The miR-20a/miR-92b Profile Is Associated with Circulating γδ T-Cell Perturbations in Mild Psoriasis
Abstract
1. Introduction
2. Results
2.1. Subject Characteristics
2.2. The γδ T-Cell Composition Is Reshaped by the CMV Infection History and the Case–Control Status
2.3. The Differential Expression of miR-20a Is Associated with Transcriptional Variations in Bulk γδ T Cells and the Altered Vδ Composition of the γδint Compartment
3. Discussion
4. Materials and Methods
4.1. Study Design and Subject Selection
4.2. The miRNA Selection
4.3. Isolation of Peripheral Blood Mononuclear Cells
4.4. Flow Cytometry Analysis and γδT Cell Sorting
4.5. cDNA Synthesis and mRNA/miRNA Expression Analysis
4.6. In Silico Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Nestle, F.O.; Kaplan, D.H.; Barker, J. Psoriasis. N. Engl. J. Med. 2009, 361, 496–509. [Google Scholar] [CrossRef]
- Prey, S.; Paul, C.; Bronsard, V.; Puzenat, E.; Gourraud, P.-A.; Aractingi, S.; Aubin, F.; Bagot, M.; Cribier, B.; Joly, P.; et al. Assessment of Risk of Psoriatic Arthritis in Patients with Plaque Psoriasis: A Systematic Review of the Literature. J. Eur. Acad. Dermatol. Venereol. JEADV 2010, 24 (Suppl. S2), 31–35. [Google Scholar] [CrossRef]
- Oliveira, M.D.F.S.P.D.; Rocha, B.D.O.; Duarte, G.V. Psoriasis: Classical and Emerging Comorbidities. An. Bras. Dermatol. 2015, 90, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Sewerin, P.; Brinks, R.; Schneider, M.; Haase, I.; Vordenbäumen, S. Prevalence and Incidence of Psoriasis and Psoriatic Arthritis. Ann. Rheum. Dis. 2019, 78, 286–287. [Google Scholar] [CrossRef] [PubMed]
- AlQassimi, S.; AlBrashdi, S.; Galadari, H.; Hashim, M.J. Global Burden of Psoriasis—Comparison of Regional and Global Epidemiology, 1990 to 2017. Int. J. Dermatol. 2020, 59, 566–571. [Google Scholar] [CrossRef]
- Laggner, U.; Meglio, P.D.; Perera, G.K.; Hundhausen, C.; Lacy, K.E.; Ali, N.; Smith, C.H.; Hayday, A.C.; Nickoloff, B.J.; Nestle, F.O. Identification of a Novel Proinflammatory Human Skin-Homing Vγ9Vδ2 T Cell Subset with a Potential Role in Psoriasis. J. Immunol. 2011, 187, 2783–2793. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Shen, X.; Ding, C.; Qi, C.; Li, K.; Li, X.; Jala, V.R.; Zhang, H.; Wang, T.; Zheng, J.; et al. Pivotal Role of Dermal IL-17-Producing Γδ T Cells in Skin Inflammation. Immunity 2011, 35, 596–610. [Google Scholar] [CrossRef]
- Qi, C.; Wang, Y.; Li, P.; Zhao, J. Gamma Delta T Cells and Their Pathogenic Role in Psoriasis. Front. Immunol. 2021, 12, 627139. [Google Scholar] [CrossRef]
- Liu, N.; Qin, H.; Cai, Y.; Li, X.; Wang, L.; Xu, Q.; Xue, F.; Chen, L.; Ding, C.; Hu, X.; et al. Dynamic Trafficking Patterns of IL-17-Producing Γδ T Cells Are Linked to the Recurrence of Skin Inflammation in Psoriasis-like Dermatitis. eBioMedicine 2022, 82, 104136. [Google Scholar] [CrossRef]
- Pantelyushin, S.; Haak, S.; Ingold, B.; Kulig, P.; Heppner, F.L.; Navarini, A.A.; Becher, B. Rorγt+ Innate Lymphocytes and Γδ T Cells Initiate Psoriasiform Plaque Formation in Mice. J. Clin. Investig. 2012, 122, 2252–2256. [Google Scholar] [CrossRef]
- Kazen, A.R.; Adams, E.J. Evolution of the V, D, and J Gene Segments Used in the Primate Gammadelta T-Cell Receptor Reveals a Dichotomy of Conservation and Diversity. Proc. Natl. Acad. Sci. USA 2011, 108, E332–E340. [Google Scholar] [CrossRef]
- Fichtner, A.S.; Ravens, S.; Prinz, I. Human Γδ TCR Repertoires in Health and Disease. Cells 2020, 9, 800. [Google Scholar] [CrossRef] [PubMed]
- Bonneville, M.; O’Brien, R.L.; Born, W.K. Γδ T Cell Effector Functions: A Blend of Innate Programming and Acquired Plasticity. Nat. Rev. Immunol. 2010, 10, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Fu, X.; Xiao, N.; Guo, Y.; Pei, Q.; Peng, Y.; Zhang, Y.; Yao, M. Involvements of ΓδT Lymphocytes in Acute and Chronic Skin Wound Repair. Inflammation 2017, 40, 1416–1427. [Google Scholar] [CrossRef] [PubMed]
- Pitard, V.; Roumanes, D.; Lafarge, X.; Couzi, L.; Garrigue, I.; Lafon, M.-E.; Merville, P.; Moreau, J.-F.; Déchanet-Merville, J. Long-Term Expansion of Effector/Memory Vδ2− Γδ T Cells Is a Specific Blood Signature of CMV Infection. Blood 2008, 112, 1317–1324. [Google Scholar] [CrossRef]
- Girard, P.; Charles, J.; Cluzel, C.; Degeorges, E.; Manches, O.; Plumas, J.; De Fraipont, F.; Leccia, M.-T.; Mouret, S.; Chaperot, L.; et al. The Features of Circulating and Tumor-Infiltrating Γδ T Cells in Melanoma Patients Display Critical Perturbations with Prognostic Impact on Clinical Outcome. OncoImmunology 2019, 8, 1601483. [Google Scholar] [CrossRef]
- Kierkels, G.J.J.; Scheper, W.; Meringa, A.D.; Johanna, I.; Beringer, D.X.; Janssen, A.; Schiffler, M.; Aarts-Riemens, T.; Kramer, L.; Straetemans, T.; et al. Identification of a Tumor-Specific Allo-HLA–Restricted ΓδTCR. Blood Adv. 2019, 3, 2870–2882. [Google Scholar] [CrossRef]
- Raverdeau, M.; Cunningham, S.P.; Harmon, C.; Lynch, L. Γδ T Cells in Cancer: A Small Population of Lymphocytes with Big Implications. Clin. Transl. Immunol. 2019, 8, e01080. [Google Scholar] [CrossRef]
- McMurray, J.L.; von Borstel, A.; Taher, T.E.; Syrimi, E.; Taylor, G.S.; Sharif, M.; Rossjohn, J.; Remmerswaal, E.B.M.; Bemelman, F.J.; Vieira Braga, F.A.; et al. Transcriptional Profiling of Human Vδ1 T Cells Reveals a Pathogen-Driven Adaptive Differentiation Program. Cell Rep. 2022, 39, 110858. [Google Scholar] [CrossRef]
- Zimmermann, J.; Kühl, A.A.; Weber, M.; Grün, J.R.; Löffler, J.; Haftmann, C.; Riedel, R.; Maschmeyer, P.; Lehmann, K.; Westendorf, K.; et al. T-Bet Expression by Th Cells Promotes Type 1 Inflammation but Is Dispensable for Colitis. Mucosal Immunol. 2016, 9, 1487–1499. [Google Scholar] [CrossRef]
- Istaces, N.; Splittgerber, M.; Lima Silva, V.; Nguyen, M.; Thomas, S.; Le, A.; Achouri, Y.; Calonne, E.; Defrance, M.; Fuks, F.; et al. EOMES Interacts with RUNX3 and BRG1 to Promote Innate Memory Cell Formation through Epigenetic Reprogramming. Nat. Commun. 2019, 10, 3306. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Fichtner, A.S.; Bruni, E.; Odak, I.; Sandrock, I.; Bubke, A.; Borchers, A.; Schultze-Florey, C.; Koenecke, C.; Förster, R.; et al. A Fetal Wave of Human Type 3 Effector Γδ Cells with Restricted TCR Diversity Persists into Adulthood. Sci. Immunol. 2021, 6, eabf0125. [Google Scholar] [CrossRef] [PubMed]
- Venken, K.; Jacques, P.; Mortier, C.; Labadia, M.E.; Decruy, T.; Coudenys, J.; Hoyt, K.; Wayne, A.L.; Hughes, R.; Turner, M.; et al. RORγt Inhibition Selectively Targets IL-17 Producing INKT and Γδ-T Cells Enriched in Spondyloarthritis Patients. Nat. Commun. 2019, 10, 9. [Google Scholar] [CrossRef]
- Kreslavsky, T.; Savage, A.K.; Hobbs, R.; Gounari, F.; Bronson, R.; Pereira, P.; Pandolfi, P.P.; Bendelac, A.; von Boehmer, H. TCR-Inducible PLZF Transcription Factor Required for Innate Phenotype of a Subset of Γδ T Cells with Restricted TCR Diversity. Proc. Natl. Acad. Sci. USA 2009, 106, 12453–12458. [Google Scholar] [CrossRef] [PubMed]
- Plužarić, V.; Štefanić, M.; Mihalj, M.; Tolušić Levak, M.; Muršić, I.; Glavaš-Obrovac, L.; Petrek, M.; Balogh, P.; Tokić, S. Differential Skewing of Circulating MR1-Restricted and Γδ T Cells in Human Psoriasis Vulgaris. Front. Immunol. 2020, 11, 572924. [Google Scholar] [CrossRef] [PubMed]
- Sanchez Sanchez, G.; Papadopoulou, M.; Azouz, A.; Tafesse, Y.; Mishra, A.; Chan, J.K.Y.; Fan, Y.; Verdebout, I.; Porco, S.; Libert, F.; et al. Identification of Distinct Functional Thymic Programming of Fetal and Pediatric Human Γδ Thymocytes via Single-Cell Analysis. Nat. Commun. 2022, 13, 5842. [Google Scholar] [CrossRef]
- Tüfekci, K.U.; Oner, M.G.; Meuwissen, R.L.J.; Genç, S. The Role of MicroRNAs in Human Diseases. Methods Mol. Biol. Clifton NJ 2014, 1107, 33–50. [Google Scholar] [CrossRef]
- Solvin, Å.Ø.; Chawla, K.; Olsen, L.C.; Hegre, S.A.; Danielsen, K.; Jenssen, M.; Furberg, A.-S.; Saunes, M.; Hveem, K.; Sætrom, P.; et al. MicroRNA Profiling of Psoriatic Skin Identifies 11 MiRNAs Associated with Disease Severity. Exp. Dermatol. 2022, 31, 535–547. [Google Scholar] [CrossRef]
- Sonkoly, E. The Expanding MicroRNA World in Psoriasis. Exp. Dermatol. 2017, 26, 375–376. [Google Scholar] [CrossRef]
- Løvendorf, M.B.; Mitsui, H.; Zibert, J.R.; Røpke, M.A.; Hafner, M.; Dyring-Andersen, B.; Bonefeld, C.M.; Krueger, J.G.; Skov, L. Laser Capture Microdissection Followed by Next-Generation Sequencing Identifies Disease-Related MicroRNAs in Psoriatic Skin That Reflect Systemic MicroRNA Changes in Psoriasis. Exp. Dermatol. 2015, 24, 187–193. [Google Scholar] [CrossRef]
- Joyce, C.E.; Zhou, X.; Xia, J.; Ryan, C.; Thrash, B.; Menter, A.; Zhang, W.; Bowcock, A.M. Deep Sequencing of Small RNAs from Human Skin Reveals Major Alterations in the Psoriasis MiRNAome. Hum. Mol. Genet. 2011, 20, 4025–4040. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Meisgen, F.; Pasquali, L.; Munkhammar, S.; Xia, P.; Ståhle, M.; Landén, N.X.; Pivarcsi, A.; Sonkoly, E. Next-Generation Sequencing Identifies the Keratinocyte-Specific MiRNA Signature of Psoriasis. J. Investig. Dermatol. 2019, 139, 2547–2550.e12. [Google Scholar] [CrossRef]
- Zibert, J.R.; Løvendorf, M.B.; Litman, T.; Olsen, J.; Kaczkowski, B.; Skov, L. MicroRNAs and Potential Target Interactions in Psoriasis. J. Dermatol. Sci. 2010, 58, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Delić, D.; Wolk, K.; Schmid, R.; Gabrielyan, O.; Christou, D.; Rieber, K.; Rolser, M.; Jakob, I.; Wiech, F.; Griesser, M.; et al. Integrated MicroRNA/MRNA Expression Profiling of the Skin of Psoriasis Patients. J. Dermatol. Sci. 2020, 97, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Ichihara, A.; Jinnin, M.; Yamane, K.; Fujisawa, A.; Sakai, K.; Masuguchi, S.; Fukushima, S.; Maruo, K.; Ihn, H. MicroRNA-Mediated Keratinocyte Hyperproliferation in Psoriasis Vulgaris. Br. J. Dermatol. 2011, 165, 1003–1010. [Google Scholar] [CrossRef]
- Alatas, E.T.; Kara, M.; Dogan, G.; Akın Belli, A. Blood MicroRNA Expressions in Patients with Mild to Moderate Psoriasis and the Relationship between MicroRNAs and Psoriasis Activity. An. Bras. Dermatol. 2020, 95, 702–707. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Yan, B.-X.; Zhou, Y.; Chen, X.-Y.; Zhang, J.; Cai, S.-Q.; Zheng, M.; Man, X.-Y. MiRNA Profiling of Extracellular Vesicles Reveals Biomarkers for Psoriasis. J. Investig. Dermatol. 2021, 141, 185–189.e4. [Google Scholar] [CrossRef]
- Seifeldin, N.S.; El Sayed, S.B.; Asaad, M.K. Increased MicroRNA-1266 Levels as a Biomarker for Disease Activity in Psoriasis Vulgaris. Int. J. Dermatol. 2016, 55, 1242–1247. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, W.; Wei, C.; Wang, L.; Zhu, G.; Shi, Q.; Li, S.; Ge, R.; Li, K.; Gao, L.; et al. Serum and Skin Levels of MiR-369-3p in Patients with Psoriasis and Their Correlation with Disease Severity. Eur. J. Dermatol. 2013, 23, 608–613. [Google Scholar] [CrossRef]
- Yi, J.Z.; McGee, J.S. Epigenetic-Modifying Therapies: An Emerging Avenue for the Treatment of Inflammatory Skin Diseases. Exp. Dermatol. 2021, 30, 1167–1176. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, J.; He, W. The Role of MicroRNAs in Γδ T Cells: A Long Way to Go. Cell. Mol. Immunol. 2021, 18, 2071–2072. [Google Scholar] [CrossRef] [PubMed]
- Reddycherla, A.V.; Meinert, I.; Reinhold, A.; Reinhold, D.; Schraven, B.; Simeoni, L. MiR-20a Inhibits TCR-Mediated Signaling and Cytokine Production in Human Naïve CD4+ T Cells. PLoS ONE 2015, 10, e0125311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, X.; Wang, W.; Cai, Y.; Li, S.; Chen, Q.; Liao, M.; Zhang, M.; Zeng, G.; Zhou, B.; et al. Down-Regulation of MiR-20a-5p Triggers Cell Apoptosis to Facilitate Mycobacterial Clearance through Targeting JNK2 in Human Macrophages. Cell Cycle 2016, 15, 2527–2538. [Google Scholar] [CrossRef]
- Zhang, W.; Yi, X.; An, Y.; Guo, S.; Li, S.; Song, P.; Chang, Y.; Zhang, S.; Gao, T.; Wang, G.; et al. MicroRNA-17-92 Cluster Promotes the Proliferation and the Chemokine Production of Keratinocytes: Implication for the Pathogenesis of Psoriasis. Cell Death Dis. 2018, 9, 567. [Google Scholar] [CrossRef]
- Steiner, D.F.; Thomas, M.F.; Hu, J.K.; Yang, Z.; Babiarz, J.E.; Allen, C.D.C.; Matloubian, M.; Blelloch, R.; Ansel, K.M. MicroRNA-29 Regulates T-Box Transcription Factors and Interferon-γ Production in Helper T Cells. Immunity 2011, 35, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Pobezinsky, L.A.; Etzensperger, R.; Jeurling, S.; Alag, A.; Kadakia, T.; McCaughtry, T.M.; Kimura, M.Y.; Sharrow, S.O.; Guinter, T.I.; Feigenbaum, L.; et al. Let-7 MicroRNAs Target the Lineage-Specific Transcription Factor PLZF to Regulate Terminal NKT Cell Differentiation and Effector Function. Nat. Immunol. 2015, 16, 517–524. [Google Scholar] [CrossRef]
- Pobezinskaya, E.L.; Wells, A.C.; Angelou, C.C.; Fagerberg, E.; Aral, E.; Iverson, E.; Kimura, M.Y.; Pobezinsky, L.A. Survival of Naïve T Cells Requires the Expression of Let-7 MiRNAs. Front. Immunol. 2019, 10, 955. [Google Scholar] [CrossRef] [PubMed]
- Wilczynska, A.; Bushell, M. The Complexity of MiRNA-Mediated Repression. Cell Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef]
- Deseke, M.; Rampoldi, F.; Sandrock, I.; Borst, E.; Böning, H.; Ssebyatika, G.L.; Jürgens, C.; Plückebaum, N.; Beck, M.; Hassan, A.; et al. A CMV-Induced Adaptive Human Vδ1+ Γδ T Cell Clone Recognizes HLA-DR. J. Exp. Med. 2022, 219, e20212525. [Google Scholar] [CrossRef]
- Halary, F.; Pitard, V.; Dlubek, D.; Krzysiek, R.; de la Salle, H.; Merville, P.; Dromer, C.; Emilie, D.; Moreau, J.-F.; Déchanet-Merville, J. Shared Reactivity of Vδ2neg Γδ T Cells against Cytomegalovirus-Infected Cells and Tumor Intestinal Epithelial Cells. J. Exp. Med. 2005, 201, 1567–1578. [Google Scholar] [CrossRef]
- Vermijlen, D.; Brouwer, M.; Donner, C.; Liesnard, C.; Tackoen, M.; Van Rysselberge, M.; Twité, N.; Goldman, M.; Marchant, A.; Willems, F. Human Cytomegalovirus Elicits Fetal Γδ T Cell Responses in Utero. J. Exp. Med. 2010, 207, 807–821. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated Analysis of Multimodal Single-Cell Data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef] [PubMed]
- Pizzolato, G.; Kaminski, H.; Tosolini, M.; Franchini, D.-M.; Pont, F.; Martins, F.; Valle, C.; Labourdette, D.; Cadot, S.; Quillet-Mary, A.; et al. Single-Cell RNA Sequencing Unveils the Shared and the Distinct Cytotoxic Hallmarks of Human TCRVδ1 and TCRVδ2 Γδ T Lymphocytes. Proc. Natl. Acad. Sci. USA 2019, 116, 11906–11915. [Google Scholar] [CrossRef] [PubMed]
- Dowd, J.B.; Palermo, T.M.; Aiello, A.E. Family Poverty Is Associated with Cytomegalovirus Antibody Titers in U.S Children. Health Psychol. Off. J. Div. Health Psychol. Am. Psychol. Assoc. 2012, 31, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Ssentongo, P.; Hehnly, C.; Birungi, P.; Roach, M.A.; Spady, J.; Fronterre, C.; Wang, M.; Murray-Kolb, L.E.; Al-Shaar, L.; Chinchilli, V.M.; et al. Congenital Cytomegalovirus Infection Burden and Epidemiologic Risk Factors in Countries With Universal Screening: A Systematic Review and Meta-Analysis. JAMA Netw. Open 2021, 4, e2120736. [Google Scholar] [CrossRef] [PubMed]
- Vilibic-Cavlek, T.; Kolaric, B.; Beader, N.; Vrtar, I.; Tabain, I.; Mlinaric-Galinovic, G. Seroepidemiology of Cytomegalovirus Infections in Croatia. Wien. Klin. Wochenschr. 2017, 129, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Déchanet, J.; Merville, P.; Lim, A.; Retière, C.; Pitard, V.; Lafarge, X.; Michelson, S.; Méric, C.; Hallet, M.-M.; Kourilsky, P.; et al. Implication of Γδ T Cells in the Human Immune Response to Cytomegalovirus. J. Clin. Investig. 1999, 103, 1437–1449. [Google Scholar] [CrossRef]
- Rampoldi, F.; Donato, E.; Ullrich, L.; Deseke, M.; Janssen, A.; Demera, A.; Sandrock, I.; Bubke, A.; Juergens, A.-L.; Swallow, M.; et al. Γδ T Cells License Immature B Cells to Produce a Broad Range of Polyreactive Antibodies. Cell Rep. 2022, 39, 110854. [Google Scholar] [CrossRef]
- Kobayashi, H.; Singer, R.H. Single-Molecule Imaging of MicroRNA-Mediated Gene Silencing in Cells. Nat. Commun. 2022, 13, 1435. [Google Scholar] [CrossRef]
- Lucasson, F.; Richette, P.; Aouad, K.; Ryussen-Witrand, A.; Wendling, D.; Fautrel, B.; Gossec, L. Prevalence and Consequences of Psoriasis in Recent Axial Spondyloarthritis: An Analysis of the DESIR Cohort over 6 Years. RMD Open 2022, 8, e001986. [Google Scholar] [CrossRef]
- Tieppo, P.; Papadopoulou, M.; Gatti, D.; McGovern, N.; Chan, J.K.Y.; Gosselin, F.; Goetgeluk, G.; Weening, K.; Ma, L.; Dauby, N.; et al. The Human Fetal Thymus Generates Invariant Effector Γδ T Cells. J. Exp. Med. 2019, 217, e20190580. [Google Scholar] [CrossRef] [PubMed]
- Guggino, G.; Orlando, V.; Saieva, L.; Ruscitti, P.; Cipriani, P.; La Manna, M.P.; Giacomelli, R.; Alessandro, R.; Triolo, G.; Ciccia, F.; et al. Downregulation of MiRNA17–92 Cluster Marks Vγ9Vδ2 T Cells from Patients with Rheumatoid Arthritis. Arthritis Res. Ther. 2018, 20, 236. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.-Y.; Wu, Q.-Y.; Zhang, C.-X.; Wang, Q.; Ling, J.; Huang, X.-T.; Sun, X.; Yuan, M.; Wu, D.; Yin, H.-F. MiR-20a Inhibits the Killing Effect of Natural Killer Cells to Cervical Cancer Cells by Downregulating RUNX1. Biochem. Biophys. Res. Commun. 2018, 505, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Penny, L.A.; Yuzefpolskiy, Y.; Sarkar, S.; Kalia, V. MicroRNA-17∼92 Regulates Effector and Memory CD8 T-Cell Fates by Modulating Proliferation in Response to Infections. Blood 2013, 121, 4473–4483. [Google Scholar] [CrossRef] [PubMed]
- Kannambath, S. Micro-RNA Feedback Loops Modulating the Calcineurin/NFAT Signaling Pathway. Non-Coding RNA 2016, 2, 3. [Google Scholar] [CrossRef]
- Nours, J.L.; Gherardin, N.A.; Ramarathinam, S.H.; Awad, W.; Wiede, F.; Gully, B.S.; Khandokar, Y.; Praveena, T.; Wubben, J.M.; Sandow, J.J.; et al. A Class of Γδ T Cell Receptors Recognize the Underside of the Antigen-Presenting Molecule MR1. Science 2019, 366, 1522–1527. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The Biochemical Basis of MicroRNA Targeting Efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. MiRDB: An Online Database for Prediction of Functional MicroRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A Decade-Long Collection of Experimentally Supported MiRNA-Gene Interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective MicroRNA Target Sites in Mammalian MRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Ye, Y.; Wang, P.; Zhou, F. MiR-489-3p Inhibits TLR4/NF-ΚB Signaling to Prevent Inflammation in Psoriasis. Exp. Ther. Med. 2021, 22, 744. [Google Scholar] [CrossRef]
- Carreras-Badosa, G.; Maslovskaja, J.; Vaher, H.; Pajusaar, L.; Annilo, T.; Lättekivi, F.; Hübenthal, M.; Rodriguez, E.; Weidinger, S.; Kingo, K.; et al. MiRNA Expression Profiles of the Perilesional Skin of Atopic Dermatitis and Psoriasis Patients Are Highly Similar. Sci. Rep. 2022, 12, 22645. [Google Scholar] [CrossRef]
- Mostafa, S.A.; Mohammad, M.H.S.; Negm, W.A.; Batiha, G.E.S.; Alotaibi, S.S.; Albogami, S.M.; Waard, M.D.; Tawfik, N.Z.; Abdallah, H.Y. Circulating MicroRNA203 and Its Target Genes’ Role in Psoriasis Pathogenesis. Front. Med. 2022, 9, 988962. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Alquicira-Hernandez, J.; Powell, J.E. Nebulosa Recovers Single-Cell Gene Expression Signals by Kernel Density Estimation. Bioinformatics 2021, 37, 2485–2487. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and Interpreting Cancer Genomics Data via the Xena Platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Lundberg, S.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. arXiv 2017, arXiv:1705.07874. [Google Scholar]
- Lipovetsky, S.; Conklin, M. Analysis of Regression in Game Theory Approach. Appl. Stoch. Models Bus. Ind. 2001, 17, 319–330. [Google Scholar] [CrossRef]
- Wodtke, G.T.; Ard, K.; Bullock, C.; White, K.; Priem, B. Concentrated Poverty, Ambient Air Pollution, and Child Cognitive Development. Sci. Adv. 2022, 8, eadd0285. [Google Scholar] [CrossRef] [PubMed]
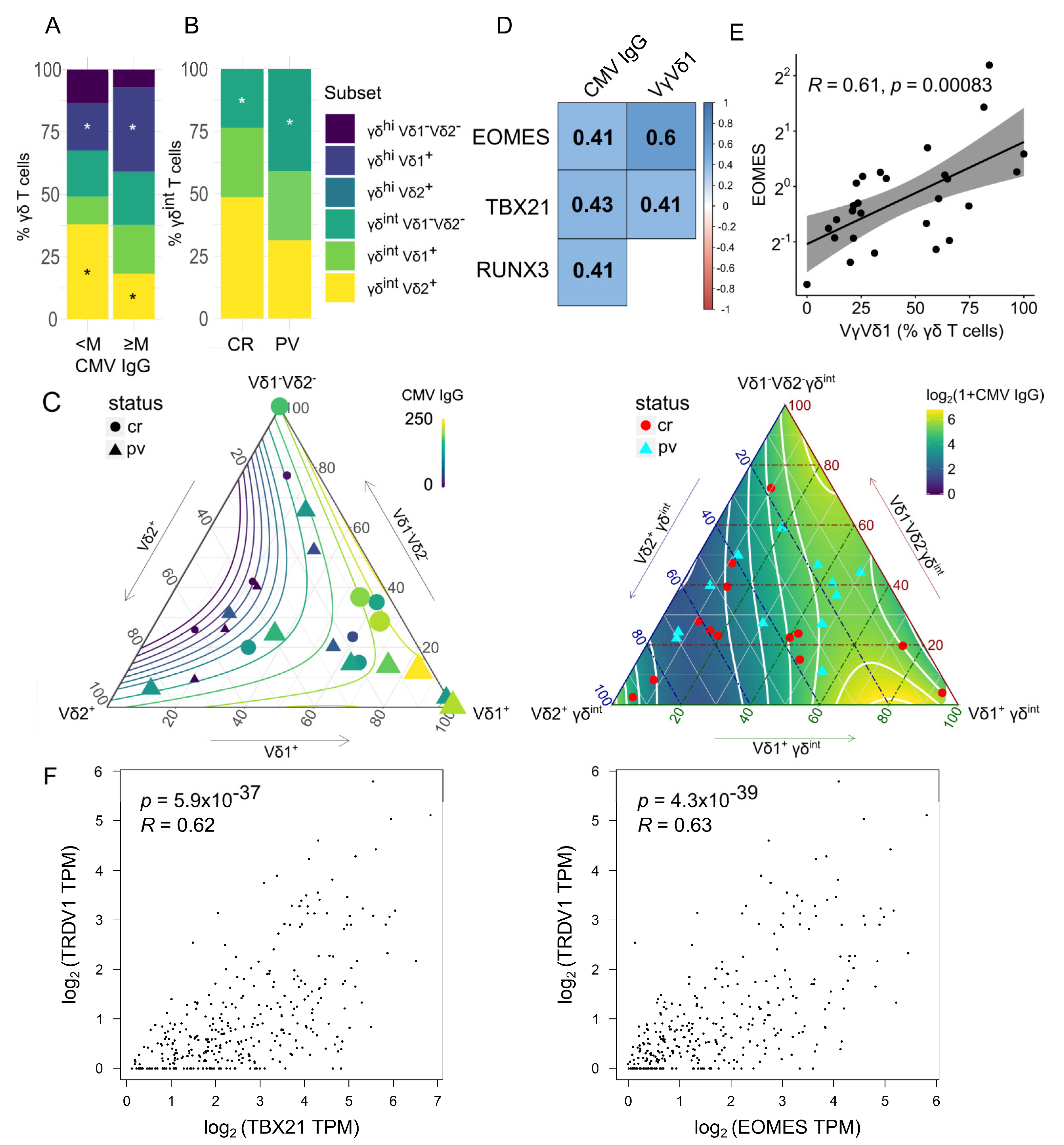
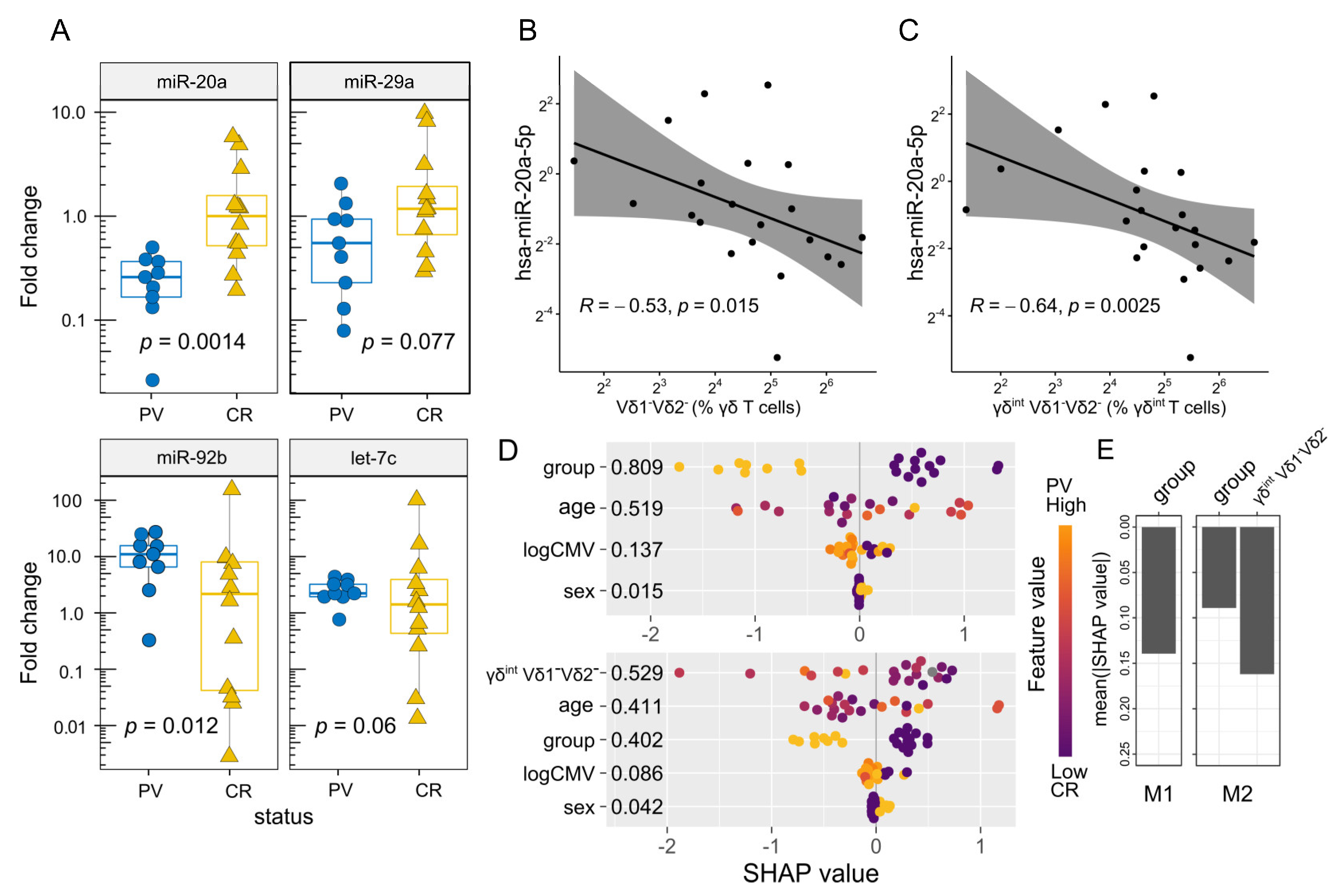

| Group | PV | Controls | p * |
|---|---|---|---|
| N (male/female) | 13 (10/3) | 14 (9/5) | 0.678 ** |
| Age (chronological, years) | 35 (28–43) | 32 (28–41) | 0.528 |
| PASI | 6.8 (5.5–11.5) | – | – |
| DLQI | 3 (0.5–6.5) | – | – |
| BMI (kg/m2) | 26.5 (21.3–30.1) | 23.9 (20–25.7) | 0.055 |
| CRP (mg/L) | 2 (0.7–2.8) | 0.7 (0.3–1.7) | 0.068 |
| Anti—CMV IgG (AU/mL) | 130 (19–178) | 135 (42–171) | 0.86 |
| Anti—CMV IgG (pos/neg) | 8/2 | 11/3 | 1 ** |
| Assay ID | miRBase ID | miRBase Accession Number | Mature miRNA Sequence |
|---|---|---|---|
| 478586_mir | hsa-miR-20a-5p | MIMAT0000075 | UAAAGUGCUUAUAGUGCAGGUAG |
| 478587_mir | hsa-miR-29a-3p | MIMAT0000086 | UAGCACCAUCUGAAAUCGGUUA |
| 478577_mir | hsa-let-7c-5p | MIMAT0000064 | UGAGGUAGUAGGUUGUAUGGUU |
| 479207_mir | hsa-miR-92b-5p | MIMAT0004792 | AGGGACGGGACGCGGUGCAGUG |
| 478327_mir | hsa-miR-423-3p | MIMAT0001340 | AGCUCGGUCUGAGGCCCCUCAGU |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tokić, S.; Jirouš, M.; Plužarić, V.; Mihalj, M.; Šola, M.; Tolušić Levak, M.; Glavaš, K.; Balogh, P.; Štefanić, M. The miR-20a/miR-92b Profile Is Associated with Circulating γδ T-Cell Perturbations in Mild Psoriasis. Int. J. Mol. Sci. 2023, 24, 4323. https://doi.org/10.3390/ijms24054323
Tokić S, Jirouš M, Plužarić V, Mihalj M, Šola M, Tolušić Levak M, Glavaš K, Balogh P, Štefanić M. The miR-20a/miR-92b Profile Is Associated with Circulating γδ T-Cell Perturbations in Mild Psoriasis. International Journal of Molecular Sciences. 2023; 24(5):4323. https://doi.org/10.3390/ijms24054323
Chicago/Turabian StyleTokić, Stana, Maja Jirouš, Vera Plužarić, Martina Mihalj, Marija Šola, Maja Tolušić Levak, Kristina Glavaš, Peter Balogh, and Mario Štefanić. 2023. "The miR-20a/miR-92b Profile Is Associated with Circulating γδ T-Cell Perturbations in Mild Psoriasis" International Journal of Molecular Sciences 24, no. 5: 4323. https://doi.org/10.3390/ijms24054323
APA StyleTokić, S., Jirouš, M., Plužarić, V., Mihalj, M., Šola, M., Tolušić Levak, M., Glavaš, K., Balogh, P., & Štefanić, M. (2023). The miR-20a/miR-92b Profile Is Associated with Circulating γδ T-Cell Perturbations in Mild Psoriasis. International Journal of Molecular Sciences, 24(5), 4323. https://doi.org/10.3390/ijms24054323







