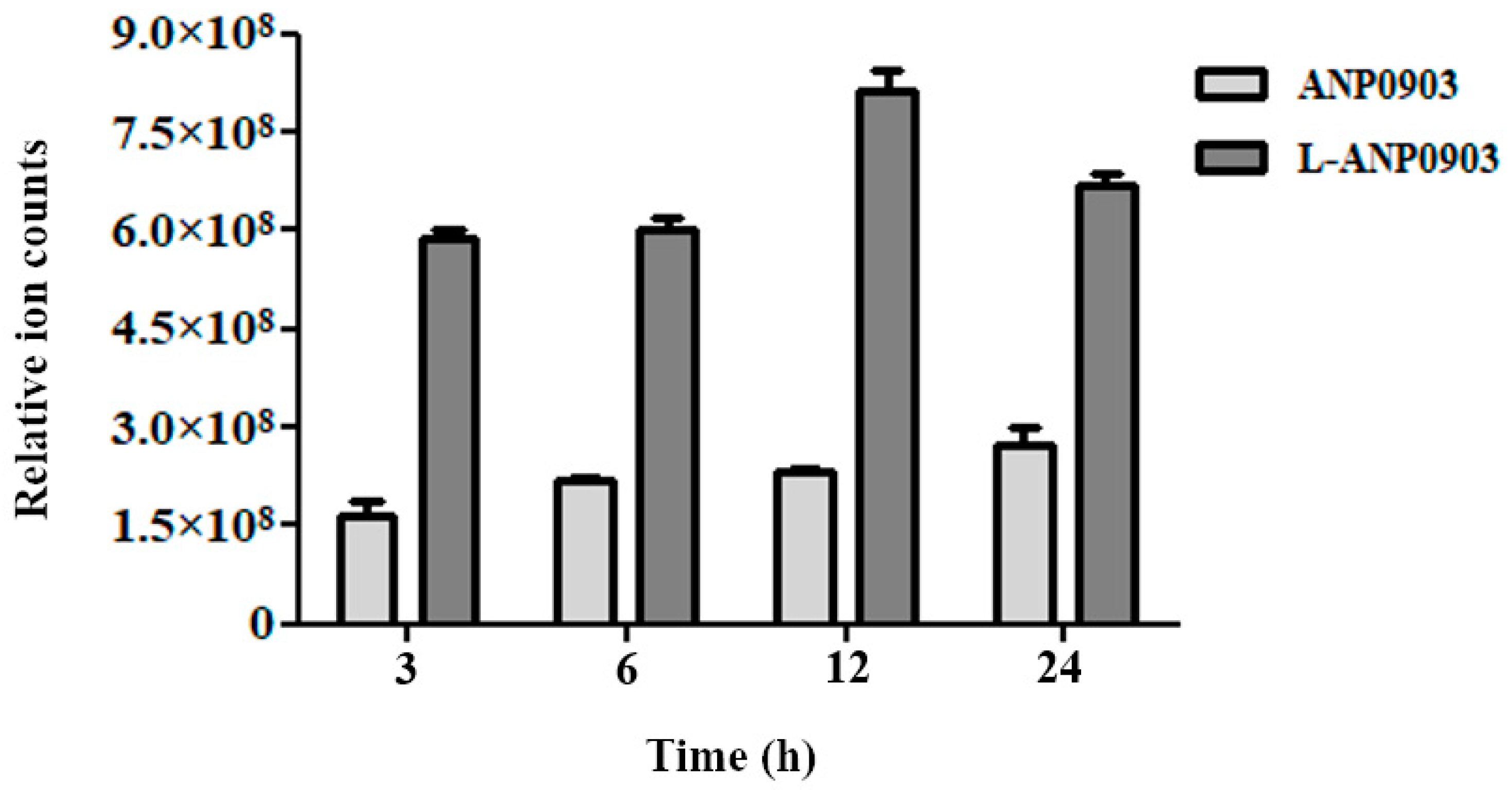
2.5. Intracellular Intake of ANP0903
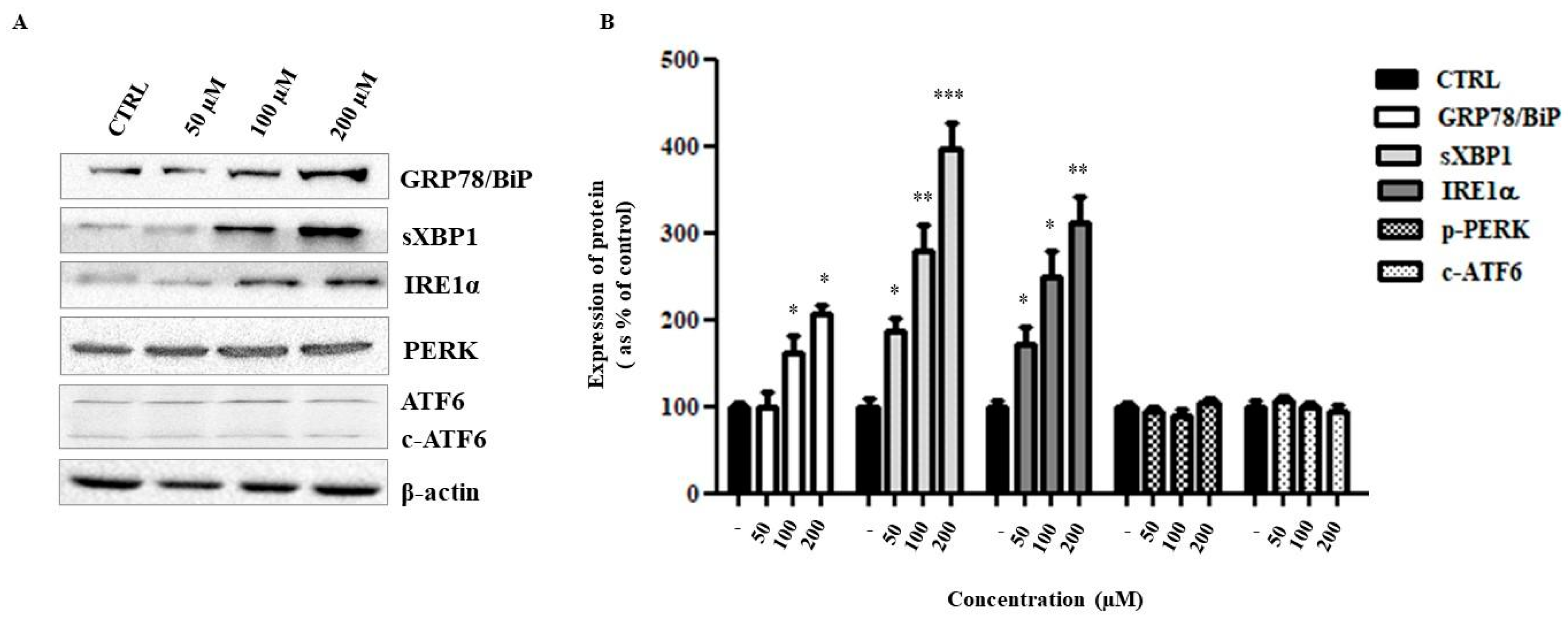
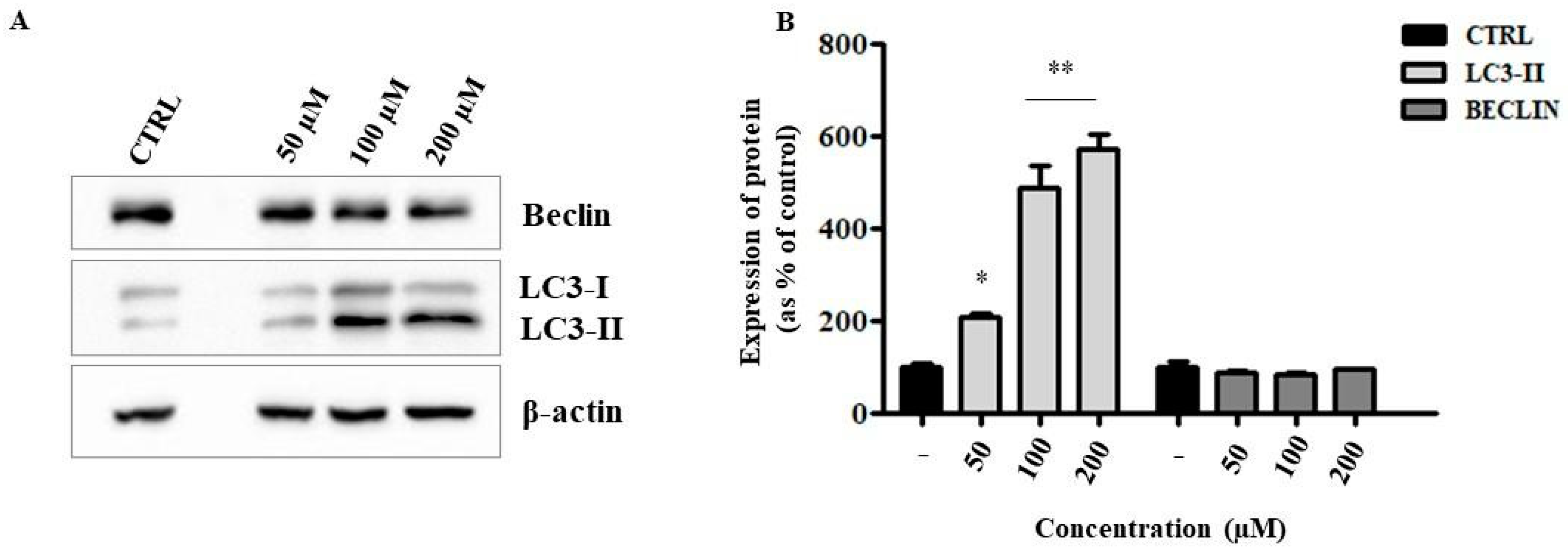
2.6. Liposomal ANP0903 Induced ER Stress and Stimulated Autophagic Mechanisms
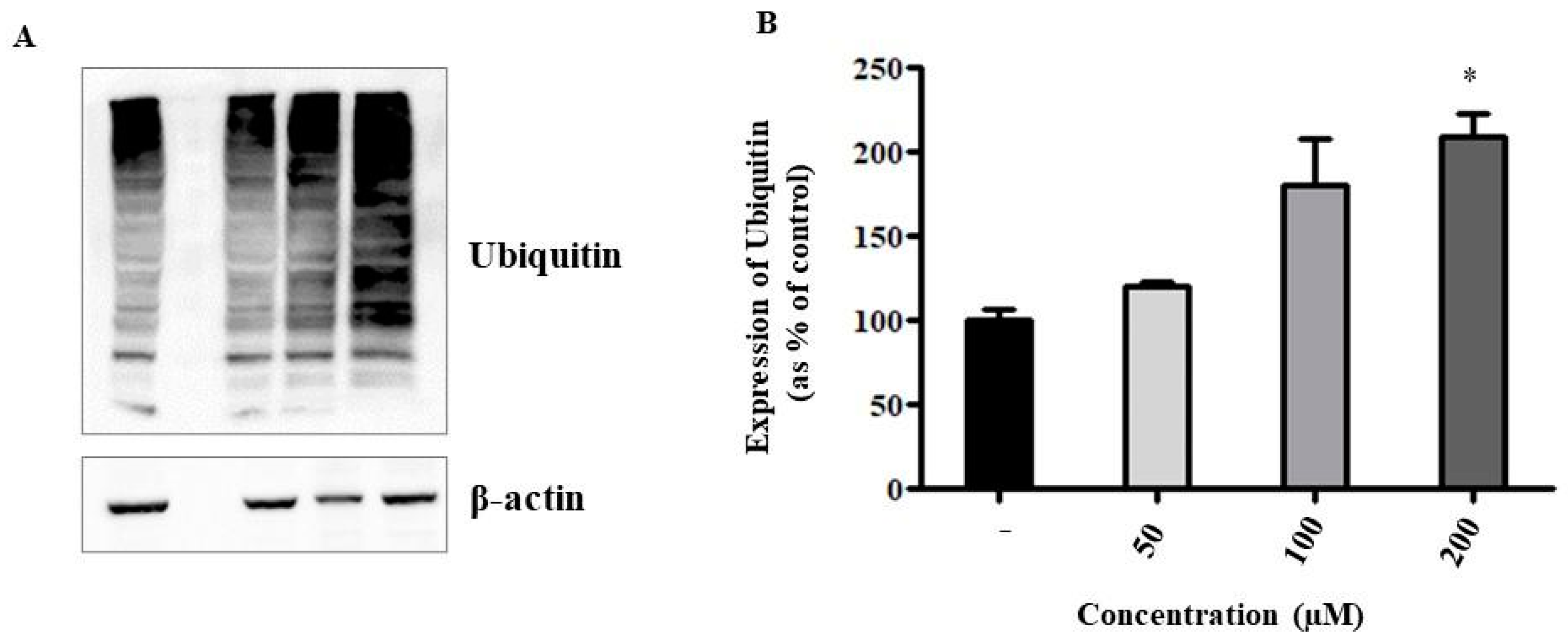
2.7. Could Liposomal ANP0903 Inhibit the Proteasome?
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Vesicle Preparation and Characterization
4.3. Stability of the Formulations during Storage and in Simulated Biological Fluid
4.4. Cell Culture and Treatments
4.5. MTT Assay
4.6. Morphological Analysis of Cells
4.7. Apoptosis Assay by Flow Cytometry
4.8. Western Blot Analysis
4.9. LC-MS/MS Analysis
4.10. Statistical Analysis of Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.Y.; Wu, K.M.; He, X.X. Advances in drug development for hepatocellular carcinoma: Clinical trials and potential therapeutic targets. J. Exp. Clin. Cancer Res. 2021, 40, 172. [Google Scholar] [CrossRef] [PubMed]
- Chidambaranathan-Reghupaty, S.; Fisher, P.B.; Sarkar, D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv. Cancer Res. 2021, 149, 1. [Google Scholar] [CrossRef] [PubMed]
- Chow, W.A.; Jiang, C.; Guan, M. Anti-HIV drugs for cancer therapeutics: Back to the future? Lancet Oncol. 2009, 10, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, W.B.; Dennis, P.A. Repositioning HIV protease inhibitors as cancer therapeutics. Curr. Opin. HIV AIDS 2008, 3, 666–675. [Google Scholar] [CrossRef]
- Narayanan, S.; Cai, C.Y.; Assaraf, Y.G.; Guo, H.Q.; Cui, Q.; Wei, L.; Huang, J.J.; Ashby, C.R.; Chen, Z.S. Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug Resist. Updat. 2020, 48, 100663. [Google Scholar] [CrossRef] [PubMed]
- Tramutola, F.; Armentano, M.F.; Berti, F.; Chiummiento, L.; Lupattelli, P.; D’Orsi, R.; Miglionico, R.; Milella, L.; Bisaccia, F.; Funicello, M. New heteroaryl carbamates: Synthesis and biological screening in vitro and in mammalian cells of wild-type and mutant HIV-protease inhibitors. Bioorganic Med. Chem. 2019, 27, 1863–1870. [Google Scholar] [CrossRef]
- Mahmoud, K.; Swidan, S.; El-Nabarawi, M.; Teaima, M. Lipid based nanoparticles as a novel treatment modality for hepatocellular carcinoma: A comprehensive review on targeting and recent advances. J. Nanobiotechnol. 2022, 20, 109. [Google Scholar] [CrossRef]
- Yang, S.; Cai, C.; Wang, H.; Ma, X.; Shao, A.; Sheng, J.; Yu, C. Drug delivery strategy in hepatocellular carcinoma therapy. Cell Commun. Signal. 2022, 20, 26. [Google Scholar] [CrossRef]
- Tian, T.; Ruan, J.; Zhang, J.; Zhao, C.-X.; Chen, D.; Shan, J. Nanocarrier-Based Tumor-Targeting Drug Delivery Systems for Hepatocellular Carcinoma Treatments: Enhanced Therapeutic Efficacy and Reduced Drug Toxicity. J. Biomed. Nanotechnol. 2022, 18, 660–676. [Google Scholar] [CrossRef]
- Graur, F.; Puia, A.; Mois, E.I.; Moldovan, S.; Pusta, A.; Cristea, C.; Cavalu, S.; Puia, C.; Al Hajjar, N. Nanotechnology in the Diagnostic and Therapy of Hepatocellular Carcinoma. Materials 2022, 15, 3893. [Google Scholar] [CrossRef]
- Wong, S.H.D.; Xu, X.; Chen, X.; Xin, Y.; Xu, L.; Lai, C.H.N.; Bian, L. Manipulation of the Nanoscale Presentation of Integrin Ligand Produces Cancer Cells with Enhanced Stemness and Robust Tumorigenicity. Nano Lett. 2021, 21, 3225–3236. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Hwang, K.; Lu, Y. Recent developments of liposomes as nanocarriers for theranostic applications. Theranostics 2016, 6, 1336–1352. [Google Scholar] [CrossRef]
- Caddeo, C.; Pucci, L.; Gabriele, M.; Carbone, C.; Fernàndez-Busquets, X.; Valenti, D.; Pons, R.; Vassallo, A.; Fadda, A.M.; Manconi, M. Stability, biocompatibility and antioxidant activity of PEG-modified liposomes containing resveratrol. Int. J. Pharm. 2018, 538, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, R.; Miglionico, R.; Nigro, I.; D’orsi, R.; Chiummiento, L.; Funicello, M.; Lupattelli, P.; Laurenzana, I.; Sgambato, A.; Monné, M.; et al. Two Novel Precursors of the HIV-1 Protease Inhibitor Darunavir Target the UPR/Proteasome System in Human Hepatocellular Carcinoma Cell Line HepG2. Cells 2021, 10, 3052. [Google Scholar] [CrossRef] [PubMed]
- Pyrko, P.; Kardosh, A.; Wang, W.; Xiong, W.; Schönthal, A.H.; Chen, T.C. HIV-1 protease inhibitors nelfinavir and atazanavir induce malignant glioma death by triggering endoplasmic reticulum stress. Cancer Res. 2007, 67, 10920–10928. [Google Scholar] [CrossRef] [PubMed]
- McLean, K.; VanDeVen, N.A.; Sorenson, D.R.; Daudi, S.; Liu, J.R. The HIV protease inhibitor saquinavir induces endoplasmic reticulum stress, autophagy, and apoptosis in ovarian cancer cells. Gynecol. Oncol. 2009, 112, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Gills, J.J.; LoPiccolo, J.; Tsurutani, J.; Shoemaker, R.H.; Best, C.J.M.; Abu-Asab, M.S.; Borojerdi, J.; Warfel, N.A.; Gardner, E.R.; Danish, M.; et al. Nelfinavir, a lead HIV protease inhibitor, is a broad-spectrum, anticancer agent that induces endoplasmic reticulum stress, autophagy, and apoptosis in vitro and in vivo. Clin. Cancer Res. 2007, 13, 5183–5194. [Google Scholar] [CrossRef]
- Rashid, H.-O.; Yadav, R.K.; Kim, H.-R.; Chae, H.-J. ER stress: Autophagy induction, inhibition and selection. Autophagy 2015, 11, 1956–1977. [Google Scholar] [CrossRef]
- Lv, Z.; Chu, Y.; Wang, Y. HIV protease inhibitors: A review of molecular selectivity and toxicity. HIV AIDS 2015, 7, 95–104. [Google Scholar] [CrossRef]
- Pajonk, F.; Himmelsbach, J.; Riess, K.; Sommer, A.; McBride, W.H. The human immunodeficiency virus (HIV)-1 protease inhibitor saquinavir inhibits proteasome function and causes apoptosis and radiosensitization in non-HIV-associated human cancer cells. Cancer Res. 2002, 62, 5230–5235. [Google Scholar] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- Onuma, A.E.; Zhang, H.; Huang, H.; Williams, T.M.; Noonan, A.; Tsung, A. Immune Checkpoint Inhibitors in Hepatocellular Cancer: Current Understanding on Mechanisms of Resistance and Biomarkers of Response to Treatment. Gene Expr. 2020, 20, 53. [Google Scholar] [CrossRef] [PubMed]
- Toschi, E.; Sgadari, C.; Malavasi, L.; Bacigalupo, I.; Chiozzini, C.; Carlei, D.; Compagnoni, D.; Bellino, S.; Bugarini, R.; Falchi, M.; et al. Human immunodeficiency virus protease inhibitors reduce the growth of human tumors via a proteasome-independent block of angiogenesis and matrix metalloproteinases. Int. J. Cancer 2011, 128, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Srirangam, A.; Mitra, R.; Wang, M.; Gorski, J.C.; Badve, S.; Baldridge, L.A.; Hamilton, J.; Kishimoto, H.; Hawes, J.; Li, L.; et al. Effects of HIV protease inhibitor ritonavir on Akt-regulated cell proliferation in breast cancer. Clin. Cancer Res. 2006, 12, 1883–1896. [Google Scholar] [CrossRef] [PubMed]
- Kraus, M.; Müller-Ide, H.; Rückrich, T.; Bader, J.; Overkleeft, H.; Driessen, C. Ritonavir, nelfinavir, saquinavir and lopinavir induce proteotoxic stress in acute myeloid leukemia cells and sensitize them for proteasome inhibitor treatment at low micromolar drug concentrations. Leuk. Res. 2014, 38, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Cerniglia, G.J.; Mick, R.; McKenna, W.G.; Muschel, R.J. HIV protease inhibitors block Akt signaling and radiosensitize tumor cells both in vitro and in vivo. Cancer Res. 2005, 65, 8256–8265. [Google Scholar] [CrossRef]
- Yin, B.; Ho, W.K.H.; Xia, X.; Chan, C.K.W.; Zhang, Q.; Ng, Y.M.; Wong, S.H.D. A Multilayered Mesoporous Gold Nanoarchitecture for Ultraeffective Near-Infrared Light-Controlled Chemo/Photothermal Therapy for Cancer Guided by SERS Imaging. Small 2023, 19, 2206762. [Google Scholar] [CrossRef]
- Sercombe, L.; Veerati, T.; Moheimani, F.; Wu, S.Y.; Sood, A.K.; Hua, S. Advances and Challenges of Liposome Assisted Drug Delivery. Front. Pharmacol. 2015, 6, 286. [Google Scholar] [CrossRef]
- Zylberberg, C.; Matosevic, S. Pharmaceutical liposomal drug delivery: A review of new delivery systems and a look at the regulatory landscape. Drug Deliv. 2016, 23, 3319–3329. [Google Scholar] [CrossRef] [PubMed]
- Nisini, R.; Poerio, N.; Mariotti, S.; De Santis, F.; Fraziano, M. The Multirole of Liposomes in Therapy and Prevention of Infectious Diseases. Front. Immunol. 2018, 9, 155. [Google Scholar] [CrossRef] [PubMed]
- Jensen, G.M.; Hodgson, D.F. Opportunities and challenges in commercial pharmaceutical liposome applications. Adv. Drug Deliv. Rev. 2020, 154–155, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Alavi, M.; Hamidi, M. Passive and active targeting in cancer therapy by liposomes and lipid nanoparticles. Drug Metab. Pers. Ther. 2019, 34, 20180032. [Google Scholar] [CrossRef]
- Daraee, H.; Etemadi, A.; Kouhi, M.; Alimirzalu, S.; Akbarzadeh, A. Application of liposomes in medicine and drug delivery. Artif. Cells Nanomed. Biotechnol. 2016, 44, 381–391. [Google Scholar] [CrossRef]
- Lee, M.K. Liposomes for Enhanced Bioavailability of Water-Insoluble Drugs: In Vivo Evidence and Recent Approaches. Pharmaceutics 2020, 12, 264. [Google Scholar] [CrossRef]
- Maksimovic-Ivanic, D.; Fagone, P.; McCubrey, J.; Bendtzen, K.; Mijatovic, S.; Nicoletti, F. HIV-protease inhibitors for the treatment of cancer: Repositioning HIV protease inhibitors while developing more potent NO-hybridized derivatives? Int. J. Cancer 2017, 140, 1713–1726. [Google Scholar] [CrossRef]
- Sun, P.; Feng, L.X.; Zhang, D.M.; Liu, M.; Liu, W.; Mi, T.; Wu, W.Y.; Jiang, B.H.; Yang, M.; Hu, L.H.; et al. Bufalin derivative BF211 inhibits proteasome activity in human lung cancer cells in vitro by inhibiting β1 subunit expression and disrupting proteasome assembly. Acta Pharmacol. Sin. 2016, 37, 908–918. [Google Scholar] [CrossRef]
- Yang, Z.; Fong, D.W.F.; Yin, L.; Wong, Y.; Huang, W. Liposomes modulate docetaxel-induced lipid oxidization and membrane damage in human hepatoma cells. J. Liposome Res. 2009, 19, 122–130. [Google Scholar] [CrossRef]
- AlQahtani, S.A.; Harisa, G.I.; Alomrani, A.H.; Alanazi, F.K.; Badran, M.M. Improved pharmacokinetic and biodistribution of 5-fluorouracil loaded biomimetic nanoerythrocytes decorated nanocarriers for liver cancer treatment. Colloids Surfaces B Biointerfaces 2021, 197, 111380. [Google Scholar] [CrossRef]
- Wei, M.; Guo, X.; Zou, Q.; Li, Q.; Tang, C.; Chen, B.; Xu, Y.; Wu, C. Lactoferrin-modified PEGylated liposomes loaded with doxorubicin for targeting delivery to hepatocellular carcinoma. Int. J. Nanomed. 2015, 10, 5123–5137. [Google Scholar] [CrossRef]
- Jain, P.; Kumar, N.; Josyula, V.R.; Jagani, H.V.; Udupa, N.; Mallikarjuna Rao, C.; Vasanth Raj, P. A study on the role of (+)-catechin in suppression of HepG2 proliferation via caspase dependent pathway and enhancement of its in vitro and in vivo cytotoxic potential through liposomal formulation. Eur. J. Pharm. Sci. 2013, 50, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Jagwani, S.; Jalalpure, S.; Dhamecha, D.; Jadhav, K.; Bohara, R. Pharmacokinetic and Pharmacodynamic Evaluation of Resveratrol Loaded Cationic Liposomes for Targeting Hepatocellular Carcinoma. ACS Biomater. Sci. Eng. 2020, 6, 4969–4984. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, D.; Yu, J.; Dou, J.; Li, X.; Mu, M.; Liang, P. Chemotherapeutic Nanoparticle-Based Liposomes Enhance the Efficiency of Mild Microwave Ablation in Hepatocellular Carcinoma Therapy. Front. Pharmacol. 2020, 11, 85. [Google Scholar] [CrossRef]
- Shin, S.H.; Park, S.S.; Lee, K.J.; Ju, E.J.; Park, J.; Ko, E.J.; Jung, J.; Kuroda, S.; Hong, S.M.; Hwang, J.J.; et al. Preclinical evaluation of cisplatin-incorporated bio-nanocapsules as chemo-radiotherapy for human hepatocellular carcinoma. Oncol. Rep. 2017, 38, 2259–2266. [Google Scholar] [CrossRef]
- Yang, H.; Yang, K.; Zhang, B. Pitting corrosion resistance of La added 316L stainless steel in simulated body fluids. Mater. Lett. 2007, 61, 1154–1157. [Google Scholar] [CrossRef]
- Bradford, M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pappalardo, I.; Santarsiero, A.; De Luca, M.; Acquavia, M.A.; Todisco, S.; Caddeo, C.; Bianco, G.; Infantino, V.; Martelli, G.; Vassallo, A. Exploiting the Anti-Inflammatory Potential of White Capsicum Extract by the Nanoformulation in Phospholipid Vesicles. Antioxidants 2021, 10, 1683. [Google Scholar] [CrossRef]
- Sinisgalli, C.; Faraone, I.; Vassallo, A.; Caddeo, C.; Bisaccia, F.; Armentano, M.F.; Milella, L.; Ostuni, A. Phytochemical Profile of Capsicum annuum L. cv Senise, Incorporation into Liposomes, and Evaluation of Cellular Antioxidant Activity. Antioxidants 2020, 9, 428. [Google Scholar] [CrossRef]
- Vassallo, A.; Santoro, V.; Pappalardo, I.; Santarsiero, A.; Convertini, P.; De Luca, M.; Martelli, G.; Infantino, V.; Caddeo, C. Liposome-Mediated Inhibition of Inflammation by Hydroxycitrate. Nanomaterials 2020, 10, 2080. [Google Scholar] [CrossRef]
- Vassallo, A.; Armentano, M.F.; Miglionico, R.; Caddeo, C.; Chirollo, C.; Gualtieri, M.J.; Ostuni, A.; Bisaccia, F.; Faraone, I.; Milella, L. Hura crepitans L. Extract: Phytochemical Characterization, Antioxidant Activity, and Nanoformulation. Pharmaceutics 2020, 12, 553. [Google Scholar] [CrossRef] [PubMed]

| Formulation | MD nm ± SD | PI ± SD | ZP mV ± SD |
|---|---|---|---|
| Empty PEG-liposomes | 87 ± 5.0 | 0.24 ± 0.03 | −19 ± 6.5 |
| ANP0903 PEG-liposomes | 86 ± 6.2 | 0.24 ± 0.03 | −17 ± 5.8 |
| Empty liposomes | 78 ± 4.8 | 0.30 ± 0.03 | * −12 ± 2.7 |
| ANP0903 liposomes | 82 ± 1.9 | 0.34 ± 0.07 | ** −8 ± 2.3 |
| Formulation | Time | MD (nm ± SD) | PI ± SD | ZP (mV ± SD) |
|---|---|---|---|---|
| Empty PEG-liposomes | t0 | 85 ± 1.5 | 0.18 ± 0.01 | −1.3 ± 0.5 |
| t24 | 80 ± 7.2 | 0.17 ± 0.01 | −1.1 ± 0.2 | |
| ANP0903 PEG-liposomes | t0 | 83 ± 8.0 | 0.21 ± 0.05 | −1.1 ± 0.3 |
| t24 | 80 ± 7.0 | 0.25 ± 0.08 | −1.6 ± 0.6 |
| Cell Line | IC50 (µM) | ||
|---|---|---|---|
| ANP0903 | L-ANP0903 | EL | |
| IHH | >400 | >400 | >400 |
| JHH6 | 174 | - | - |
| HuH7 | 161.7 | - | - |
| HepG2 | 150.1 | 102.5 | >400 |
| Formulation | DPPC | P90G | MPEG-5000-DPPE | ANP0903 | EtOH | H2O |
|---|---|---|---|---|---|---|
| Empty PEG-liposomes | 20 mg | 20 mg | 2 mg | -- | 50 µL | 950 µL |
| ANP0903 PEG-liposomes | 20 mg | 20 mg | 2 mg | 2.4 mg (5 mM) | 50 µL | 950 µL |
| Empty liposomes | 20 mg | 20 mg | -- | -- | 50 µL | 950 µL |
| ANP0903 liposomes | 20 mg | 20 mg | -- | 2.4 mg (5 mM) | 50 µL | 950 µL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caddeo, C.; Miglionico, R.; Rinaldi, R.; Nigro, I.; Lamorte, D.; Chiummiento, L.; Lupattelli, P.; Funicello, M.; D’Orsi, R.; Valenti, D.; et al. PEGylated Liposomes Loaded with Carbamate Inhibitor ANP0903 Trigger Apoptosis by Enhancing ER Stress in HepG2 Cancer Cells. Int. J. Mol. Sci. 2023, 24, 4552. https://doi.org/10.3390/ijms24054552
Caddeo C, Miglionico R, Rinaldi R, Nigro I, Lamorte D, Chiummiento L, Lupattelli P, Funicello M, D’Orsi R, Valenti D, et al. PEGylated Liposomes Loaded with Carbamate Inhibitor ANP0903 Trigger Apoptosis by Enhancing ER Stress in HepG2 Cancer Cells. International Journal of Molecular Sciences. 2023; 24(5):4552. https://doi.org/10.3390/ijms24054552
Chicago/Turabian StyleCaddeo, Carla, Rocchina Miglionico, Roberta Rinaldi, Ilaria Nigro, Daniela Lamorte, Lucia Chiummiento, Paolo Lupattelli, Maria Funicello, Rosarita D’Orsi, Donatella Valenti, and et al. 2023. "PEGylated Liposomes Loaded with Carbamate Inhibitor ANP0903 Trigger Apoptosis by Enhancing ER Stress in HepG2 Cancer Cells" International Journal of Molecular Sciences 24, no. 5: 4552. https://doi.org/10.3390/ijms24054552
APA StyleCaddeo, C., Miglionico, R., Rinaldi, R., Nigro, I., Lamorte, D., Chiummiento, L., Lupattelli, P., Funicello, M., D’Orsi, R., Valenti, D., Santoro, V., Fadda, A. M., Bisaccia, F., Vassallo, A., & Armentano, M. F. (2023). PEGylated Liposomes Loaded with Carbamate Inhibitor ANP0903 Trigger Apoptosis by Enhancing ER Stress in HepG2 Cancer Cells. International Journal of Molecular Sciences, 24(5), 4552. https://doi.org/10.3390/ijms24054552













