Synthesis of 3-(2-Alkylthio-4-chloro-5-methylbenzenesulfonyl)-2-(1-phenyl-3-arylprop-2-enylideneamino)guanidine Derivatives with Pro-Apoptotic Activity against Cancer Cells
Abstract
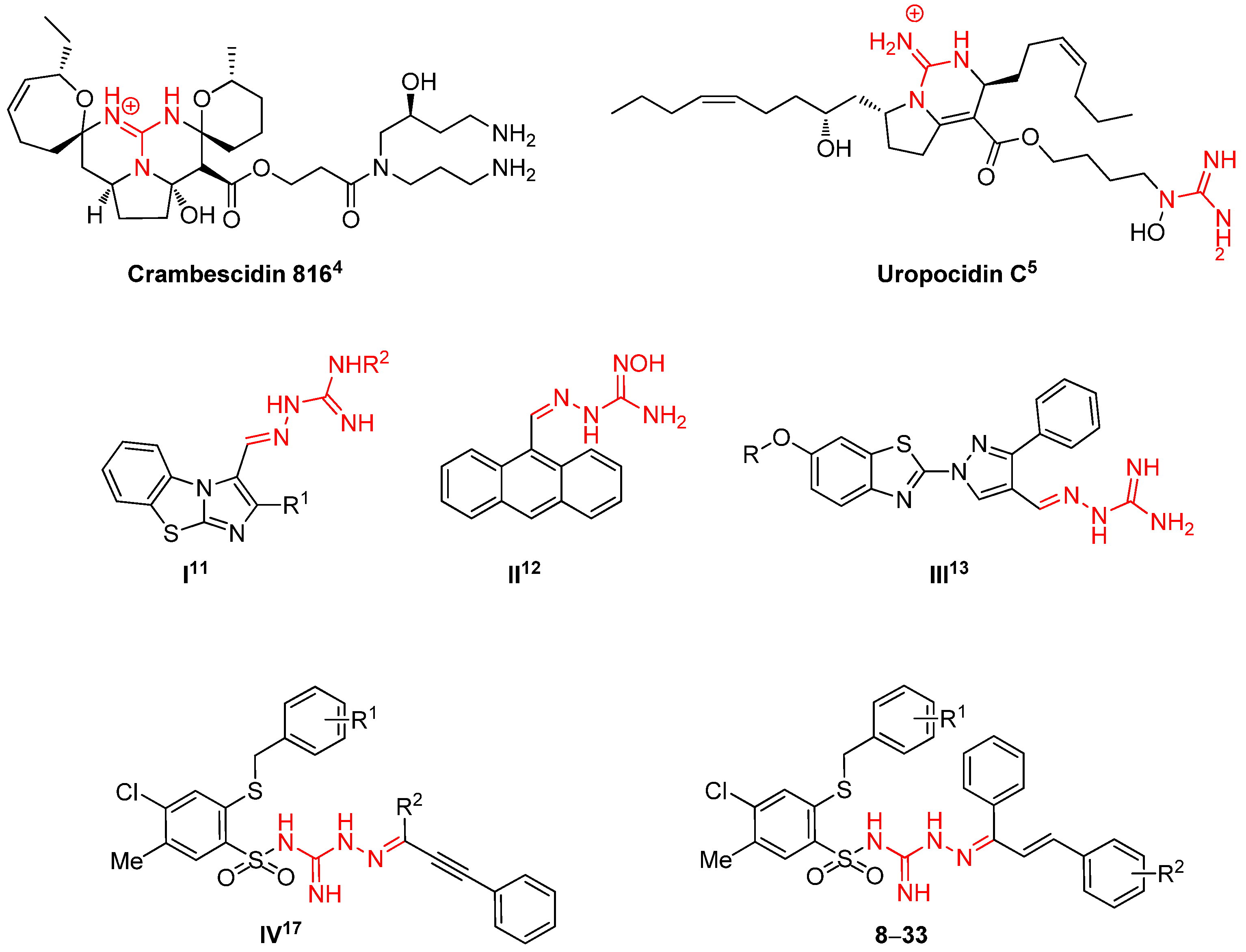
1. Introduction
2. Results and Discussion
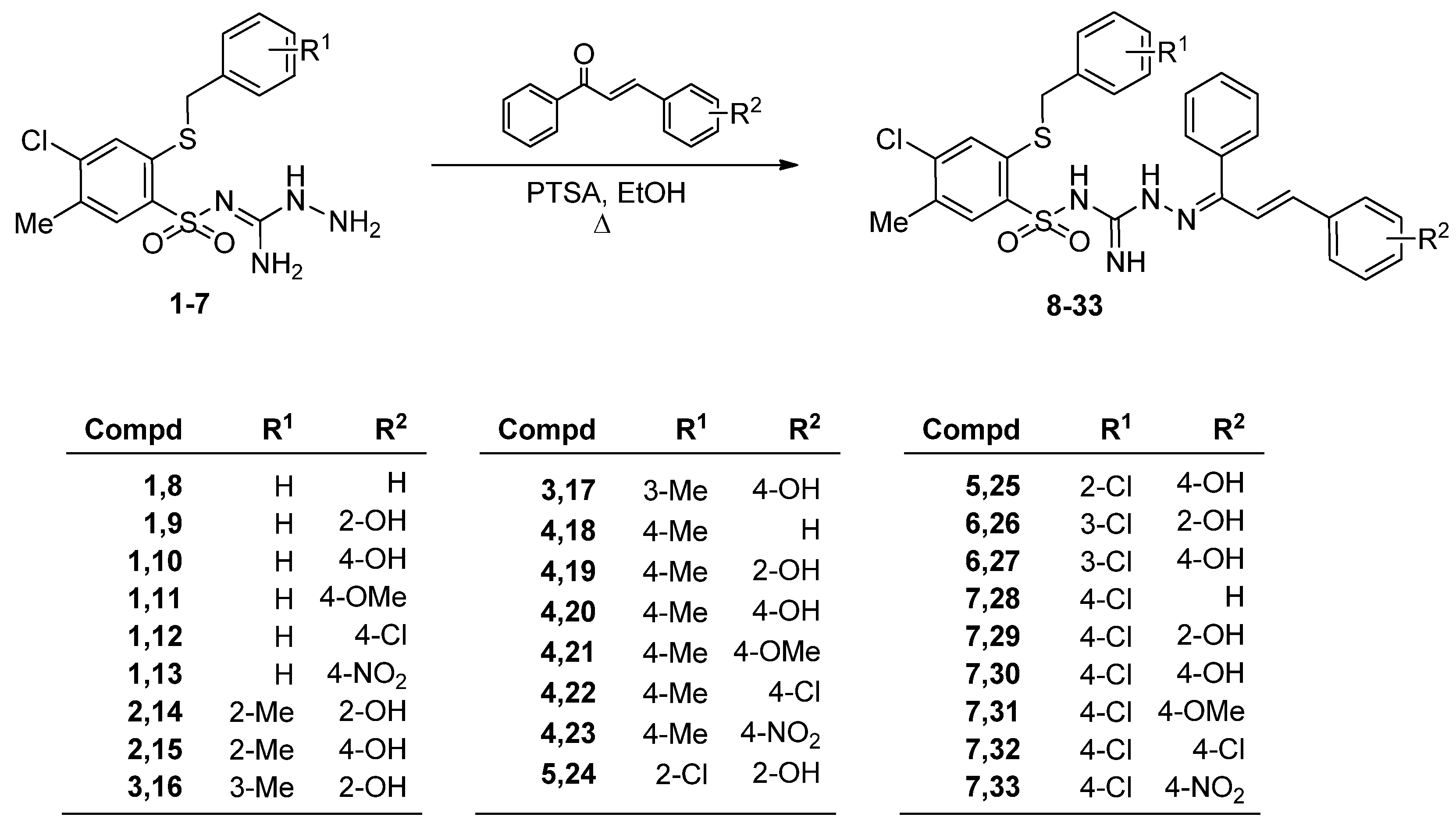
2.1. Chemistry
2.2. Cytotoxic Activity
2.3. Apoptotic Activity
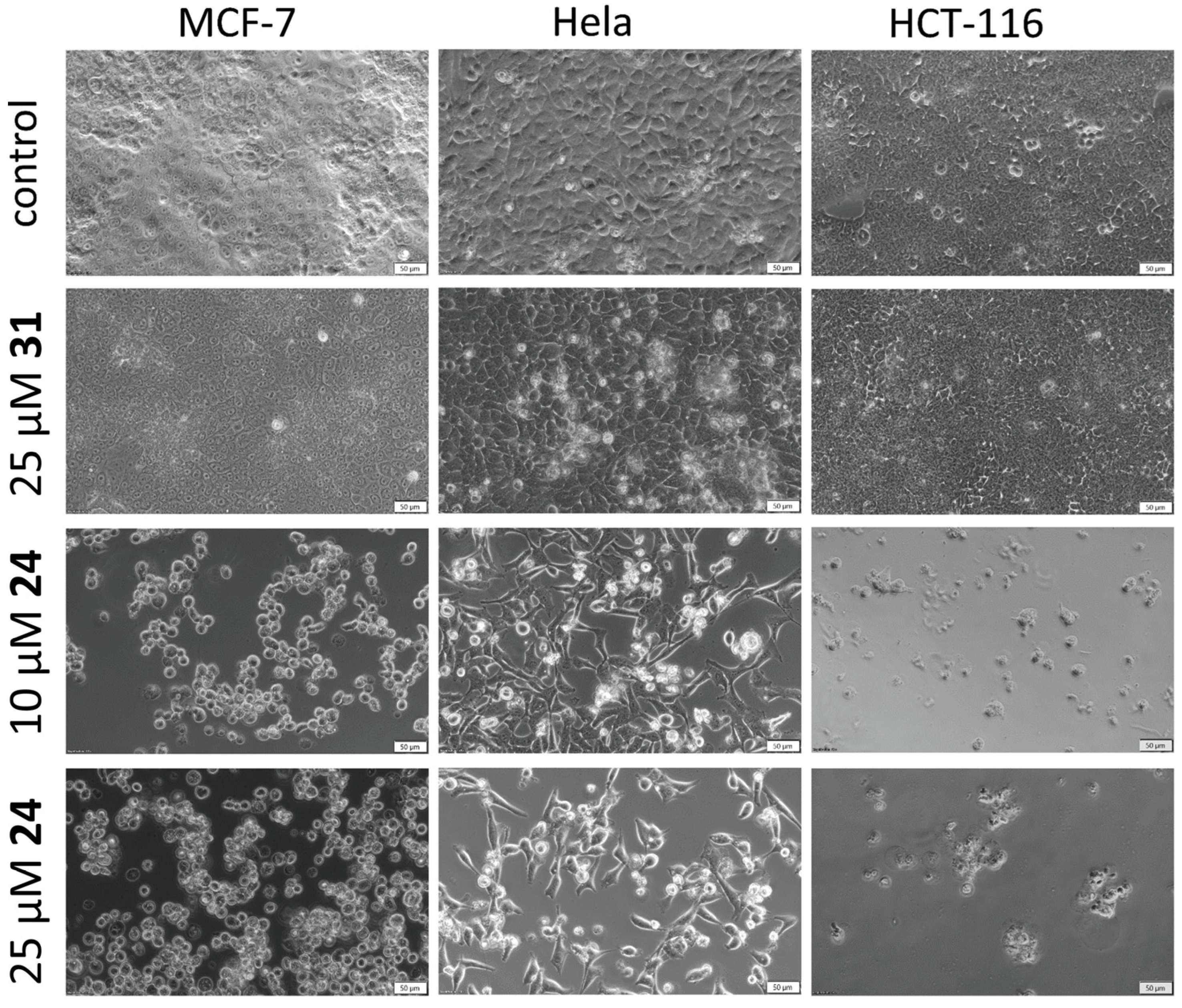
2.3.1. Cell Morphology
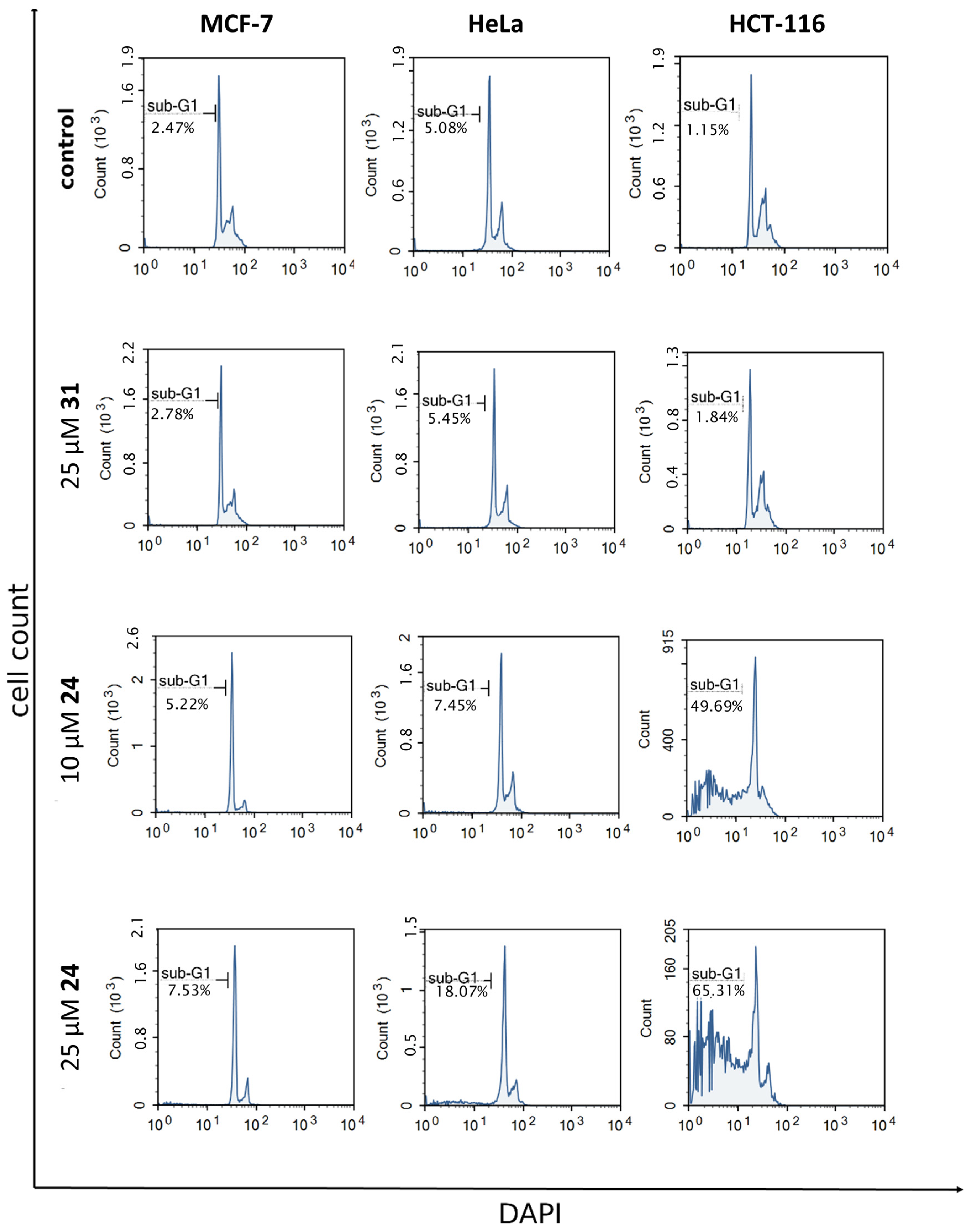
2.3.2. Cell Cycle Analysis
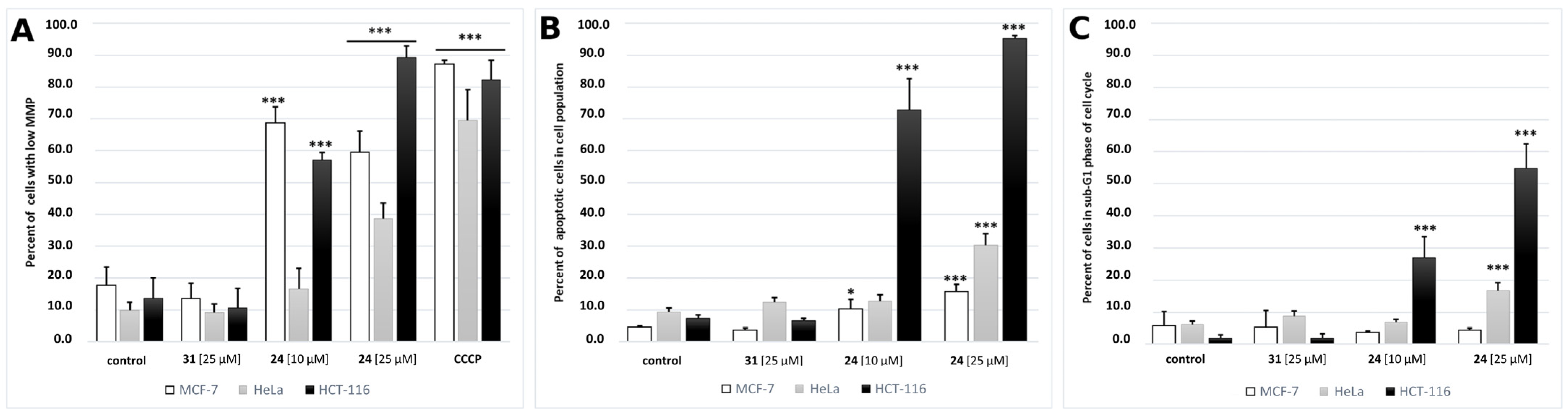
2.3.3. Mitochondrial Membrane Potential (ΔΨm) Analysis
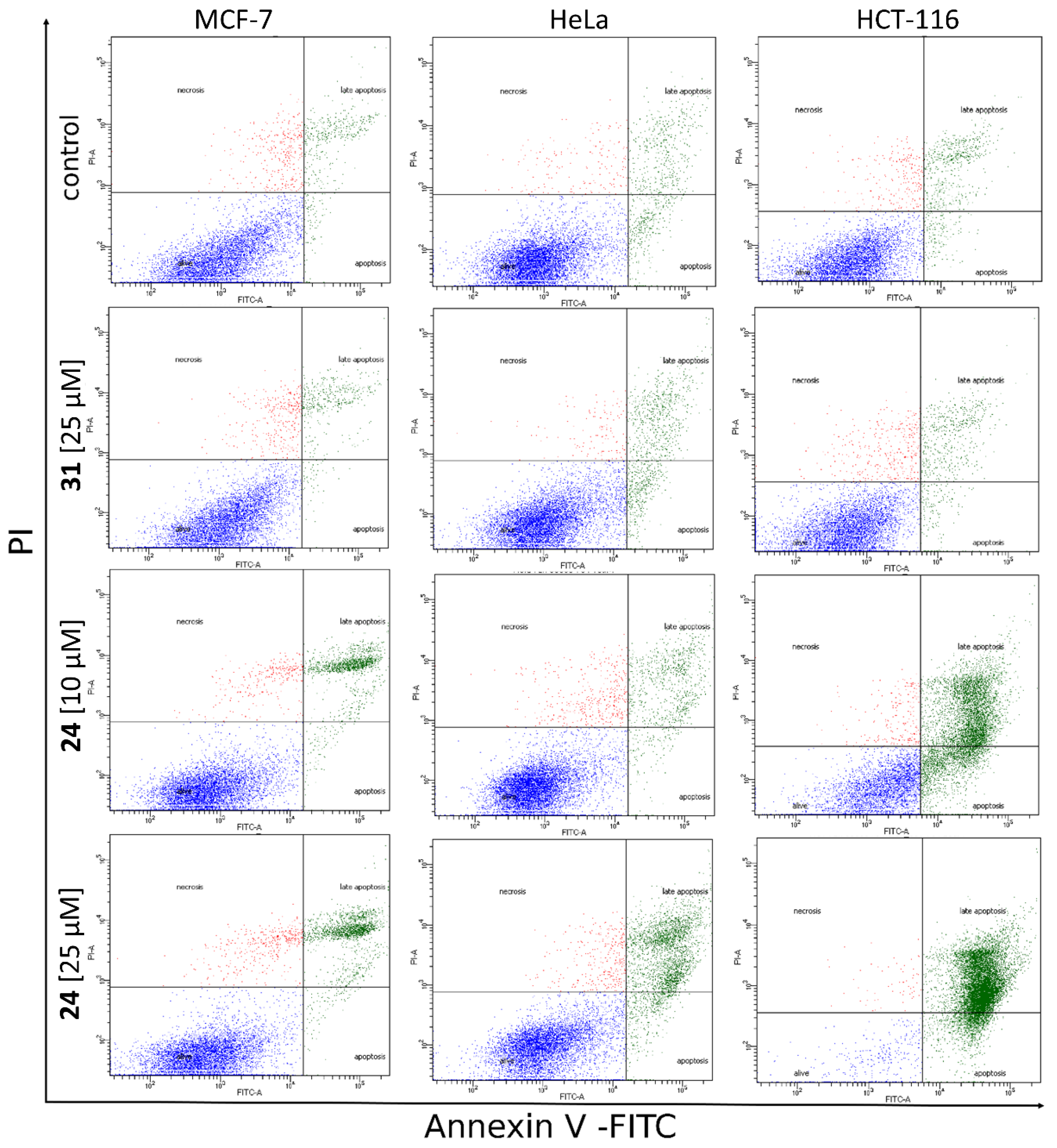
2.3.4. Translocation of Phosphatidylserine to Outer Leaflet of Cell Membrane
3. Materials and Methods
3.1. Synthesis
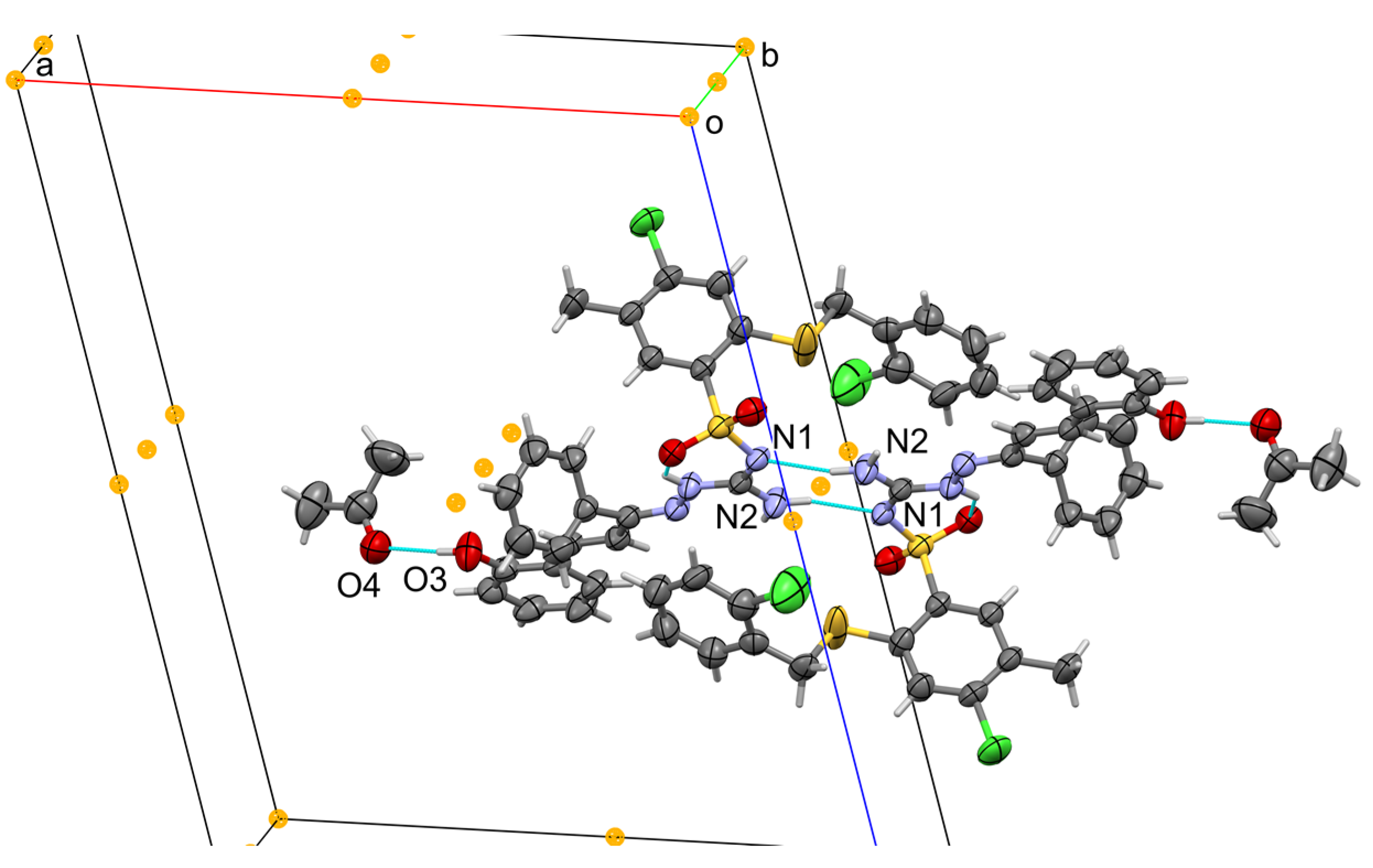
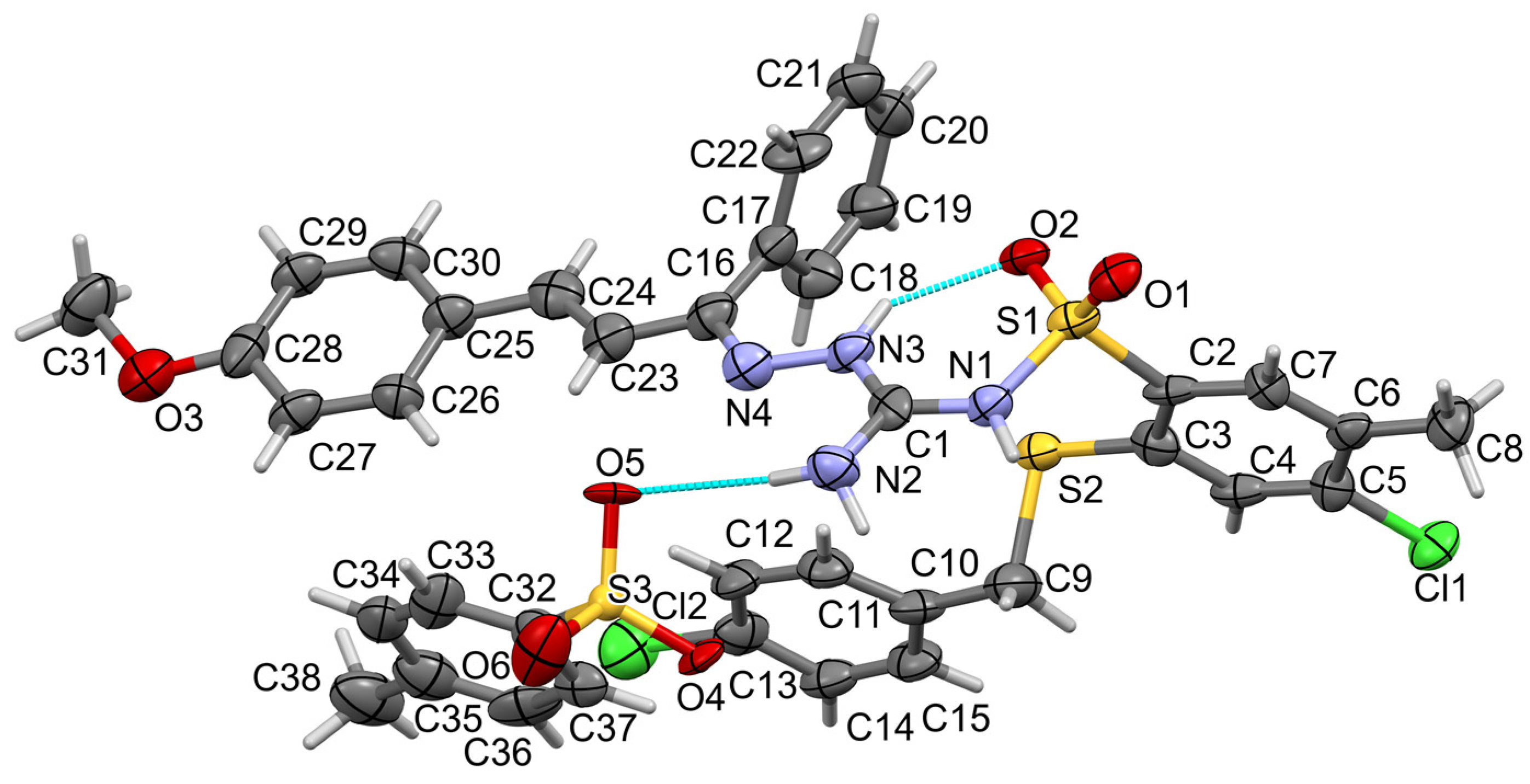
3.2. Crystallographic Details
3.3. Cell Culture and Cell Viability Assay
3.3.1. Cell Morphology
3.3.2. Mitochondrial Membrane Potential (Δψm) Analysis
3.3.3. Translocation of Phosphatidylserine to the Outer Leaflet of Cell Membrane
3.3.4. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Agency for Research on Cancer. Available online: https://gco.iarc.fr/tomorrow/en/dataviz/isotype?types=0&single_unit=500000 (accessed on 27 June 2022).
- A Few Q&As about Small Molecule Inhibitors. Available online: https://www.pharmiweb.com/article/a-few-qas-about-small-molecule-inhibitors (accessed on 27 June 2022).
- Liang, X.; Wu, P.; Yang, Q.; Xie, Y.; He, C.; Yin, L.; Yin, Z.; Yue, G.; Zou, Y.; Li, L.; et al. An update of new small-molecule anticancer drugs approved from 2015 to 2020. Eur. J. Med. Chem. 2021, 220, 113473. [Google Scholar] [CrossRef] [PubMed]
- Roel, M.; Rubiolo, J.A.; Guerra-Varela, J.; Silva, S.B.L.; Thomas, O.P.; Cabezas-Sainz, P.; Sánchez, L.; López, R.; Botana, L.M. Marine guanidine alkaloids crambescidins inhibit tumor growth and activate intrinsic apoptotic signaling inducing tumor regression in a colorectal carcinoma zebrafish xenograft model. Oncotarget 2016, 7, 83071–83087. [Google Scholar] [CrossRef] [PubMed]
- Dyshlovoy, S.A.; Kudryashova, E.K.; Kaune, M.; Makarieva, T.N.; Shubina, L.K.; Busenbender, T.; Denisenko, V.A.; Popov, R.S.; Hauschild, J.; Fedorov, S.N.; et al. Urupocidin C: A new marine guanidine alkaloid which selectively kills prostate cancer cells via mitochondria targeting. Sci. Rep. 2020, 10, 9764. [Google Scholar] [CrossRef] [PubMed]
- Habashneh, A.Y.; El-Abadelah, M.M.; Bardaweel, S.K.; Taha, M. Synthesis and Structure-Activity Relationship; Exploration of some Potent Anti-Cancer Phenyl Amidrazone Derivatives. Med. Chem. 2018, 14, 468–477. [Google Scholar] [CrossRef]
- Abu-Aisheh, M.N.; Mustafa, M.S.; El-Abadelah, M.M.; Naffa, R.G.; Ismail, S.I.; Zihlif, M.A.; Taha, M.O.; Mubarak, M.S. Synthesis and biological activity assays of some new N1-(flavon-7-yl)amidrazone derivatives and related congeners. Eur. J. Med. Chem. 2012, 54, 65–74. [Google Scholar] [CrossRef]
- Sabbah, D.A.; Hajjo, R.; Sweidan, K.; Zhong, H.A. An Integrative Informatics Approach to Explain the Mechanism of Action of N1-(Anthraquinon-2-yl) Amidrazones as BCR/ABL Inhibitors, Curr. Comput. Aided-Drug Des. 2020, 17, 817–830. [Google Scholar] [CrossRef]
- Al-Qtaitat, M.A.; El-Abadelah, M.M.; Sabbah, D.A.; Bardaweel, S.; Sweidan, K.; Sabri, S.S.; Mubarak, M.S. Synthesis, characterization, and bioactivity of new bisamidrazone derivatives as possible anticancer agents. Med. Chem. Res. 2018, 27, 1419–1431. [Google Scholar] [CrossRef]
- Qian, Y.; Zhang, H.-J.; Lv, P.-C.; Zhu, H.-L. Synthesis, molecular modeling and biological evaluation of guanidine derivatives as novel antitubulin agents. Bioorg. Med. Chem. 2010, 18, 8218–8225. [Google Scholar] [CrossRef]
- Andreani, A.; Granaiola, M.; Leoni, A.; Locatelli, A.; Morigi, R.; Rambaldi, M.; Varoli, L.; Lannigan, D.; Smith, J.; Scudiero, D.; et al. Imidazo[2,1-b]thiazole guanylhydrazones as RSK2 inhibitors. Eur. J. Med. Chem. 2011, 46, 4311–4323. [Google Scholar] [CrossRef]
- Basu, A.; Sinha, B.N.; Saiko, P.; Graser, G.; Szekeres, T. N-Hydroxy-N′-aminoguanidines as anti-cancer lead molecule: QSAR, synthesis and biological evaluation. Bioorg. Med. Chem. Lett. 2011, 21, 3324–3328. [Google Scholar] [CrossRef]
- Liu, D.C.; Gao, M.J.; Huo, Q.; Ma, T.; Wang, Y.; Wu, C.Z. Design, synthesis, and apoptosis-promoting effect evaluation of novel pyrazole with benzo[d]thiazole derivatives containing aminoguanidine units. J. Enzym. Inhib. Med. Chem. 2019, 34, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Sielaff, F.; Than, M.E.; Bevec, D.; Lindberg, I.; Steinmetzer, T. New furin inhibitors based on weakly basic amidinohydrazones. Bioorg. Med. Chem. Lett. 2011, 21, 836–840. [Google Scholar] [CrossRef] [PubMed]
- Żołnowska, B.; Sławiński, J.; Szafrański, K.; Angeli, A.; Supuran, C.T.; Kawiak, A.; Wieczór, M.; Zielińska, J.; Bączek, T.; Bartoszewska, S. Novel 2-(2-arylmethylthio-4-chloro-5-methylbenzenesulfonyl)-1-(1,3,5-triazin-2-ylamino)guanidine derivatives: Inhibition of human carbonic anhydrase cytosolic isozymes I and II and the transmembrane tumor-associated isozymes IX and XII, anticancer activity, and molecular modeling studies. Eur. J. Med. Chem. 2018, 143, 1931–1941. [Google Scholar] [PubMed]
- Żołnowska, B.; Sławiński, J.; Pogorzelska, A.; Szafrański, K.; Kawiak, A.; Stasiłojć, G.; Belka, M.; Zielińska, J.; Bączek, T. Synthesis, QSAR studies, and metabolic stability of novel 2-alkylthio-4-chloro-N-(5-oxo-4,5-dihydro-1,2,4-triazin-3-yl)benzenesulfonamide derivatives as potential anticancer and apoptosis-inducing agents. Chem. Biol. Drug Des. 2017, 90, 380–396. [Google Scholar] [CrossRef] [PubMed]
- Pogorzelska, A.; Sławiński, J.; Żołnowska, B.; Szafrański, K.; Kawiak, A.; Chojnacki, J.; Ulenberg, S.; Zielińska, J.; Bączek, T. Novel 2-(2-alkylthiobenzenesulfonyl)-3-(phenylprop-2-ynylideneamino)guanidine derivatives as potent anticancer agents: Synthesis, molecular structure, QSAR studies and metabolic stability. Eur. J. Med. Chem. 2017, 138, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Sławiński, J.; Szafrański, K.; Pogorzelska, A.; Żołnowska, B.; Kawiak, A.; Macur, K.; Belka, M.; Bączek, T. Novel 2-benzylthio-5-(1,3,4-oxadiazol-2-yl)benzenesulfonamides with anticancer activity: Synthesis, QSAR study, and metabolic stability. Eur. J. Med. Chem. 2017, 132, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Sławiński, J.; Brożewicz, K.; Fruziński, A.; Główka, M.L. Synthesis and antitumor activity of novel N′-(2-benzylthiobenzenesulfonyl)-1H-pyrazole-1-amidine derivatives. Heterocycles 2011, 83, 1093–1109. [Google Scholar] [CrossRef]
- Pogorzelska, A.; Sławiński, J.; Kawiak, A.; Żołnowska, B.; Chojnacki, J.; Stasiłojć, G.; Ulenberg, S.; Szafrański, K.; Bączek, T. Synthesis, molecular structure, and metabolic stability of new series of N′-(2-alkylthio-4-chloro-5-methylbenzenesulfonyl)-1-(5-phenyl-1H-pyrazol-1-yl)amidine as potential anti-cancer agents. Eur. J. Med. Chem. 2018, 155, 670–680. [Google Scholar] [CrossRef]
- Sławiński, J.; Bednarski, P.; Grünert, R.; Reszka, P. Syntheses of a new series of N-amino-N″-(benzenesulphonyl)guanidine derivatives with potential antitumor activity. Polish J. Chem. 2003, 77, 53–64. [Google Scholar]
- Sławiński, J.; Pogorzelska, A.; Żołnowska, B.; Kędzia, A.; Ziółkowska-Klinkosz, M.; Kwapisz, E. Synthesis and anti-yeast evaluation of novel 2-alkylthio-4-chloro-5-methyl-N-[imino-(1-oxo-(1H)-phthalazin-2-yl)methyl]benzenesulfonamide derivatives. Molecules 2014, 19, 13704–13723. [Google Scholar] [CrossRef]
- Żołnowska, B.; Sławiński, J.; Pogorzelska, A.; Szafrański, K.; Kawiak, A.; Stasiłojć, G.; Belka, M.; Ulenberg, S.; Bączek, T.; Chojnacki, J. Novel 5-substituted 2-(arylmethylthio)-4-chloro-N-(5-aryl-1,2,4-triazin-3-yl)benzenesulfonamides: Synthesis, molecular structure, anticancer activity, apoptosis-inducing activity, and metabolic stability. Molecules 2016, 21, 808. [Google Scholar] [CrossRef] [PubMed]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. A 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystaqllogr. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Stasiłojć, G.; Pinto, S.; Wyszkowska, R.; Wejda, M.; Słomińska, E.M.; Filipska, M.; Koszałka, P.; Swierczyński, J.; O’Connor, J.E.; Bigda, J.J. U937 variant cells as a model of apoptosis without cell disintegration. Cell. Mol. Biol. Lett. 2013, 18, 249–262. [Google Scholar] [CrossRef]

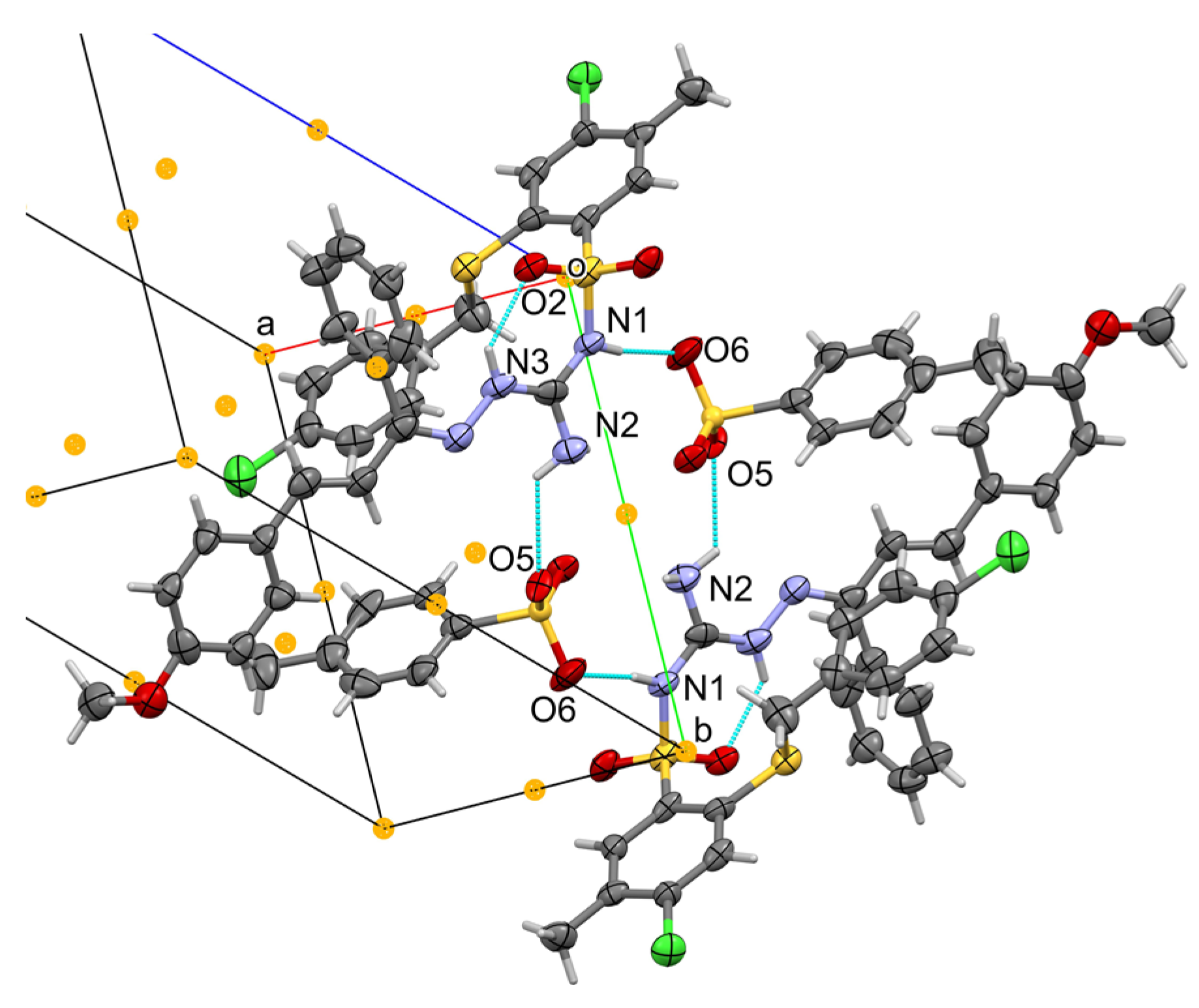

| Compd | R1 | R2 | IC50 [μM] | |||
|---|---|---|---|---|---|---|
| MCF-7 | HeLa | HCT-116 | HaCaT | |||
| 8 | H | H | 130 ± 8 | 120 ± 8 | 167 ± 7 | - |
| 9 | H | 2-OH | 18 ± 1 | 17 ± 1 | 11 ± 0.5 | 35 ± 1 |
| 10 | H | 4-OH | 15 ± 1 | 15 ± 1 | 9 ± 0.1 | 31 ± 1 |
| 11 | H | 4-OMe | 330 ± 17 | nd | 300 ± 21 | - |
| 12 | H | 4-Cl | 210 ± 13 | 190 ± 10 | 113 ± 6 | - |
| 13 | H | 4-NO2 | 370 ± 20 | 340 ± 31 | 320 ± 10 | - |
| 14 | 2-MeC6H4 | 2-OH | 13 ± 0.6 | 18 ± 1 | 9 ± 0.1 | 31 ± 2 |
| 15 | 2-MeC6H4 | 4-OH | 13 ± 0.4 | 18 ± 0.5 | 10 ± 0.6 | 33 ± 2 |
| 16 | 3-MeC6H4 | 2-OH | 20 ± 0.5 | 36 ± 2 | 17 ± 0.1 | 41 ± 2 |
| 17 | 3-MeC6H4 | 4-OH | 17 ± 1 | 21 ± 1 | 12 ± 0.6 | 35 ± 2 |
| 18 | 4-MeC6H4 | H | 260 ± 15 | 200 ± 10 | 160 ± 5 | - |
| 19 | 4-MeC6H4 | 2-OH | 14 ± 1 | 23 ± 1 | 11 ± 0.2 | 36 ± 2 |
| 20 | 4-MeC6H4 | 4-OH | 15 ± 1 | 15 ± 1 | 8.5 ± 0.2 | 31 ± 1 |
| 21 | 4-MeC6H4 | 4-OMe | 150 ± 10 | 250 ± 12 | 235 ± 5 | - |
| 22 | 4-MeC6H4 | 4-Cl | 193 ± 10 | 705 ± 35 | 183 ± 13 | - |
| 23 | 4-MeC6H4 | 4-NO2 | 280 ± 17 | 170 ± 10 | nd | - |
| 24 | 2-ClC6H4 | 2-OH | 12 ± 0.6 | 17 ± 0.5 | 9 ± 0.5 | 33 ± 2 |
| 25 | 2-ClC6H4 | 4-OH | 19 ± 1 | 18 ± 0.5 | 15 ± 0.6 | 41 ± 2 |
| 26 | 3-ClC6H4 | 2-OH | 13 ± 0.6 | 19 ± 0.5 | 10 ± 0.4 | 38 ± 2 |
| 27 | 3-ClC6H4 | 4-OH | 16 ± 1 | 20 ± 0.5 | 12 ± 0.1 | 37 ± 2 |
| 28 | 4-ClC6H4 | H | 230 ± 12 | 20 ± 1 | 220 ± 11 | 99 ± 3 |
| 29 | 4-ClC6H4 | 2-OH | 19 ± 1 | 41 ± 2 | 18 ± 0.4 | 45 ± 3 |
| 30 | 4-ClC6H4 | 4-OH | 18 ± 1 | 37 ± 1 | 8 ± 0.2 | 85 ± 2 |
| 31 | 4-ClC6H4 | 4-OMe | 230 ± 13 | 110 ± 5 | 365 ± 11 | - |
| 32 | 4-ClC6H4 | 4-Cl | 198 ± 10 | nd | 204 ± 12 | - |
| 33 | 4-ClC6H4 | 4-NO2 | 220 ± 12 | 110 ± 3 | 450 ± 27 | - |
| Cisplatin | 3.0 ± 0.1 | 2.2 ± 0.1 | 3.8 ± 0.2 | 7.7 ± 0.2 | ||
| Compd | R1 | R2 | Selectivity Index | ||
|---|---|---|---|---|---|
| MCF-7/HaCaT | HeLa/HaCaT | HCT-116/HaCaT | |||
| 9 | H | 2-OH | 1.9 | 2.1 | 3.2 |
| 10 | H | 4-OH | 2.1 | 2.1 | 3.4 |
| 14 | 2-MeC6H4 | 2-OH | 2.4 | 1.7 | 3.4 |
| 15 | 2-MeC6H4 | 4-OH | 2.5 | 1.8 | 3.3 |
| 16 | 3-MeC6H4 | 2-OH | 2.0 | ns | 2.4 |
| 17 | 3-MeC6H4 | 4-OH | 2.1 | 1.7 | 2.9 |
| 19 | 4-MeC6H4 | 2-OH | 1.9 | 1.6 | 3.3 |
| 20 | 4-MeC6H4 | 4-OH | 2.1 | 2.1 | 3.6 |
| 24 | 2-ClC6H4 | 2-OH | 2.7 | 1.9 | 3.7 |
| 25 | 2-ClC6H4 | 4-OH | 2.2 | 2.3 | 2.7 |
| 26 | 3-ClC6H4 | 2-OH | 2.9 | 2.0 | 3.8 |
| 27 | 3-ClC6H4 | 4-OH | 2.3 | 1.8 | 3.1 |
| 29 | 4-ClC6H4 | 2-OH | 2.4 | ns | 2.5 |
| 30 | 4-ClC6H4 | 4-OH | 4.7 | 2.3 | 10.6 |
| Cisplatin | 2.6 | 3.5 | 2.0 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pogorzelska, A.; Sławiński, J.; Kawiak, A.; Stasiłojć, G.; Chojnacki, J. Synthesis of 3-(2-Alkylthio-4-chloro-5-methylbenzenesulfonyl)-2-(1-phenyl-3-arylprop-2-enylideneamino)guanidine Derivatives with Pro-Apoptotic Activity against Cancer Cells. Int. J. Mol. Sci. 2023, 24, 4436. https://doi.org/10.3390/ijms24054436
Pogorzelska A, Sławiński J, Kawiak A, Stasiłojć G, Chojnacki J. Synthesis of 3-(2-Alkylthio-4-chloro-5-methylbenzenesulfonyl)-2-(1-phenyl-3-arylprop-2-enylideneamino)guanidine Derivatives with Pro-Apoptotic Activity against Cancer Cells. International Journal of Molecular Sciences. 2023; 24(5):4436. https://doi.org/10.3390/ijms24054436
Chicago/Turabian StylePogorzelska, Aneta, Jarosław Sławiński, Anna Kawiak, Grzegorz Stasiłojć, and Jarosław Chojnacki. 2023. "Synthesis of 3-(2-Alkylthio-4-chloro-5-methylbenzenesulfonyl)-2-(1-phenyl-3-arylprop-2-enylideneamino)guanidine Derivatives with Pro-Apoptotic Activity against Cancer Cells" International Journal of Molecular Sciences 24, no. 5: 4436. https://doi.org/10.3390/ijms24054436
APA StylePogorzelska, A., Sławiński, J., Kawiak, A., Stasiłojć, G., & Chojnacki, J. (2023). Synthesis of 3-(2-Alkylthio-4-chloro-5-methylbenzenesulfonyl)-2-(1-phenyl-3-arylprop-2-enylideneamino)guanidine Derivatives with Pro-Apoptotic Activity against Cancer Cells. International Journal of Molecular Sciences, 24(5), 4436. https://doi.org/10.3390/ijms24054436









