Amblyopinae Mitogenomes Provide Novel Insights into the Paraphyletic Origin of Their Adaptation to Mudflat Habitats
Abstract
1. Introduction
2. Results
2.1. Characteristics of Amblyopinae Mitogenomes
2.2. Phylogenetic Analyses
2.3. Positive Selection Analyses
3. Discussion
4. Materials and Methods
4.1. DNA Samples and Sequence Determination
4.2. Sequence Analysis
4.3. Phylogenetic Construction
4.4. Analysis of Selective Pressures
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- You, X.X.; Bian, C.; Zan, Q.J.; Xu, X.; Liu, X.; Chen, J.M.; Wang, J.T.; Qiu, Y.; Li, W.J.; Zhang, X.H.; et al. Mudskipper genome provide insights into the terrestrial adaptation of amphibious fishes. Nat. Commun. 2014, 5, 5594. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Schloissnig, S.; Franchini, P.; Du, K.; Woltering, J.; Irisarri, I.; Wong, W.Y.; Nowoshilow, S.; Kneitz, S.; Kawaguchi, A.; et al. Giant lungfish genome elucidates the conquest of land by vertebrates. Nature 2021, 590, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, J.; Zhu, C.L.; Yang, L.D.; Ren, Y.D.; Ruan, J.; Fan, G.Y.; Hu, J.; Xu, W.J.; Bi, X.P.; et al. African lungfish genome sheds light on the vertebrate water-to-land transition. Cell 2021, 184, 1362–1376. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.L.M. Time and tide wait for no fish: Intertidal fishes out of water. Environ. Biol. Fish. 1995, 44, 165–181. [Google Scholar] [CrossRef]
- Ord, T.J.; Cooke, G.M. Repeated evolution of amphibious behavior in fifish and its implications for the colonization of novel environments. Evolution 2016, 70, 1747–1759. [Google Scholar] [CrossRef]
- Gonzales, T.T.; Katoh, M.; Ishimatsu, A. Air breathing of aquatic burrow-dwelling eel goby, Odontamblyopus lacepedii (Gobiidae: Amblyopinae). J. Exp. Biol. 2006, 209, 1085–1092. [Google Scholar] [CrossRef]
- Gonzales, T.T.; Katoh, M.; Ishimatsu, A. Respiratory vasculatures of the intertidal air-breathing eel goby, Odontamblyopus lacepedii (Gobiidae: Amblyopinae). Environ. Biol. Fish. 2007, 82, 341–351. [Google Scholar] [CrossRef]
- Gonzales, T.T.; Katoh, M.; Ishimatsu, A. Intertidal burrows of the air-breathing eel goby, Odontamblyopus lacepedii (Gobiidae: Amblyopinae). Ichthyol. Res. 2008, 55, 303–306. [Google Scholar] [CrossRef]
- Maeda, K.; Hanahara, N.; Uehara, M.; Tachihara, K. Larval study revealed diversity and life-history traits of crypto-benthic eel gobies. J. Fish Biol. 2022, 101, 1411–1427. [Google Scholar] [CrossRef]
- Kurita, T.; Yoshino, T. Cryptic diversity of the eel goby, genus Taenioides (Gobiidae: Amblyopinae), in Japan. Zool. Sci. 2012, 29, 538–545. [Google Scholar] [CrossRef]
- Ishimatsu, A. Evolution of the cardiorespiratory system in air-breathing fishes. Aqua-BioSci. Monogr. 2012, 5, 1–28. [Google Scholar] [CrossRef]
- Romero, P.E.; Weigand, A.M.; Pfenninger, M. Positive selection on panpulmonate mitogenomes provide new clues on adaptations to terrestrial life. BMC Evol. Biol. 2016, 16, 164. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, I. Mitochondrial adaptations to variable environments and their role in animals’ stress tolerance. Integr. Comp. Biol. 2018, 58, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Gan, H.M.; Tan, M.H.; Lee, Y.P.; Schultz, M.B.; Horwitz, P.; Burnham, Q.; Austin, C.M. More evolution underground: Accelerated mitochondrial substitution rate in Australian burrowing freshwater crayfifishes (Decapoda: Parastacidae). Mol. Phylogenet. Evol. 2018, 118, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Dhar, D.; Dey, D.; Basu, S.; Helena, F. Insight into the adaptive evolution of mitochondrial genomes in intertidal chitons. J. Mollus. Stud. 2021, 87, eyab018. [Google Scholar] [CrossRef]
- Seutin, G.; Lang, B.F.; Mindell, D.P.; Morais, R. Evolution of the WANCY region in amniote mitochondrial DNA. Mol. Biol. Evol. 1994, 11, 329–340. [Google Scholar]
- Alcock, A.W. Natural history notes from H.M. Indian marine survey steamer Investigator, Commander, R.F.; Hoskyn, R.N., commanding. No. 20. On some unde scribed shorefishes from the Bay of Bengal. Annu. Mag. Nat. Hist. 1890, 6, 425–443. [Google Scholar] [CrossRef]
- Steindachner, F. Ichthyologische Notizen, vierte Folge. Anzeiger der Kaiserlichen Akademie der Wissenschaften, Mathematisch-Naturwissenschaftlichen Classe. FishBase 1867, 4, 63–64. [Google Scholar]
- Bleeker, P. Dertiende bijdrage tot de kennis der vischfauna van Borneo. Acta Soc. Sci. Indo Neerl. 1860, 8, 1–64. [Google Scholar]
- Cuvier, G.; Valenciennes, A. Tome douzième. Suite du livre quatorzième. Gobioïdes, Livre quinzième. Acanthoptérygiens à pectorales pédiculées. Hist. Nat. Des Poisson. 1837, 12, 1–507. [Google Scholar]
- Linnaeus, C. Systema Naturae: Editio Decima Reformata; Impensis Laurentii Salvii Press: Holmiae, Sweden, 1758. [Google Scholar]
- Steppan, S.J.; Meyer, A.A.; Barrow, L.N.; Alhajeri, B.H.; Al-Zaidan, A.S.Y.; Gignac, P.M.; Erickson, G.M. Phylogenetics and the evolution of terrestriality in mudskippers (Gobiidae: Oxudercinae). Mol. Phylogenet. Evol. 2022, 169, 107416. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kocot, K.M.; Schander, C.; Santos, S.R.; Thornhill, D.J.; Halanych, K.M. Mitogenomics reveals phylogeny and repeated motifs in control regions of the deep-sea family Siboglinidae (Annelida). Mol. Phylogenet. Evol. 2015, 85, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.J.; Dai, W.; Gao, Z.M.; Yan, G.Y.; Gan, X.N.; He, S.P. Molecular phylogeny and divergence time estimates using the mitochondrial genome for the hadal snailfish from the Mariana trench. Sci. Bull. 2017, 62, 1106–1108. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lv, Y.Y.; Wen, Z.Y.; Bian, C.; Zhang, X.H.; Guo, S.T.; Shi, Q.; Li, D.Q. The complete mitochondrial genome of the intertidal spider (Desis jiaxiangi) provides novel insights into the adaptive evolution of the mitogenome and the evolution of spiders. BMC Evol. Biol. 2021, 21, 72. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.E.; Sha, Z.L.; Wang, Y.R. The complete mitochondrial genomes of two vent squat lobsters, Munidopsis lauensis and M. verrilli: Novel gene arrangements and phylogenetic implications. Ecol. Evol. 2019, 9, 12390–12407. [Google Scholar] [CrossRef]
- Rand, D.M. Endotherms, Ectotherms, and Mitochondrial Genome-Size Variation. J. Mol. Evol. 1993, 37, 281–295. [Google Scholar] [CrossRef]
- Thacker, C.E. Molecular phylogeny of the gobioid fishes (Teleostei: Perciformes: Gobioidei). Mol. Phylogenet. Evol. 2003, 26, 354–368. [Google Scholar] [CrossRef]
- Kuang, T.; Tornabene, L.; Li, J.Y.; Jiang, J.M.; Chakrabarty, P.; Sparks, J.S.; Naylor, G.J.P.; Li, C.H. Phylogenomic analysis on the exceptionally diverse fish clade Gobioidei (Actinopterygii: Gobiiformes) and data-filtering based on molecular clocklikeness. Mol. Phylogenet. Evol. 2018, 128, 192–202. [Google Scholar] [CrossRef]
- Ballard, J.W.O.; Whitlock, M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef]
- da Fonseca, R.R.; Johnson, W.E.; O’Brien, S.J.; Ramos, M.J.; Antunes, A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genom. 2008, 9, e119. [Google Scholar] [CrossRef]
- Yu, L.; Wang, X.P.; Ting, N.; Zhang, Y.P. Mitogenomic analysis of Chinese snub-nosed monkeys: Evidence of positive selection in NADH dehydrogenase genes in high altitude adaptation. Mitochondrion 2011, 11, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.C.; Shen, X.J.; Irwin, D.M.; Zhang, Y.P. Mitogenomic analyses propose positive selection in mitochondrial genes for high-altitude adaptation in galliform birds. Mitochondrion 2014, 18, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Stager, M.; Cerasale, D.J.; Dor, R.; Winkler, D.W.; Cheviron, Z.A. Signatures of natural selection in the mitochondrial genomes of Tachycineta swallows and their implications for latitudinal patterns of the ‘pace of life’. Gene 2014, 546, 104–111. [Google Scholar] [CrossRef]
- Lamb, A.M.; Gan, H.M.; Greening, C.; Joseph, L.; Lee, Y.P.; Morán-Ordóñez, A.; Sunnucks, P.; Pavlova, A. Climate-driven mitochondrial selection: A test in Australian songbirds. Mol. Ecol. 2018, 27, 898–918. [Google Scholar] [CrossRef] [PubMed]
- Fiedorczuk, K.; Letts, J.A.; Degliesposti, G.; Kaszuba, K.; Skehel, M.; Sazanov, L.A. Atomic structure of the entire mammalian mitochondrial complex I. Nature 2016, 538, 406–410. [Google Scholar] [CrossRef]
- Zhu, J.; Vinothkumar, K.R.; Hirst, J. Structure of mammalian respiratory complex I. Nature 2016, 536, 354–358. [Google Scholar] [CrossRef]
- Wang, Z.F.; Shi, X.J.; Sun, L.X.; Bai, Y.Z.; Zhang, D.Z.; Tang, B.P. Evolution of mitochondrial energy metabolism genes associated with hydrothermal vent adaption of Alvinocaridid shrimps. Genes Genom. 2017, 39, 1367–1376. [Google Scholar] [CrossRef]
- Wang, X.B.; Zhou, S.Y.; Wu, X.Y.; Wei, Q.G.; Shang, Y.Q.; Sun, G.L.; Mei, X.; Dong, Y.; Sha, W.; Zhang, H. High-altitude adaptation in vertebrates as revealed by mitochondrial genome analyses. Ecol. Evol. 2021, 11, 15077–15084. [Google Scholar] [CrossRef]
- Lehninger, A.L. Mitochondria and calcium ion transport. Biochem. J. 1970, 119, 129–138. [Google Scholar] [CrossRef]
- Jin, Y.; Wo, Y.; Tong, H.; Song, S.; Zhang, L.X.; Brown, R.P. Evolutionary analysis of mitochondrially encoded proteins of toad-headed lizards, Phrynocephalus, along an altitudinal gradient. BMC Genom. 2018, 19, 185. [Google Scholar] [CrossRef]
- Yang, M.; Dong, D.; Li, X.Z. The complete mitogenome of Phymorhynchus sp. (Neogastropoda, Conoidea, Raphitomidae) provides insights into the deep-sea adaptive evolution of Conoidea. Ecol. Evol. 2021, 11, 7518–7531. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Meng, G.; Li, Y.; Yang, C.; Liu, S.L. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE on-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, 54–57. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of phyml 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.D.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efcient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 4, 1586–1591. [Google Scholar] [CrossRef]

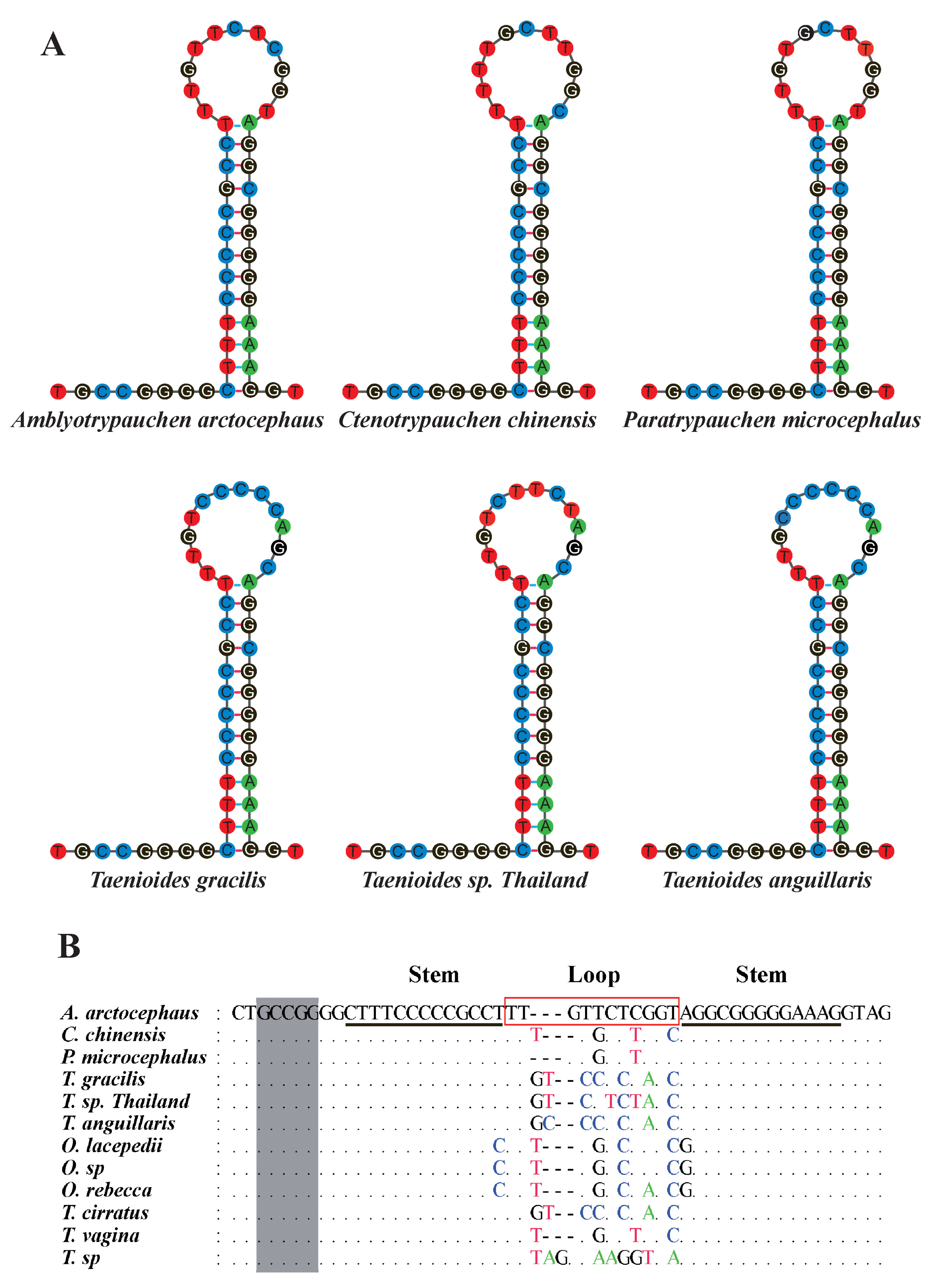


| Species | Sampling Locality | Mitogenome Size | Accession No. |
|---|---|---|---|
| Amblyotrypauchen arctocephalus Alcock, 1890 [17] | South China Sea; Yangjiang, Guangdong | 17,133 bp | MK541896 |
| Ctenotrypauchen chinensis Steindachner, 1867 [18] | South China Sea; Guangzhou, Guangdong | 16,552 bp | MK541901 |
| Paratrypauchen microcephalus Bleeker, 1860 [19] | Yellow Sea; Dandong, Liaoning | 17,086 bp | MK541897 |
| Taenioides gracilis Cuvier, 1837 [20] | South China Sea; Haikou, Hainan | 16,710 bp | MK541898 |
| Taenioides anguillaris Linnaeus, 1758 [21] | East China Sea; Wenling, Zhejiang | 16,973 bp | OL625024 |
| Taenioides sp. Thailand Kurita, 2012 [10] | South China Sea; Haikou, Hainan | 16,718 bp | MW682859 |
| Species | Starting Point * | Sequences of Perfect Repeat (Length) | Number of Repeats | Sequences of Imperfect Repeat | Number of Repeats |
|---|---|---|---|---|---|
| Amblyotrypauchen arctocephalus | 1157 | AAACAGGAAAGACTCGAGCTAGGAATTGCATGCCCCAAACTATGTGTATATACATTATTACCAATGATTTCCCTTTGTAATAAAGCGCACATCATTCATTCAACCCTAAATACTTCAACAATTTTATAGCAGAAGCCTATATCTAA (146 bp) | 4 | AAACAGGAAAGCTTCGAGCTAGAAATTGCATGCCCCAAACTATGTGTATATACATTATTACAATGATTTC | 1 |
| Ctenotrypauchen chinensis | 1156 | GAAACAGGAAAGCCTCGAGCTAGGAACTACATGCCCCAAATTGCATGTATATACATTATTACAATAATACCAATACAACAAATTATTTTTTAAACCAACAACCCTAAAATCAGGAAACCTGTCAAATTTTTAAGACAGTTATGCCCCCCA (150 bp) | 1 | GAAACAGGAAAGCCTCGAGCTAAGAACTTCATGCCCCAAATTGCATGCATATACATTATTGCAATGATTCT | 1 |
| Paratrypauchen microcephalus | 1157 | AAACAGGAAAGCCTCGAGCTAGGAATTAACATGCCCCAAACTGTGAATACACACATTATTACAGTAATTCTGTTTGTAAACAACACAATTCAAAAATTCAACCCTTAAAAGTTTGTTATATTTATAGCACACCTCTTTCATAAA (144 bp) | 4 | AAACAGGAAAACCTCGAGCTAGAAATTAACATACCCCAAACGTGAATATACACACTATTATAATAATTT | 1 |
| Taenioides gracilis | 1157 | AAACAGGAAAGCCTCGAGCATTAAAATTAACTGCCCACAAATTACCACATATACATTATTGCAAAAATTGCTACATATACATTATTACAATAATTCTTTTACTTAAATAATATTTTATAAACATTCAACCCTCAAGCCCCTATCAACCCCTTCCCCCAAAAAG (163 bp) | 1 | AACAAAAAATCCTCGAGTATAAAAACCCACTGCCTACAAATTTCTACATATTCATCATTACAGACAG | 1 |
| Taenioides anguillaris | 1156 | AAAACAGGATAAGCCTCGAGCATAGGGACCACACCCCAAATTGCAACATATACATTATTGCAATAATTCTTTTACCTAAGCAGCAAACTATAGGCACCCCACCCTTAAAACTTCTATCAGCTTCTTTTACACTCTGACTTCACA (144 bp) | 3 | AAAACAGGATAAGCCTCGAGCATAGGGACCACACCCCAAATTGCAACATATACATTATTGCAAACAT | 1 |
| Taenioides sp. Thailand | 1157 | AAACAGGAAAGCCTCGAGTATTAAAACTTACTACTCATAAATTGCTACATATACATTATTACAAAAATTGCTACATATATATTATTACAATAATTCTTTTACTTAAATAATAAAATATAAACACTCAACCCTAAAACTTCTAACACCTCTTTACACCCCTAA (162 bp) | 1 | AAACAAAAAAGCCTCGAGTATAGAAACTTGCACCTGCAAACTGCTAAATATATATTATTACAGACAG | 1 |
| Trypauchen vagina | 1152 | CCCGGAAACAGGAAAGCCTCGAGTTAGGAATTATGTACCCCAAGTTACATACCTATACACTATTGCAATAATACCCACACAATAGACTATCTTTAAAAATTCAACCCTGAAAACAATATCAAATTTTTTATACATTTTAAA (141 bp) | 2 | CCCGGAAACAGGAAAGCCTCGAGTTAGGAATTATGTACCCCAAGTTACATACCTATACACTATTGCAATAATTCT | 1 |
| Taenioides cirratus | 1156 | AAAACAGGATAAGTCTCGAGCATAGGGACCACGCCCCAAATTGCAACATATACATTATTGCAATAATTCTTTTACCTAAGCAGCAAACTATAGGCACCCCACCCTTAAAACTTCTATCAGCTTCTTTTACACTCTGACTTCAC (143 bp) | 3 | AAAACAGGATAAGTCTCGAGCATAGGGACCACGCCCCAAATTGCAACATATACATTATTGCAAACAT | 1 |
| Trypauchenopsis sp. | 1157 | AAACAGGAAAGTCTCGAGCTAGGGACAAAACACCCCCAATTGCATAAATATACATTATTGCAATAATACTTTTATTGTTTTAATAATTCTATAATAGCGTGCCGTCAACTTACTATCATATTTTTTCCCCCGCCTAAAAGCCGTA (145 bp) | 1 | ATTTTTTATTGAAGAAAAGTAAAAATTCTTCAAGTGAACCTGCACCCCCACTTAACTCTTAAACATTATAATATAACTAA | 1 |
| Odontamblyopus lacepedii | 1298 | TGAAATCAAAGATTTCCAAGTTATATGTACACATTATTACAATAATTCACTTTTTTATAATTTAAAACAACTAATAAGCCCGTCAAAACAATCTTAAAGCAACCCCGATAAAACTTTATA (120 bp) | 5 | TGAAATCAAAGATTTCCAAGTTATATGTACACATTATTACAATAATTCACTT | 1 |
| Odontamblyopus sp. | 1306 | AAATCAAAGATTTCCAAGTTATATGTACACATTATTACAATAATTCATTTTTTTATAATTTAAAACAACTAACAAGCCCGTCCAAACAATCTTAAAGCGACCCCGATAAAACTTTATATG (120 bp) | 3 | AAATCAAAGATTTCCAAGTTATATGTACACATTATTACAATGATTCACTT | 1 |
| Odontamblyopus rebecca | 1306 | ATGTTAAAGATTTACAAGTTATATGCACACATTATTACAATAATTCACTTTTTTTATGATTTAAAACAATCAACAAGCCCGTCCAAATAGTCTTAAAGTAACCCAGATAAAACTTTATATA (121 bp) | 3 | ATGTTAAAGATTTACAAGTTATATGCACACATTATTACAATAATTCACTT | 1 |
| Boleophthalmus pectinirostris | 1159 | ACAGGAAAGTCTCGAGCAAGGGCCACATGCCCAAAGTTGTGTCAAATATATTACAATAATTCACTTATTAATATACAAAATAAAAGCACCCAACCCTTACTTAGCCCTAACACATCTCACCCTTTCCTAG (130 bp) | 5 | ACAGGAAAGTCTCGAGCAAGGGCCACATGCCCAGAGTTGTGTCAAATATATTATTATGATACTTACT | 1 |
| Boleophthalmus sp. | 1159 | ACAGGAAAGCCTCGAGTGAGGGCCAAAAACCCAAAGTTGTGTCAAAGATATTACAATAATTCACTTACTAATATACAAAATAAAAGACACCCAACCCCGACAGAGCCCTCACACATTTTATTGCTCCAACC (131 bp) | 5 | ACAGGAAAGCCTCGAGTGAGGGCCAAAAACCCAAAGTTGTGTCAAAGATACTATTATGATATTTTCT | 1 |
| Boleophthalmus boddarti | 1152 | CCCGGAAACAGGAAAGCCTCGAGCAAAGGGCACATACCCAAAGTTGTGTTAAACATATTACAATAATTCACTTGCTAATGTACAAAATAAGAGACACCCAACCCAGACTTAACCGTCACACACTTTTAATT (131 bp) | 2 | CCCGGAAACAGGAAAGCCTCGAGCAAAGGGCACATACCCAAAGTTGTGTTAAACATATTATTATGATTACTTTCT | 1 |
| Scartelaos gigas | 1157 | AAACAGGAAAGCCTCGAGTTAGGGACCATGTGCTAAAAATGTACACATACACATTATTACAATAATTCACTTATCACACAAAATAAAGATAGAAACATTTAACCCAGCATTTTCTATCATCTTTTTAACCCCCCTCCTTCCAACCGTGGACTTTTATTCACCCGGAACCTT (171 bp) | 1 | AACAAAACCTAGTCTAAAGTTAGAAACCACATGCTCCAAAATGCGTGTATTTACATTATTGCAATAGTTCACTT | 1 |
| Oxuderces dentatus | 1189 | ACCTCTCAAGTGTTTATGTACCAATTATTACAATAATTCATTTATTTGTACAAAAAATAATAAACCTCCGCCCTAAATAAACTAACAATTAAAAAATTACTATATAACTAATATCCCTCGATATAGATTTAACCAACACATGTAAATCGC (150 bp) | 4 | AGCT | 1 |
| Parapocryptes serperaster | 1156 | GAAACAGGAAAGCCTCGAGCTAGAAATCATGCCCCCAAGTTGCATGCACAAATTATTACAATAATTCACTTATTAACTCACCCCACCCATCTCACCCCAACCGAAAAATCCCTGCCAACTTTGAATCCCCCGACCCCTAAACTCCTAGAGTAATATTCACCGTTAACCCAAACCCCAA (178 bp) | 4 | GAAACAGGAAAGCCTCGAGCTAGAAATCATGCCCCCAAGTTGCACGGCCCCCCACCT | 1 |
| Periophthalmus argentilineatus | ----- | ----- | ----- | ----- | ----- |
| Periophthalmus modestus | ----- | ----- | ----- | ----- | ----- |
| Periophthalmus minutus | ----- | ----- | ----- | ----- | ----- |
| Periophthalmus magnuspinnatus | ----- | ----- | ----- | ----- | ----- |
| Periophthalmodon schlosseri | ----- | ----- | ----- | ----- | ----- |
| Gene | Model | lnL | LRT | Parameter |
|---|---|---|---|---|
| atp6 | M0 | −12,288.396 | ω0 = 0.036 | |
| M2 | −12,288.359 | 0.074 | ω0 = 0.035; ω1 = 0.040 | |
| atp8 | M0 | −2884.378 | ω0 = 0.116 | |
| M2 | −2884.367 | 0.021 | ω0 = 0.116; ω1 = 0.128 | |
| cox1 | M0 | −21,414.251 | ω0 = 0.011 | |
| M2 | −21,410.655 | 7.192 ** | ω0 = 0.011; ω1 = 0.055 | |
| cox2 | M0 | −8871.766 | ω0 = 0.017 | |
| M2 | −8871.123 | 1.285 | ω0 = 0.017; ω1 = 0.038 | |
| cox3 | M0 | −10,794.094 | ω0 = 0.024 | |
| M2 | −10,791.750 | 4.688 * | ω0 = 0.024; ω1 = 0.089 | |
| cytb | M0 | −18,249.518 | ω0 = 0.021 | |
| M2 | −18,248.658 | 1.719 | ω0 = 0.021; ω1 = 0.080 | |
| nad1 | M0 | −15,830.249 | ω0 = 0.021 | |
| M2 | −15,824.827 | 10.844 ** | ω0 = 0.021; ω1 = 0.338 | |
| nad2 | M0 | −21,447.715 | ω0 = 0.053 | |
| M2 | −21,437.677 | 20.076 ** | ω0 = 0.053; ω1 = 999.000 | |
| nad3 | M0 | −6097.474 | ω0 = 0.041 | |
| M2 | −6095.969 | 3.010 | ω0 = 0.040; ω1 = 999.000 | |
| nad4 | M0 | −25,867.666 | ω0 = 0.041 | |
| M2 | −25,862.873 | 9.585 ** | ω0 = 0.041; ω1 = 0.198 | |
| nad4l | M0 | −4790.108 | ω0 = 0.032 | |
| M2 | −4789.945 | 0.325 | ω0 = 0.031; ω1 = 999.000 | |
| nad5 | M0 | −33,909.683 | ω0 = 0.044 | |
| M2 | −33,907.749 | 3.867 * | ω0 = 0.043; ω1 = 0.095 | |
| nad6 | M0 | −10,277.916 | ω0 = 0.043 | |
| M2 | −10,275.634 | 4.564 * | ω0 = 0.043; ω1 = 0.912 |
| Gene | Model | lnL | LRT | Parameter | Positive Selected Site |
|---|---|---|---|---|---|
| atp6 | Null model | −12,265.068 | P0 = 0.940; P1 = 0.030; P2a = 0.029; P2b = 0.001; ω0 = 0.032; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| Model A | −12,264.833 | 0.471 | P0 = 0.955; P1 = 0.030; P2a = 0.015; P2b = 0.000; ω0 = 0.032; ω1 = 1.000; ω2a = 3.021; ω2b = 3.021 | ||
| atp8 | Null model | −2874.537 | P0 = 0.842; P1 = 0.087; P2a = 0.064; P2b = 0.007; ω0 = 0.103; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| Model A | −2874.010 | 1.053 | P0 = 0.860; P1 = 0.089; P2a = 0.046; P2b = 0.005; ω0 = 0.102; ω1 = 1.000; ω2a = 4.512; ω2b = 4.512 | ||
| cox1 | Null model | −21,265.614 | P0 = 0.979; P1 = 0.008; P2a = 0.013; P2b = 0.000; ω0 = 0.008; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| Model A | −21,265.614 | 0.000 | P0 = 0.979; P1 = 0.008; P2a = 0.013; P2b = 0.000; ω0 = 0.008; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| cox2 | Null model | −8871.244 | P0 = 0.975; P1 = 0.000; P2a = 0.025; P2b = 0.000; ω0 = 0.017; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| Model A | −8871.244 | 0.000 | P0 = 0.975; P1 = 0.000; P2a = 0.025; P2b = 0.000; ω0 = 0.017; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| cox3 | Null model | −10,731.495 | P0 = 0.946; P1 = 0.028; P2a = 0.025; P2b = 0.001; ω0 = 0.018; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| Model A | −10,728.961 | 5.067 * | P0 = 0.965; P1 = 0.029; P2a = 0.006; P2b = 0.000; ω0 = 0.018; ω1 = 1.000; ω2a = 31.441; ω2b = 31.441 | 162 (0.988) | |
| cytb | Null model | −18,282.716 | P0 = 0.899; P1 = 0.014; P2a = 0.085; P2b = 0.001; ω0 = 0.019; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| Model A | −18,282.716 | 0.000 | P0 = 0.899; P1 = 0.014; P2a = 0.085; P2b = 0.001; ω0 = 0.019; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| nad1 | Null model | −15,810.905 | P0 = 0.662; P1 = 0.004; P2a = 0.331; P2b = 0.002; ω0 = 0.020; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| Model A | −15,810.905 | 0.000 | P0 = 0.662; P1 = 0.004; P2a = 0.331; P2b = 0.002; ω0 = 0.020; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| nad2 | Null model | −21,254.298 | P0 = 0.327; P1 = 0.018; P2a = 0.620; P2b = 0.035; ω0 = 0.045; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
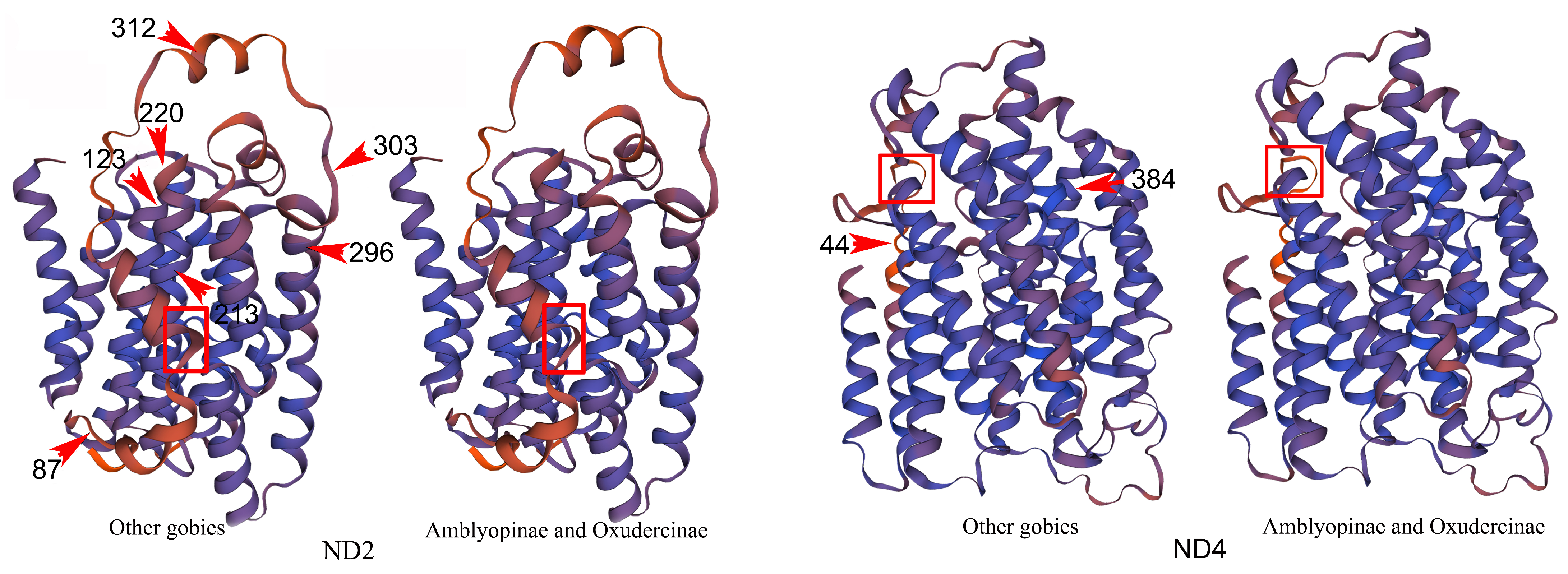
| Model A | −21,248.610 | 11.376 ** | P0 = 0.900; P1 = 0.050 P2a = 0.047; P2b = 0.003; ω0 = 0.045; ω1 = 1.000; ω2a = 999.0; ω2b = 999.0 | 87(0.966); 123 (0.987); 213(0.984); 220 (0.989); 296 (0.986); 303 (0.960); 312 (0.975) | |
| nad3 | Null model | −5981.539 | P0 = 0.000; P1 = 0.000 P2a = 0.915; P2b = 0.085; ω0 = 0.023; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| Model A | −5981.466 | 0.146 | P0 = 0.000; P1 = 0.000 P2a = 0.915; P2b = 0.085; ω0 = 0.023; ω1 = 1.000; ω2a = 64.038; ω2b = 64.038 | ||
| nad4 | Null model | −25,655.501 | P0 = 0.877; P1 = 0.057; P2a = 0.062; P2b = 0.004; ω0 = 0.030; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| Model A | −25,653.475 | 4.053 * | P0 = 0.928; P1 = 0.060; P2a = 0.012; P2b = 0.001; ω0 = 0.030; ω1 = 1.000; ω2a = 11.628; ω2b = 11.628 | 44 (0.991); 384 (0.988) | |
| nad4l | Null model | −4771.271 | P0 = 0.885; P1 = 0.028; P2a = 0.084; P2b = 0.003; ω0 = 0.028; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| Model A | −4771.271 | 0.000 | P0 = 0.969; P1 = 0.031; P2a = 0.000; P2b = 0.000; ω0 = 0.028; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| nad5 | Null model | −33,633.451 | P0 = 0.916; P1 = 0.052; P2a = 0.030; P2b = 0.002; ω0 = 0.033; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| Model A | −33,633.451 | 0.000 | P0 = 0.916; P1 = 0.052; P2a = 0.030; P2b = 0.002; ω0 = 0.033; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| nad6 | Null model | −10,198.030 | P0 = 0.785; P1 = 0.074; P2a = 0.129; P2b = 0.012; ω0 = 0.033; ω1 = 1.000; ω2a = 1.000; ω2b = 1.000 | ||
| Model A | −10,195.755 | 4.550 * | P0 = 0.866; P1 = 0.082; P2a = 0.047; P2b = 0.004; ω0 = 0.033; ω1 = 1.000; ω2a = 999.0; ω2b = 999.0 | 78 (0.951) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lü, Z.; Liu, Y.; Zhao, S.; Fang, J.; Zhu, K.; Liu, J.; Gong, L.; Liu, L.; Liu, B. Amblyopinae Mitogenomes Provide Novel Insights into the Paraphyletic Origin of Their Adaptation to Mudflat Habitats. Int. J. Mol. Sci. 2023, 24, 4362. https://doi.org/10.3390/ijms24054362
Lü Z, Liu Y, Zhao S, Fang J, Zhu K, Liu J, Gong L, Liu L, Liu B. Amblyopinae Mitogenomes Provide Novel Insights into the Paraphyletic Origin of Their Adaptation to Mudflat Habitats. International Journal of Molecular Sciences. 2023; 24(5):4362. https://doi.org/10.3390/ijms24054362
Chicago/Turabian StyleLü, Zhenming, Yantao Liu, Shijie Zhao, Jiaqi Fang, Kehua Zhu, Jing Liu, Li Gong, Liqin Liu, and Bingjian Liu. 2023. "Amblyopinae Mitogenomes Provide Novel Insights into the Paraphyletic Origin of Their Adaptation to Mudflat Habitats" International Journal of Molecular Sciences 24, no. 5: 4362. https://doi.org/10.3390/ijms24054362
APA StyleLü, Z., Liu, Y., Zhao, S., Fang, J., Zhu, K., Liu, J., Gong, L., Liu, L., & Liu, B. (2023). Amblyopinae Mitogenomes Provide Novel Insights into the Paraphyletic Origin of Their Adaptation to Mudflat Habitats. International Journal of Molecular Sciences, 24(5), 4362. https://doi.org/10.3390/ijms24054362






